Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention
Abstract
:1. Introduction
Global Burden and Geographic Distribution of HIV
2. HIV-1 Antiretroviral Therapy: Current Status and Challenges
2.1. Constraints to Current ART/HAART Regime
2.2. Preventive Measures
3. Host–Viral Interactomes Offering New Drug Targets
3.1. Advances in Viral-Host Interactomics
3.2. Drug Discovery Now Targets Host Factors
4. HIV-1-Host Interactors: Evaluating the Therapeutic Potential
4.1. Host Restriction Factors
4.2. Druggable Host Factors Involved during HIV-1 Entry
4.3. Other HIV-1-Cellular Interactors as Possible Targets
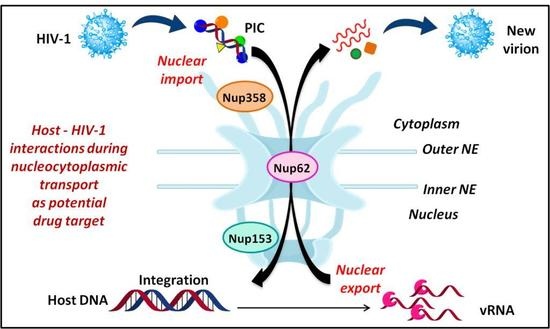
5. Nuclear Entry and Egress of HIV-1
5.1. Nuclear Import of PIC
5.1.1. Interaction of Nup358 with HIV-1 CA
5.1.2. Interaction of Nup153 with Different HIV-1 Proteins
5.1.3. Role of Nup62
5.2. Nuclear Export of Viral mRNAs
6. Significance of Targeting Nup-HIV-1 Interactions as Drug Targets
7. Challenges and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIDS | Acquired Immunodeficiency Syndrome |
| APOBEC3G | Apolipoprotein B mRNA Editing Enzyme Catalytic polypeptide-like 3G |
| CA | Capsid |
| CHD | Cypa Homology Domain |
| CPSF6 | Cleavage and Polyadenylation Specific Factor 6 |
| CypA | Cyclophillin A |
| eIF-5A | Eukaryotic translation Initiation Factor-5A |
| ESCRT-I | Endosomal Sorting Complex Required for Transport I |
| FDA | Food and Drug Association |
| HAART | Highly Advanced AntiRetroviral Therapy |
| HIV | Human Immunodeficiency Virus |
| IN | Integrase |
| Crm1 | Chromosome Region Maintenance-1 |
| LEDGF | Lens Epithelium Derived Growth Factor |
| LTR | Long Terminal Repeat |
| MX2 | human Myxovirus resistance 2 |
| NPC | Nuclear Pore Complex |
| Nup | Nucleoporin |
| NTR | Nuclear Transport Receptor |
| NLS | Nuclear Localisation Signal |
| NES | Nuclear Export Signal |
| PIC | Pre Initiation Complex |
| PPI | Protein-Protein Interaction |
| pTEFb | positive Transcription Elongation Factor b |
| Rev | regulator of virion expression |
| RT | Reverse Transcriptase |
| SAMHD1 | SAM domain and HD domain-containing protein 1 |
| TNPO3 | transportin 3 |
| Tat | Transactivator of Transmembrane |
| TRIM5α | Tripartite Motif-containing protein 5α- isoform |
| TSG101 | Tumor Susceptibility Gene 101 |
| UNAIDS | joint United Nations program for Acquired Immunodeficiency Syndrome |
| V-H interactome | Viral Host Interactome |
| Vif | Viral Infectivity Factor |
| Vpr | Viral Protein R |
| Vpu | Viral Protein U |
References
- Centers for Disease Control (CDC). Pneumocystis Pneumonia - Los Angeles. Morb. Mortal. Wkly Rep. 1981, 30, 250–252. [Google Scholar]
- Hahn, B.H.; Shaw, G.M.; de Cock, K.M.; Sharp, P.M. AIDS as a Zoonosis: Scientific and Public Health Implications. Science 2000, 287, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemey, P.; Pybus, O.G.; Wang, B.; Saksena, N.K.; Salemi, M.; Vandamme, A.M. Tracing the Origin and History of the HIV-2 Epidemic. Proc. Natl. Acad. Sci. USA 2003, 100, 6588–6592. [Google Scholar] [CrossRef] [PubMed]
- McCutchan, F.E. Understanding the Genetic Diversity of HIV-1. AIDS 2000, 14, S31–S44. [Google Scholar] [PubMed]
- Peeters, M. The Genetic Variability of HIV-1 and Its Implications. Transfus. Clin. Biol. 2001, 8, 222–225. [Google Scholar] [CrossRef]
- Levy, J.A. HIV and the Pathogenesis of AIDS, 3rd ed.; ASM Press: Washington, DC, USA, 2007; p. 588. [Google Scholar]
- UNAIDS. Global HIV and AIDS Statistics—2019 Fact Sheet. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 10 September 2019).
- UNAIDS. 2018 Global AIDS Update Slides. Available online: https://www.unaids.org/en/resources/documents/2018/2018-global-aids-update-slides-part1 (accessed on August 2018).
