- 1Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2School of Medicine, Zhejiang University, Hangzhou, China
- 3NHC Key Laboratory of Combined Multi-organ Transplantation, Hangzhou, China
- 4Key Laboratory of the Diagnosis and Treatment of Organ Transplantation, Research Unit of Collaborative Diagnosis and Treatment For Hepatobiliary and Pancreatic Cancer, Chinese Academy of Medical Sciences (2019RU019), Hangzhou, China
- 5Key Laboratory of Organ Transplantation, Research Center for Diagnosis and Treatment of Hepatobiliary Diseases, Hangzhou, China
Chemokine-like factor (CKLF)–like MARVEL transmembrane domain-containing family (CMTMs) is a new gene family, consisting of CKLF and CMTM1 to CMTM8, which plays an important role in hematopoiesis system, autoimmune diseases, male reproduction etc. Abnormal expression of CMTMs is also associated with tumor genesis, development and metastasis. In this review, we briefly describe the characteristics of CMTM family, outline its functions in multiple kinds of carcinomas, and summarize the latest research on their roles in hepatocellular carcinoma which are mainly related to the expression, prognostic effect, potential functions, and mechanism of action. The CMTM family is expected to provide new ideas and targets for HCC diagnosis and treatment.
Introduction
Chemokine-like factor (CKLF)–like MARVEL transmembrane domain-containing family (CMTMs) as a new gene family, consists of nine genes totally, which includes CKLF and CMTM1 until CMTM8 genes. CKLF1 and its three variants were first discovered and reported by Han et al. from Peking University Human Disease Gene Research Center in 2001 (1). They also identified CMTM1 to CMTM8 genes by reverse transcription polymerase chain reaction (PCR) techniques in the subsequent studies (1, 2). The genes of CMTM families are located on different chromosomes. CKLF and CMTM1–4 form a gene cluster on chromosome 16q, CMTM5 is located on chromosome 14q11.2, and CMTM6–8 form another gene cluster on chromosome 3p22.3 (2). They have different alternative RNA splicing forms respectively. Their coding products are mostly distributed both in the cytomembrane and cytoplasm. The functional characteristics of the gene products lie between classical chemokines and members of the transmembrane 4 super family (TM4SF). These characteristics partly owe to the special molecular structure of CMTMs protein which contains the MARVEL domain with four transmembrane-helix architecture and is closely linked with vesicle transport and membrane binding related events (3–5).
The members of CMTM family are widely expressed in human tissues and involve multiple biological systems, such as immune (5–9), male reproductive (10–12), hematopoiesis (13, 14), circulatory (15, 16), and muscular systems (1, 17). The abnormal expressions of CMTMs are associated with various diseases. CKLF1 mediates the immune inflammatory reaction process in rheumatoid arthritis and atopic dermatitis (18, 19). CKLF1 helps to promote the migration and proliferation of vascular smooth muscle and skeletal muscle cells (1, 17). Additionally, CKLF1 is also a potential target for the treatment of focal cerebral ischemia and cardiopulmonary complications (16, 20). CMTM1, CMTM2, CMTM3, and CMTM4 play a crucial role in the spermatogenesis process or testicular development, and these CMTMs could be used as potential molecular markers for diagnosis or treatment of male infertility (10–12, 21). CMTM2 exhibits a negative regulatory effect on human immunodeficiency virus type-1 transcription by inhibiting the AP-1 and CREB pathways (22). The promoter hypermethylation of CMTM2 could distinguish Sézary syndrome from erythrodermic inflammatory dermatosis (7). Both CMTM3 and CMTM4 are associated with angiogenesis by regulating cell surface availability of VE-cadherin (13, 14). Methylation levels in CMTM4 and CMTM5 are significantly different in the case of systemic lupus erythematosus and rheumatoid arthritis (8). CMTM7 contributes to B-1a cell development where the regulation of B-cell antigen receptors expression takes place (23). The polymorphisms of CMTM7 gene are associated with heart failure mortality among the European patients and increase the risk of obesity in Han Chinese male children (15, 24). CMTM8 promotes bone marrow-derived mesenchymal stem/stromal cells proliferation and migration via the epidermal growth factor receptor (EGFR) signaling, which provides a new research direction in bone regeneration and tissue engineering (25).