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and Function of HIV-1 Reverse Transcriptase: Molecular Mechanisms of Polymerization and Inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, T.K.; Davies, D.R. Structure and Function of HIV-1 Integrase. Curr. Top Med. Chem. 2004, 4, 965–977. [Google Scholar] [CrossRef]
- Brik, A.; Wong, C.-H. HIV-1 Protease: Mechanism and Drug Discovery. Org. Biomol. Chem. 2003, 1, 5–14. [Google Scholar] [CrossRef]
- Fiorentini, S.; Marini, E.; Caracciolo, S.; Caruso, A. Functions of the HIV-1 Matrix Protein P17. New Microbiol. 2006, 29, 1–10. [Google Scholar]
- Campbell, E.M.; Hope, T.J. HIV-1 Capsid: The Multifaceted Key Player in HIV-1 Infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef]
- Levin, J.G.; Mitra, M.; Mascarenhas, A.; Musier-Forsyth, K. Role of HIV-1 Nucleocapsid Protein in HIV-1 Reverse Transcription. RNA Biol. 2010, 7, 754–774. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV Gp120 Envelope Glycoprotein in Complex with the CD4 Receptor and a Neutralizing Human Antibody. Nature 1998, 393, 648. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Desrosiers, R.C. The Tale of the Long Tail: The Cytoplasmic Domain of HIV-1 Gp41. J. Virol. 2013, 87, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.P. The HIV-1 Tat Protein: Mechanism of Action and Target for HIV-1 Cure Strategies. Curr. Pharm. Des. 2017, 23, 4098–4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malim, M.H.; Pollard, V.W. The HIV-1 Rev Protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar]
- Pereira, E.A.; da Silva, L.L.P. HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 2016, 17, 976–996. [Google Scholar] [CrossRef]
- Solbak, S.M.O.; Reksten, T.R.; Hahn, F.; Wray, V.; Henklein, P.; Henklein, P.; Halskau, O.; Schubert, U.; Fossen, T. HIV-1 P6 - A Structured to Flexible Multifunctional Membrane-Interacting Protein. Biochim. Biophys. Acta-Biomembranes 2013, 1828, 816–823. [Google Scholar] [CrossRef]
- Rose, K.M.; Marin, M.; Kozak, S.L.; Kabat, D. The Viral Infectivity Factor (Vif) of HIV-1 Unveiled. Trends Mol. Med. 2004, 10, 291–297. [Google Scholar] [CrossRef]
- González, M. The HIV-1 Vpr Protein: A Multifaceted Target for Therapeutic Intervention. Int. J. Mol. Sci. 2017, 18, 126. [Google Scholar] [CrossRef]
- Pandey, R.C.; Datta, D.; Mukerjee, R.; Srinivasan, A.; Mahalingam, S.; Sawaya, B.E. HIV-1 Vpr: A Closer Look at the Multifunctional Protein from the Structural Perspective. Curr. HIV Res. 2009, 7, 114–128. [Google Scholar] [CrossRef]
- González, M. Vpu Protein: The Viroporin Encoded by HIV-1. Viruses 2015, 7, 4352–4368. [Google Scholar] [CrossRef] [Green Version]
- Pomerantz, R.J.; Horn, D.L. Twenty Years of Therapy for HIV-1 Infection. Nat. Med. 2003, 9, 867–873. [Google Scholar] [CrossRef]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining Morbidity and Mortality among Patients with HIV Infection: HIV Outpatient Study Investigators. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef]
- Lu, D.Y. High Active Anti-Retroviral Therapy for HIV/AIDS, Progresses and Drawback. Adv. Pharmacoepidemiol. Drug Saf. 2012, 1, e115. [Google Scholar] [CrossRef]
- Larder, B.A.; Darby, G.; Richman, D.D. HIV with Reduced Sensitivity to Zidovudine (AZT) Isolated during Prolonged Therapy. Science 1989, 243, 1731–1734. [Google Scholar] [CrossRef]
- Larder, B. Mechanisms of HIV-1 Drug Resistance. AIDS 2001, 15, S27–S34. [Google Scholar] [CrossRef]
- USFDA. HIV and AIDS Activities - Antiretroviral Drugs Used in the Treatment of HIV Infection. Available online: https://www.fda.gov/patients/hiv-treatment/antiretroviral-drugs-used-treatment-hiv-infection (accessed on 3 July 2019).
- Ray, N.; Doms, R.W. HIV-1 Coreceptors and Their Inhibitors. Curr. Top Microbiol. Imm. 2006, 303, 97–120. [Google Scholar]
- Eggink, D.; Berkhout, B.; Sanders, R.W. Inhibition of HIV-1 by Fusion Inhibitors. Curr. Pharm. Des. 2010, 16, 3716–3728. [Google Scholar] [CrossRef]
- Zhan, P.; Chen, X.; Li, D.; Fang, Z.; De Clercq, E.; Liu, X. HIV-1 NNRTIs: Structural Diversity, Pharmacophore Similarity, and Impliations for Drug Design. Med. Res. Rev. 2013, 33, E1–E72. [Google Scholar] [CrossRef]
- Waller, D.G.; Sampson, A.P. Chemotherapy of infections. Medical Pharmacology and Therapeutics, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 581–629. [Google Scholar]
- Flexner, C. HIV-Protease Inhibitors. N. Engl. J. Med. 1998, 338, 1281–1293. [Google Scholar] [CrossRef]
- Hazuda, D.J.; Felock, P.; Witmer, M.; Wolfe, A.; Stillmock, K.; Grobler, J.A.; Espeseth, A.; Gabryelski, L.; Schleif, W.; Blau, C.; et al. Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science 2000, 287, 646–650. [Google Scholar] [CrossRef]
- Kaufmann, D.E.; Lichterfeld, M.; Altfeld, M.; Addo, M.M.; Johnston, M.N.; Lee, P.K.; Wagner, B.S.; Kalife, E.T.; Strick, D.; Rosenberg, E.S.; et al. Limited Durability of Viral Control Following Treated Acute HIV Infection. PLoS Med. 2004, 137–148. [Google Scholar] [CrossRef]
- Vervoort, S.C.; Borleffs, J.C.; Hoepelman, A.I.; Grypdonck, M.H. Adherence in Antiretroviral Therapy: A Review of Qualitative Studies. AIDS 2007, 21, 271–281. [Google Scholar] [CrossRef]
- Brinkman, K.; ter Hofstede, H.J.M.; Burger, D.M.; Smeitink, J.A.M.; Koopmans, P.P. Adverse Effects of Reverse Transcriptase Inhibitors. AIDS 1998, 12, 1735–1744. [Google Scholar] [CrossRef]
- Brown, T.; Qaqish, R. Antiretroviral Therapy and the Prevalence of Osteopenia and Osteoporosis. AIDS 2006, 20, 2165–2174. [Google Scholar] [CrossRef]
- Croxford, S.; Kitching, A.; Desai, S.; Kall, M.; Edelstein, M.; Skingsley, A.; Burns, F.; Copas, A.; Brown, A.E.; Sullivan, A.K. Mortality and Causes of Death in People Diagnosed with HIV in the Era of Highly Active Antiretroviral Therapy Compared with The General Population: An Analysis Of A National Observational Cohort. Lancet Public Health 2017, 2, e35–e46. [Google Scholar] [CrossRef]
- Ganji, R.; Dhali, S.; Rizvi, A.; Sankati, S.; Vemula, M.H.; Mahajan, G.; Rapole, S.; Banerjee, S. Proteomics Approach to Understand Reduced Clearance of Mycobacteria and High Viral Titers during HIV--Mycobacteria Co-Infection. Cell Microbiol. 2016, 18, 355–368. [Google Scholar] [CrossRef]
- Ganji, R.; Dhali, S.; Rizvi, A.; Rapole, S.; Banerjee, S. Understanding HIV-Mycobacteria Synergism through Comparative Proteomics of Intra-Phagosomal Mycobacteria during Mono-and HIV Co-Infection. Sci. Rep. 2016, 6, 22060. [Google Scholar] [CrossRef]
- Johnston, M.I.; Fauci, A.S. An HIV Vaccine - Evolving Concepts. N. Engl. J. Med. 2007, 356, 2073–2081. [Google Scholar] [CrossRef]
- Escolano, A.; Dosenovic, P.; Nussenzweig, M.C. Progress toward Active or Passive HIV-1 Vaccination. J. Exp. Med. 2017, 214, 3–16. [Google Scholar] [CrossRef]
- Cohen, M.S.; Hellmann, N.; Levy, J.A.; Decock, K.; Lange, J. The Spread, Treatment, and Prevention of HIV-1: Evolution of a Global Pandemic. J. Clin. Invest. 2008, 118, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Perreau, M.; Banga, R.; Pantaleo, G. Targeted Immune Interventions for an HIV-1 Cure. Trends Mol. Med. 2017, 23, 945–961. [Google Scholar] [CrossRef] [PubMed]
- CDC. CDC HIV Basics. Available online: https://www.cdc.gov/hiv/basics/ (accessed on 30 April 2019).
- NIH News Release. Available online: https://www.nih.gov/news-events/news-releases/nih-trial-evaluates-long-acting-hiv-medication-unable-adhere-strict-daily-regimens (accessed on 31 May 2019).
- Gibson, T.J.; Davey, N.E.; Trave, G. How Viruses Hijack Cell Regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar]
- Hagai, T.; Azia, A.; Babu, M.M.; Andino, R. Use of Host-like Peptide Motifs in Viral Proteins Is a Prevalent Strategy in Host-Virus Interactions. Cell Rep. 2014, 7, 1729–1739. [Google Scholar] [CrossRef] [Green Version]
- Meyniel-Schicklin, L.; de Chassey, B.; André, P.; Lotteau, V. Viruses and Interactomes in Translation. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef]
- de Chassey, B.; Meyniel-Schicklin, L.; Vonderscher, J.; André, P.; Lotteau, V. Virus-Host Interactomics: New Insights and Opportunities for Antiviral Drug Discovery. Genome Med. 2014, 6, 115. [Google Scholar] [CrossRef]
- Dolan, P.T.; Zhang, C.; Khadka, S.; Arumugaswami, V.; Vangeloff, A.D.; Heaton, N.S.; Sahasrabudhe, S.; Randall, G.; Sun, R.; Lacount, D.J. Identification and Comparative Analysis of Hepatitis C Virus-Host Cell Protein Interactions. Mol. BioSys. 2013, 9, 3199–3209. [Google Scholar] [CrossRef] [PubMed]
- Mairiang, D.; Zhang, H.; Sodja, A.; Murali, T.; Suriyaphol, P.; Malasit, P.; Limjindaporn, T.; Finley, R.L. Identification of New Protein Interactions between Dengue Fever Virus and Its Hosts, Human and Mosquito. PLoS ONE 2013, 8, e53535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, L.; Li, S. Influenza A Virus-Host Protein Interactions Control Viral Pathogenesis. Int. J. Mol. Sci. 2017, 18, 1673. [Google Scholar] [CrossRef] [PubMed]
- Pizzorno, A.; Padey, B.; Terrier, O.; Rosa-Calatrava, M. Drug Repurposing Approaches for the Treatment of Influenza. Front Immunol. 2019, 10, 531. [Google Scholar] [CrossRef]
- De Chassey, B.; Meyniel-Schicklin, L.; Aublin-Gex, A.; André, P.; Lotteau, V. New Horizons for Antiviral Drug Discovery from Virus-Host Protein Interaction Networks. Curr. Opin. Virol. 2012, 2, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Scheuch, G.; Canisius, S.; Nocker, K.; Hofmann, T.; Naumann, R.; Pleschka, S.; Ludwig, S.; Welte, T.; Planz, O. Targeting Intracellular Signaling as an Antiviral Strategy: Aerosolized LASAG for the Treatment of Influenza in Hospitalized Patients. Emerg. Microbes Infect. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Sayana, S.; Khanlou, H. Maraviroc: A New CCR5 Antagonist. Expert Rev. Anti Infect. Ther. 2009, 7, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Chen, Z.J. Intrinsic Antiviral Immunity. Nat. Immunol. 2012, 13, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Hultquist, J.F.; Evans, D.T. The Restriction Factors of Human Immunodeficiency Virus. J. Biol. Chem. 2012, 287, 40875–40883. [Google Scholar] [CrossRef] [Green Version]