With the in-depth study, researchers found that the CMTMs do not only play important biological roles in these diseases as mentioned above, but also have been implicated in various cancers, involving tumorigenesis, development and metastasis (26). Different CMTMs have different effects on tumors, some of them have become potential therapeutic targets or prognostic indicators of tumors. Hepatocellular Carcinoma (HCC) is one of the most common malignancies worldwide with high recurrence rate and poor prognosis. Alpha-fetoprotein is the main serum biomarkers to diagnosis of HCC over past years, but the specificity and sensitivity is not satisfactory. There is a pressing need to explore novel biomarkers to detect early onset HCC and identify prognostic biomarkers for HCC to improve clinical outcomes. In addition, effective molecular therapeutic target is lacking which limit the roles of molecular targeted therapy and immunotherapy in HCC. Recent researches have revealed that CMTMs might also be closely associated with HCC. CMTM members expected to represent promising targets for HCC diagnosis and treatment. Herein, we briefly describe the characteristics of the CMTM family, outline the functions of CMTM family in diverse tumors, and focus on summarizing the latest studies on the detailed functions and underlying molecular mechanisms of CMTMs in HCC (Table 1).
CMTM Family Members and HCC
CKLF
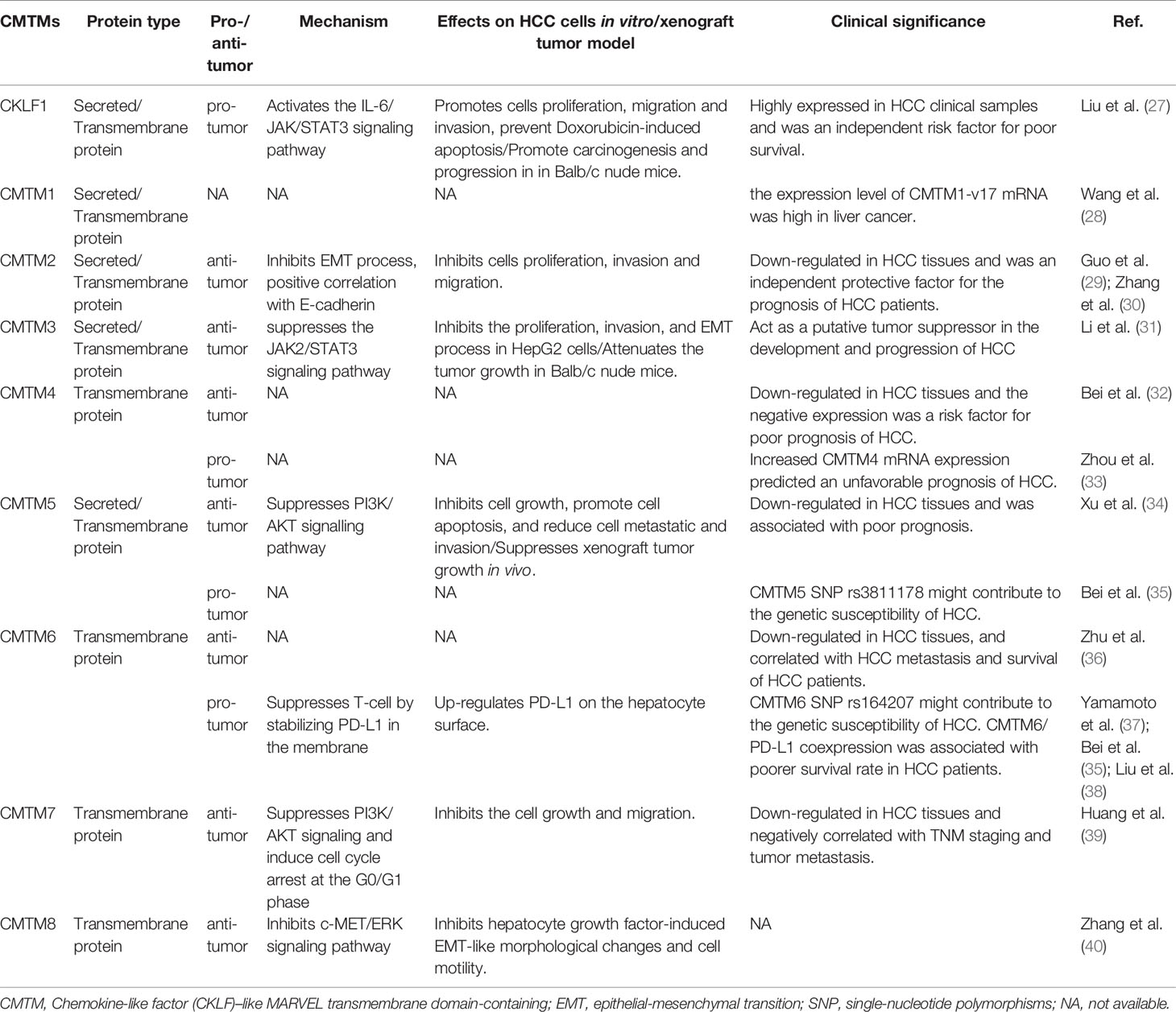
CKLF is the first identified member of CMTM family which located at chromosome 16q21. It has at least four alternative RNA splicing forms: CKLF1, CKLF2, CKLF3, CKLF4, in which CKLF2 is a full length cDNA product (1). CKLF1 and CKLF3 are secreted isoform, whereas CKLF2 and CKLF4 are transmembrane isoform. CKLFs show the most similarity to chemokines in CMTM family. These proteins have broad-spectrum chemotactic activity by interaction with human CC chemokine receptor 4 (CCR4) through two C-terminal peptides, C19 and C27 (41). Currently, a large number of studies have already revealed