- Malim, M.H.; Bieniasz, P.D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb. Perspect Med. 2012, 2, a006940. [Google Scholar] [CrossRef]
- Mbisa, J.L.; Barr, R.; Thomas, J.A.; Vandegraaff, N.; Dorweiler, I.J.; Svarovskaia, E.S.; Brown, W.L.; Mansky, L.M.; Gorelick, R.J.; Harris, R.S.; et al. Human Immunodeficiency Virus Type 1 CDNAs Produced in the Presence of APOBEC3G Exhibit Defects in Plus-Strand DNA Transfer and Integration. J. Virol. 2007, 81, 7099–7110. [Google Scholar] [CrossRef]
- Bishop, K.N.; Verma, M.; Kim, E.Y.; Wolinsky, S.M.; Malim, M.H. APOBEC3G Inhibits Elongation of HIV-1 Reverse Transcripts. PLoS Pathog. 2008, 4, e1000231. [Google Scholar] [CrossRef]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific Recognition and Accelerated Uncoating of Retroviral Capsids by the TRIM5alpha Restriction Factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef]
- Evans, D.T.; Serra-Moreno, R.; Singh, R.K.; Guatelli, J.C. BST-2 Tetherin a New Component of the Innate Immune Response to Enveloped Viruses. Trends Microbiol. 2010, 18, 388–396. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 Is the Dendritic- and Myeloid-Cell-Specific HIV-1 Restriction Factor Counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Dicks, M.D.J.; Betancor, G.; Jimenez-Guardeño, J.M.; Pessel-Vivares, L.; Apolonia, L.; Goujon, C.; Malim, M.H. Multiple Components of the Nuclear Pore Complex Interact with the Amino-Terminus of MX2 to Facilitate HIV-1 Restriction. PLoS Pathog. 2018, 14, e1007408. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Emerman, M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe. 2008, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a Human Gene That Inhibits HIV-1 Infection and Is Suppressed by the Viral Vif Protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin Inhibits Retrovirus Release and Is Antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Lobritz, M.A.; Ratcliff, A.N.; Arts, E.J. HIV-1 Entry, Inhibitors, and Resistance. Viruses 2010, 2, 1069–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilton, J.C.; Doms, R.W. Entry Inhibitors in the Treatment of HIV-1 Infection. Antiviral Res. 2010, 85, 91–100. [Google Scholar] [CrossRef]
- Arhel, N.; Kirchhoff, F. Host Proteins Involved in HIV Infection: New Therapeutic Targets. Biochim. Biophys. Acta-Mol. Basis Dis. 2010, 1802, 313–321. [Google Scholar] [CrossRef]
- Klibanov, O.M. Vicriviroc, a CCR5 Receptor Antagonist for the Potential Treatment of HIV Infection. Curr. Opin. Investig. Drugs 2009, 10, 845–859. [Google Scholar]
- Zhou, Q.; Yik, J.H.N. The Yin and Yang of P-TEFb Regulation: Implications for Human Immunodeficiency Virus Gene Expression and Global Control of Cell Growth and Differentiation. Microbiol. Mol. Biol. Rev. 2006, 70, 646–659. [Google Scholar] [CrossRef] [Green Version]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B.R. Recruitment of Cyclin T1/P-TEFb to an HIV Type 1 Long Terminal Repeat Promoter Proximal RNA Target Is Both Necessary and Sufficient for Full Activation of Transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 7791–7796. [Google Scholar] [CrossRef]
- Ishaq, M.; Hu, J.; Wu, X.; Fu, Q.; Yang, Y.; Liu, Q.; Guo, D. Knockdown of Cellular RNA Helicase DDX3 by Short Hairpin RNAs Suppresses HIV-1 Viral Replication without Inducing Apoptosis. Mol. Biotechnol. 2008, 39, 231–238. [Google Scholar] [CrossRef]
- Stuchell, M.D.; Garrus, J.E.; Müller, B.; Stray, K.M.; Ghaffarian, S.; McKinnon, R.; Kräusslich, H.G.; Morham, S.G.; Sundquist, W.I. The Human Endosomal Sorting Complex Required for Transport (ESCRT-I) and Its Role in HIV-1 Budding. J. Biol. Chem. 2004, 279, 36059–36071. [Google Scholar] [CrossRef] [Green Version]
- Garrus, J.E.; Von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Fujii, K.; Hurley, J.H.; Freed, E.O. Beyond Tsg101: The Role of Alix in “ESCRTing” HIV-1. Nat. Rev. Microbiol. 2007, 5, 912–916. [Google Scholar] [CrossRef]
- Hütter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müssig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kücherer, C.; Blau, O.; et al. Long-Term Control of HIV by CCR5 Delta32/Delta32 Stem-Cell Transplantation. Pediatrics 2009, 360, 692–698. [Google Scholar]
- Brown, T.R. I Am the Berlin Patient: A Personal Reflection. AIDS Res. Hum. Retrov. 2015, 31, 2–3. [Google Scholar] [CrossRef]
- Gupta, R.K.; Abdul-Jawad, S.; McCoy, L.E.; Mok, H.P.; Peppa, D.; Salgado, M.; Martinez-Picado, J.; Nijhuis, M.; Wensing, A.M.J.; Lee, H.; et al. HIV-1 Remission Following CCR5Δ32/Δ32 Haematopoietic Stem-Cell Transplantation. Nature 2019, 568, 244–248. [Google Scholar] [CrossRef]
- Brass, A.L.; Elledge, S.J.; Xavier, R.J.; Dykxhoorn, D.M.; Yan, N.; Lieberman, J.; Benita, Y.; Engelman, A. Identification of Host Proteins Required for HIV Infection through a Functional Genomic Screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef]
- König, R.; De Jesus, P.D.; Opaluch, A.M.; Chanda, S.K.; Elleder, D.; Seidel, S.; Young, J.A.; Lilley, C.E.; Weitzman, M.D.; Bandyopadhyay, S.; et al. Global Analysis of Host-Pathogen Interactions That Regulate Early-Stage HIV-1 Replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, M.; Huang, Q.; Gates, A.T.; Zhang, X.D.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.J.; et al. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host Microbe. 2008, 4, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Sanders-Beer, B.E.; Katz, K.S.; Maglott, D.R.; Pruitt, K.D.; Ptak, R.G. Human Immunodeficiency Virus Type 1, Human Protein Interaction Database at NCBI. Nucleic Acids Res. 2009, 37, D417–D422. [Google Scholar] [CrossRef]
- Ptak, R.G.; Fu, W.; Sanders-Beer, B.E.; Dickerson, J.E.; Pinney, J.W.; Robertson, D.L.; Rozanov, M.N.; Katz, K.S.; Maglott, D.R.; Pruitt, K.D.; et al. Cataloguing the HIV-1 human protein interaction network. AIDS Res. Hum. Retrovir. 2008, 24, 1497–1502. [Google Scholar] [CrossRef]
- Pinney, J.W.; Dickerson, J.E.; Fu, W.; Sanders-Beer, B.E.; Ptak, R.G.; Robertson, D.L. HIV-Host Interactions: A Map of Viral Perturbation of the Host System. AIDS 2009, 23, 549–554. [Google Scholar] [CrossRef]
- Fassati, A. HIV Infection of Non-Dividing Cells: A Divisive Problem. Retrovirology 2006, 3, 74. [Google Scholar] [CrossRef]
- Kabachinski, G.; Schwartz, T.U. The Nuclear Pore Complex - Structure and Function at a Glance. J. Cell Sci. 2015, 128, 423–429. [Google Scholar] [CrossRef]
- Hurt, E.; Beck, M. Towards Understanding Nuclear Pore Complex Architecture and Dynamics in the Age of Integrative Structural Analysis. Curr. Opin. Cell Biol. 2015, 34, 31–38. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The Nuclear Pore Complex: Understanding Its Function through Structural Insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Von Appen, A.; Beck, M. Structure Determination of the Nuclear Pore Complex with Three-Dimensional Cryo Electron Microscopy. J. Mol. Biol. 2016, 428, 2001–2010. [Google Scholar] [CrossRef]
- Wälde, S.; Kehlenbach, R.H. The Part and the Whole: Functions of Nucleoporins in Nucleocytoplasmic Transport. Trends Cell Biol. 2010, 20, 461–469. [Google Scholar] [CrossRef]
- Jamali, T.; Jamali, Y.; Mehrbod, M.; Mofrad, M.R.K. Nuclear Pore Complex. Biochemistry and Biophysics of Nucleocytoplasmic Transport in Health and Disease. Int. Rev. Cell Mol. Biol. 2011, 287, 233–286. [Google Scholar]
- Hoelz, A.; Debler, E.W.; Blobel, G.; Fahrenkrog, B.; Aebi, U. The Nuclear Pore Complex: Nucleocytoplasmic Transport and Beyond. Nat. Rev. Mol. Cell Biol. 2003, 4, 757–766. [Google Scholar]
- Monette, A.; Panté, N.; Mouland, A.J. HIV-1 Remodels the Nuclear Pore Complex. J. Cell Biol. 2011, 193, 619–631. [Google Scholar] [CrossRef]
- Nunzio, F.D.; Danckaert, A.; Fricke, T.; Perez, P.; Fernandez, J.; Perret, E.; Roux, P.; Shorte, S.; Charneau, P.; Diaz-Griffero, F.; et al. Human Nucleoporins Promote HIV-1 Docking at the Nuclear Pore, Nuclear Import and Integration. PLoS ONE 2012, 7, e46037. [Google Scholar] [CrossRef]
- Matreyek, K.A.; Engelman, A. Viral and Cellular Requirements for the Nuclear Entry of Retroviral Preintegration Nucleoprotein Complexes. Viruses 2013, 5, 2483–2511. [Google Scholar] [CrossRef] [Green Version]
- Jayappa, K.D.; Ao, Z.; Yao, X. The HIV-1 Passage from Cytoplasm to Nucleus: The Process Involving a Complex Exchange between the Components of HIV-1 and Cellular Machinery to Access Nucleus and Successful Integration. Int. J. Biochem. Mol. Biol. 2012, 3, 70–85. [Google Scholar]
- Suzuki, Y.; Craigie, R. The Road to Chromatin - Nuclear Entry of Retroviruses. Nat. Rev. Microbiol. 2007, 5, 187–196. [Google Scholar] [CrossRef]
- Burkinsky, M.I.; Haggerty, S.; Sharova, N.; Adzhubei, A.; Spitz, L.; Lewis, P.; Goldfarb, D.; Emerman, M.; Stevenson, M. A Nuclear Localization Signal within HIV-1 Matrix Protein That Governs Infection of Non-Dividing Cells. Nature 1993, 365, 666–669. [Google Scholar]
- Von Schwedler, U.; Kornbluth, R.S.; Trono, D. The Nuclear Localization Signal of the Matrix Protein of Human Immunodeficiency Virus Type 1 Allows the Establishment of Infection in Macrophages and Quiescent T Lymphocytes. Proc. Natl. Acad. Sci. USA 2006, 6992–6996. [Google Scholar] [CrossRef]
- Jenkins, Y.; McEntee, M.; Weis, K.; Greene, W.C. Characterization of HIV-1 Vpr Nuclear Import: Analysis of Signals and Pathways. J. Cell Biol. 1998, 91, 875–885. [Google Scholar] [CrossRef]