that CKLFs play an important role in inflammatory and autoimmune diseases. CKLFs may also have effects on both malignant and benign tumors (42). CKLF1 was upregulated in ovarian cancer tissues but downregulated in lung cancer tissues (42). Overexpressed CKLF1 was related to the formation of keloid and abdominal aortic aneurysms (43, 44). Recently, Liu et al. analyzed the expression level, prognostic value and potential function of CKLF1 in HCC (27). They found that CKLF1 was highly expressed in HCC tissues and related to the vascular invasion and tumor size. CKLF1 could activate the IL-6/STAT3 signaling pathway and up-regulation of Bcl-xl, MYC, and cyclins D1 to enhance HCC development and metastasis, and resist the apoptosis induced by Doxorubicin. Their findings were consistent with the results of the Human Protein Atlas which showed that the higher CMTM1 expression was, the poorer prognosis of HCC would be (Figure 1). Therefore, CKLF1 may be a pivotal modulator in the occurrence and development of HCC. CKLF1 could affect the biological behavior and prognosis of HCC, and is expected to become a potential target for the diagnosis and treatment of HCC.
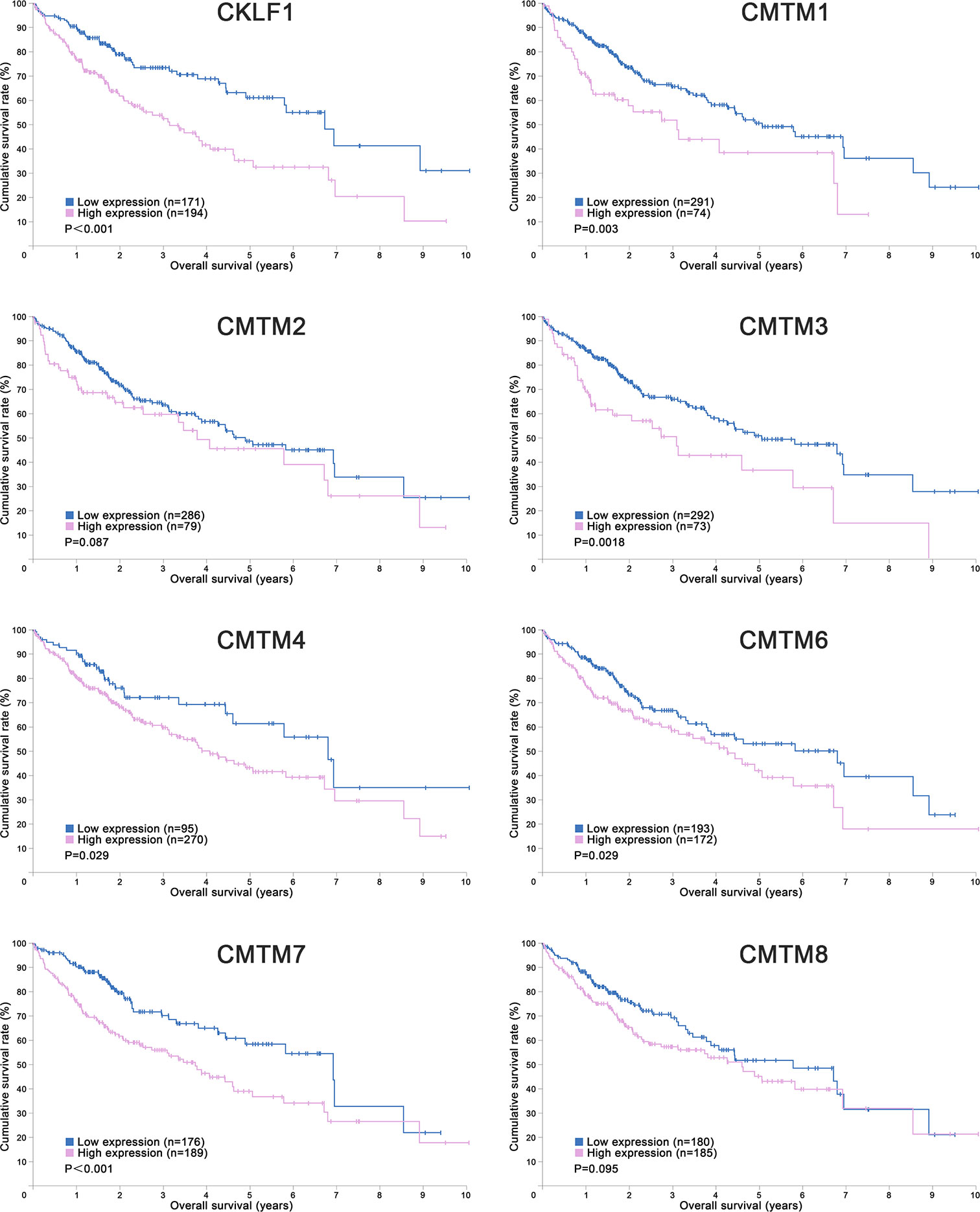
Figure 1 Kaplan-Meier Curves of CMTMs in Hepatocellular Carcinoma. Image credit: the Human Protein Atlas database. https://www.proteinatlas.org/.
CMTM1
CMTM1 is located at chromosome 16q21 and consists of 23 isoforms (CMTM1 v1-v23). The proteins of CMTM1 v1-16 and CMTM1 v17-23 are encoded by open reading frame 1 and open reading frame 2 respectively. The expression of CMTM1 is tissue specific and is predominantly expressed in testis tissue. Via PCR techniques, Wang et al. found that CMTM1-v17 was also highly expressed in many kinds of tumors, such as breast, kidney, lung, ovarian and liver cancers (28). The study demonstrated that CMTM1-v17 enhanced the cellular proliferation of breast cancer and prevented TNF-α-induced apoptosis by activating the NF-κB pathway. Si et al. discovered that CMTM1-v17 levels in non-small cell