- Karni, O.; Friedler, A.; Zakai, N.; Gilon, C.; Loyter, A. A Peptide Derived from the N-Terminal Region of HIV-1 Vpr Promotes Nuclear Import in Permeabilized Cells: Elucidation of the NLS Region of the Vpr. FEBS Lett. 1998, 429, 421–425. [Google Scholar] [CrossRef]
- Gallay, P.; Hope, T.; Chin, D.; Trono, D. HIV-1 Infection of Nondividing Cells through the Recognition of Integrase by the Importin/Karyopherin Pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 9825–9830. [Google Scholar] [CrossRef]
- Depienne, C.; Leh, H.; Le Rouzic, E.; Dormont, D.; Benichou, S.; Dargemont, C. Characterization of the Nuclear Import Pathway for HIV-1 Integrase. J. Biol. Chem. 2001, 276, 18102–18107. [Google Scholar] [CrossRef] [Green Version]
- Hamid, F.B.; Kim, J.; Shin, C.-G. Cellular and Viral Determinants of Retroviral Nuclear Entry. Can. J. Microbiol. 2016, 62, 1–15. [Google Scholar] [CrossRef]
- Bhargava, A.; Lahaye, X.; Manel, N. Let Me in: Control of HIV Nuclear Entry at the Nuclear Envelope. Cytokine Growth Factor Rev. 2018, 40, 59–67. [Google Scholar] [CrossRef]
- Ao, Z.; Jayappa, K.D.; Wang, B.; Zheng, Y.; Wang, X.; Peng, J.; Yao, X. Contribution of Host Nucleoporin 62 in HIV-1 Integrase Chromatin Association and Viral DNA Integration. J. Biol. Chem. 2012, 287, 10544–10555. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Matunis, M.J.; Kraemer, D.; Blobel, G.; Coutavas, E. Nup358, a Cytoplasmically Exposed Nucleoporin with Peptide Repeats, Ran- GTP Binding Sites, Zinc Fingers, a Cyclophilin a Homologous Domain, and a Leucine-Rich Region. J. Biol. Chem. 1995, 270, 14209–14213. [Google Scholar] [CrossRef]
- Yokoyama, N.; Hayashi, N.; Seki, T.; Pante, N.; Ohba, T.; Nishii, K.; Kuma, K.; Hayashida, T.; Miyata, T.; Aebi, U.; et al. A Giant Nucleopore Protein That Binds Ran/TC4. Nature 1995, 376, 184–188. [Google Scholar] [CrossRef]
- Francis, A.C.; Melikyan, G.B. Live-Cell Imaging of Early Steps of Single HIV-1 Infection. Viruses 2018, 10, 275. [Google Scholar] [CrossRef]
- Rawle, D.J.; Harrich, D. Toward the “Unravelling” of HIV: Host Cell Factors Involved in HIV-1 Core Uncoating. PLoS Pathog. 2018, 14, e1007270. [Google Scholar] [CrossRef]
- Bichel, K.; Price, A.J.; Schaller, T.; Towers, G.J.; Freund, S.M.; James, L.C. HIV-1capsid undergoes coupled binding and isomerization by the nuclear pore proteinNUP358. Retrovirology 2013, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, F. New Insights in the Role of Nucleoporins: A Bridge Leading to Concerted Steps from HIV-1 Nuclear Entry until Integration. Virus Res. 2013, 178, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Schaller, T.; Ocwieja, K.E.; Rasaiyaah, J.; Price, A.J.; Brady, T.L.; Roth, S.L.; Hué, S.; Fletcher, A.J.; Lee, K.E.; KewalRamani, V.N.; et al. HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency. PLoS Pathog. 2011, 7, e1002439. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.H.; Zimmermann, S.; Stuwe, T.; Stuwe, E.; Hoelz, A. Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2. J. Mol. Biol. 2013, 425, 1318–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakielny, S. Nup153 Is an M9-Containing Mobile Nucleoporin with a Novel Ran-Binding Domain. EMBO J. 1999, 18, 1982–1995. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, F.; Souque, P.; Charneau, P.; Fricke, T.; Valle-Casuso, J.C.; Perez, P.; Diaz-Griffero, F.; Miccio, A.; Mavilio, F.; Rizzi, E.; et al. Nup153 and Nup98 Bind the HIV-1 Core and Contribute to the Early Steps of HIV-1 Replication. Virology 2013, 440, 8–18. [Google Scholar] [CrossRef]
- Paulillo, S.M.; Phillips, E.M.; Köser, J.; Sauder, U.; Ullman, K.S.; Powers, M.A.; Fahrenkrog, B. Nucleoporin Domain Topology Is Linked to the Transport Status of the Nuclear Pore Complex. J. Mol. Biol. 2005, 351, 784–798. [Google Scholar] [CrossRef]
- Ball, J.R.; Ullman, K.S. Versatility at the Nuclear Pore Complex: Lessons Learned from the Nucleoporin Nup153. Chromosoma 2005, 114, 319–330. [Google Scholar] [CrossRef]
- Bayliss, R.; Littlewood, T.; Stewart, M. Structural basis for the interaction between FXFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 2000, 102, 99–108. [Google Scholar] [CrossRef]
- Woodward, C.L.; Prakobwanakit, S.; Mosessian, S.; Chow, S.A. Integrase Interacts with Nucleoporin NUP153 To Mediate the Nuclear Import of Human Immunodeficiency Virus Type 1. J. Virol. 2009, 83, 6522–6533. [Google Scholar] [CrossRef] [Green Version]
- Matreyek, K.A.; Engelman, A. The Requirement for Nucleoporin NUP153 during Human Immunodeficiency Virus Type 1 Infection Is Determined by the Viral Capsid. J. Virol. 2011, 85, 7818–7827. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Ng, S.B.H.; Mahalingam, S.; Balasundaram, D.; Dorairajoo, D.S.K.; Gandotra, S.; Varadarajan, P. The Functionally Conserved Nucleoporins Nup124p from Fission Yeast and the Human Nup153 Mediate Nuclear Import and Activity of the Tf1 Retrotransposon and HIV-1 Vpr. Mol. Biol. Cell 2005, 16, 1823–1838. [Google Scholar] [Green Version]
- Dewangan, P.S.; Sonawane, P.J.; Chouksey, A.R.; Chauhan, R. The Nup62 Coiled-Coil Motif Provides Plasticity for Triple-Helix Bundle Formation. Biochemistry 2017, 56, 2803–2811. [Google Scholar] [CrossRef]
- Chopra, K.; Bawaria, S.; Chauhan, R. Evolutionary Divergence of the Nuclear Pore Complex from Fungi to Metazoans. Prot. Sci. 2019, 8, 571–586. [Google Scholar] [CrossRef]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Koh, Y.; Wu, X.; Ferris, A.L.; Matreyek, K.A.; Smith, S.J.; Lee, K.; KewalRamani, V.N.; Hughes, S.H.; Engelman, A. Differential Effects of Human Immunodeficiency Virus Type 1 Capsid and Cellular Factors Nucleoporin 153 and LEDGF/P75 on the Efficiency and Specificity of Viral DNA Integration. J. Virol. 2013, 87, 648–658. [Google Scholar] [CrossRef]
- Emerman, M. HIV-1 Regulatory/Accessory Genes: Keys to Unraveling Viral and Host Cell Biology. Science 1998, 280, 1880–1884. [Google Scholar] [CrossRef]
- Ptak, R.G. HIV-1 Regulatory Proteins: Targets for Novel Drug Development. Expert Opin. Investigl. Drugs 2005, 11, 1099–1115. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B.R. The Arginine-Rich Domains Present in Human Immunodeficiency Virus Type 1 Tat and Rev Function as Direct Importin β-Dependent Nuclear Localization Signals. Mol. Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef]
- Barboric, M.; Fujinaga, K. The Two Sides of Tat. ELife 2016, 19, e12686. [Google Scholar] [CrossRef]
- Dayton, A.I. Within You, without You: HIV-1 Rev and RNA Export. Retrovirology 2004, 1, 35. [Google Scholar] [CrossRef]
- Szebeni, A.; Baumann, A.; Olson, M.O.J.; Mehrotra, B.; Adam, S.A.; Wingfield, P.T. Nucleolar Protein B23 Stimulates Nuclear Import of the HIV-1 Rev Protein and NLS-Conjugated Albumin. Biochemistry 1997, 36, 3941–3949. [Google Scholar] [CrossRef]
- Zolotukhin, A.S.; Felber, B.K. Nucleoporins Nup98 and Nup214 Participate in Nuclear Export of Human Immunodeficiency Virus Type 1 Rev. J. Virol. 1999, 73, 120–127. [Google Scholar] [Green Version]
- Hutten, S.; Kehlenbach, R.H. Nup214 Is Required for CRM1-Dependent Nuclear Protein Export In Vivo. Mol. Cell Biol. 2006, 26, 6772–6785. [Google Scholar] [CrossRef] [Green Version]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.; Rosbash, M. The Importin-Beta Family Member Crm1p Bridges the Interaction between Rev and the Nuclear Pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Hofmann, W.; Reichart, B.; Ewald, A.; Müller, E.; Schmitt, I.; Stauber, R.H.; Lottspeich, F.; Jockusch, B.M.; Scheer, U.; Hauber, J.; et al. Cofactor Requirements for Nuclear Export of Rev Response Element (RRE)- and Constitutive Transport Element (CTE)-Containing Retroviral RNAs: An Unexpected Role for Actin. J. Cell Biol. 2001, 152, 895–910. [Google Scholar] [CrossRef]
- Strebel, K. Virus-Host Interactions: Role of HIV Proteins Vif, Tat, and Rev. AIDS 2003, 17, S25–S34. [Google Scholar] [CrossRef]
- Hezwani, M.; Fahrenkrog, B. The Functional Versatility of the Nuclear Pore Complex Proteins. Semin. Cell Dev. Biol. 2017, 68, 2–9. [Google Scholar] [CrossRef]
- Jühlen, R.; Fahrenkrog, B. Moonlighting Nuclear Pore Proteins: Tissue-Specific Nucleoporin Function in Health and Disease. Histochem. Cell Biol. 2018, 150, 593–605. [Google Scholar] [CrossRef]
- Borlido, J.; D’Angelo, M.A. Nup62-mediated nuclear import of p63 in squamous cell carcinoma. EMBO Rep. 2018, 19, 3–4. [Google Scholar] [CrossRef]
- Rodriguez-Bravo, V.; Pippa, R.; Song, W.; Carceles-Cordon, M.; Dominguez-Andres, A.; Fujiwara, N.; Woo, J.; Koh, A.P.; Ertel, A.; Lokareddy, R.K.; et al. Nuclear Pores Promote Lethal Prostate Cancer by Increasing POM121-Driven E2F1, MYC, and AR Nuclear Import. Cell 2018, 174, 1200–1215. [Google Scholar] [CrossRef]
- Chan, E.Y.; Diamond, D.L.; Katze, M.G.; Qian, W.-J.; Liu, T.; Gritsenko, M.A.; Monroe, M.E.; Camp II, D.G.; Smith, R.D. Quantitative Analysis of Human Immunodeficiency Virus Type 1-Infected CD4+ Cell Proteome: Dysregulated Cell Cycle Progression and Nuclear Transport Coincide with Robust Virus Production. J. Virol. 2007, 88, 7571–7583. [Google Scholar] [CrossRef]
- Chan, E.Y.; Sutton, J.N.; Jacobs, J.M.; Bondarenko, A.; Smith, R.D.; Katze, M.G. Dynamic Host Energetics and Cytoskeletal Proteomes in Human Immunodeficiency Virus Type 1-Infected Human Primary CD4 Cells: Analysis by Multiplexed Label-Free Mass Spectrometry. J. Virol. 2009, 83, 9283–9295. [Google Scholar] [CrossRef] [Green Version]
- Ori, A.; Banterle, N.; Iskar, M.; Andrés-Pons, A.; Escher, C.; Khanh, H.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P. Cell Type-Specific Nuclear Pores: A Case in Point For Context-Dependent Stoichiometry Of Molecular Machines. Mol. Sys. Biol. 2013, 9, 648. [Google Scholar] [CrossRef]
- Kane, M.; Rebensburg, S.V.; Takata, M.A.; Zang, T.M.; Yamashita, M.; Kvaratskhelia, M.; Bieniasz, P.D. Nuclear Pore Heterogeneity Influences HIV-1 Infection and the Antiviral Activity of MX2. ELife 2018, 7, e35738. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Gomez-Cavazos, J.S.; Mei, A.; Lackner, D.H.; Hetzer, M.W. A Change in Nuclear Pore Complex Composition Regulates Cell Differentiation. Dev. Cell 2012, 22, 446–458. [Google Scholar] [CrossRef] [Green Version]
- Raices, M.; D’Angelo, M.A. Nuclear Pore Complex Composition: A New Regulator of Tissue-Specific and Developmental Functions. Nat. Rev. Mol. Cell Biol. 2012, 13, 687–699. [Google Scholar] [CrossRef]
- Gorjánácz, M. Nuclear Assembly as a Target for Anti-Cancer Therapies. Nucleus 2014, 5, 47–55. [Google Scholar] [CrossRef]
- Kosyna, F.; Depping, R. Controlling the Gatekeeper: Therapeutic Targeting of Nuclear Transport. Cells 2018, 7, 221. [Google Scholar] [CrossRef]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B Inactivates CRM1/Exportin 1 by Covalent Modification at a Cysteine Residue in the Central Conserved Region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef]
- Dickmanns, A.; Monecke, T.; Ficner, R. Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells 2015, 4, 538–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newlands, E.S.; Rustin, G.J.; Brampton, M.H. Phase I trial of elactocin. Br. J. Cancer 1996, 74, 648–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, A.; Loyter, A.; Bukrinsky, M. Strategies to Inhibit Viral Protein Nuclear Import: HIV-1 as a Target. Biochim. Biophys. Acta-Mol. Cell Res. 2011, 1813, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Armon-Omer, A.; Rosenbluh, J.; Melamed-Book, N.; Graessmann, A.; Waigmann, E.; Loyter, A. Inhibition of HIV-1 Integrase Nuclear Import and Replication by a Peptide Bearing Integrase Putative Nuclear Localization Signal. Retrovirology 2009, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Hayouka, Z.; Levin, A.; Maes, M.; Hadas, E.; Shalev, D.E.; Volsky, D.J.; Loyter, A.; Friedler, A. Mechanism of Action of the HIV-1 Integrase Inhibitory Peptide LEDGF. Biochem. Biophys. Res. Comm. 2010, 394, 260–265. [Google Scholar] [CrossRef] [PubMed]








| Class | Protein | Function |
|---|---|---|
| Viral enzymes (encoded by pol) | Reverse transcriptase (RT) | builds a DNA copy of the viral RNA genome; reverse transcription and RNAse H activity [9] |
| Integrase (IN) | takes the DNA copy of the viral genome inside nucleus and inserts it into the infected cellular genome [10] | |
| Protease (PR) | essential for cleavage of polypeptides and maturation of HIV particles [11] | |
| Structural proteins (encoded by gag and env) | Matrix (MA) | forms a coat on the inner surface of the viral membrane [12] |
| Capsid (CA) | forms a cone-shaped coat around the viral RNA, delivering it into the cell during infection [13] | |
| Nucleocapsid (NC) | forms a stable complex with the viral RNA to protect it [14] | |
| Gp120 (SU envelope protein) | bind to receptors on the surface of host cell and then penetrate the cell surface making way for fusion [15,16] | |
| Gp41 (TM envelope protein) | ||
| Accessory proteins | Tat | transactivator of transmembrane, enhances transcription of viral mRNA [17] |
| Rev (regulator of virion expression) | regulates the splicing and nuclear export of viral RNA [18] | |
| Nef (negative regulatory factor) | inhibits host’s defences; pleiotropic effects, can increase or decrease virus replication [19] | |
| P6 | incorporation of Vpr into new viruses; virion budding [20] | |
| Vif (viral infectivity factor) | increases virus infectivity by inhibiting host’s defence proteins; helps in virion assembly [21] | |
| Vpr (viral protein r) | nuclear import of viral DNA [22,23] | |
| Vpu (viral protein u) | Helps in virus budding and release [24] |
| Drug Classes | Mode of Action |
|---|---|
| Receptor/co-receptor antagonist | Inhibits HIV fusion to host cells (gp120/CCR5/CXCR4) [31] |
| Fusion Inhibitors | Blocks virus penetration through host cell membrane [32] |
| NNRTI | Non-nucleoside reverse transcriptase inhibitors; binds at position distant from active sites of RT [33] |
| NRTI | Nucleoside reverse transcriptase inhibitors; competitive inhibitor [34] |
| PR inhibitors | Inhibits protease; prevents virion maturation [35] |
| INSTI | Integrase strand transfer inhibitor; prevents vDNA integration into the host genome [36] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, E.; Chauhan, R. Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention. Cells 2019, 8, 1155. https://doi.org/10.3390/cells8101155
Shukla E, Chauhan R. Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention. Cells. 2019; 8(10):1155. https://doi.org/10.3390/cells8101155
Chicago/Turabian StyleShukla, Ekta, and Radha Chauhan. 2019. "Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention" Cells 8, no. 10: 1155. https://doi.org/10.3390/cells8101155