lung carcinoma groups was much higher than in parocarcinoma tissue, which might promote the chemoresistance and lead to poor prognosis (45). Additionally, CMTM1 promoted glioblastoma cells (A172 and U251MG) proliferation and invasion which have already been verified through in vitro experiments (46). Mays et al. found that CMTM1 was highly expressed in salivary adenoid cystic carcinoma (SACC) cells (SACC-83) (47). When the tumor tissues without recurrence/metastasis was compared with SACC-LM and tumor tissues with recurrence/metastasis, the result showed that CMTM1 might have enhanced the effectively of anti-tumor metastasis in SACC. A new study found that CMTM1-v5 specifically induced the human lymphoma cells apoptosis and may be a novel therapeutic for lymphoma treatment (48). Except for Wang et al. reported that CMTM1-v17 mRNA level was higher in liver tumor tissues than normal tissues which was consistent with the data from UALCAN (Figure 2), there was no other study has been taken up to investigate the relationship between CMTM1 and HCC so far. The Human Protein Atlas showed that the higher level of CMTM1 expression was associated with a lower survival probability in HCC (Figure 1). Combined with the results of PCR and bioinformatics analysis, we speculate that CMTM1 may serve as a potential tumor promoting role in HCC progression. The function and mechanism of CMTM1 in HCC should be elaborated in vitro and in vivo experiments in further study.
Figure 2 Expression profiles of CMTMs in Hepatocellular Carcinoma and the corresponding normal tissues. Image credit: UALCAN database. http://ualcan.path.uab.edu/.
CMTM2
CMTM2 gene is closely linked with and CMTM1 on chromosome 16q21, the distance between CMTM1 and CMTM2 is only 311 bp. They share a high level of amino acid sequence identity. Similar with CMTM1, the expression of CMTM2 is also high in the human testicular tissues, which can be secreted further into the seminiferous tubules (49). Moreover, the expression level of CMTM2 in SACC patients with tumor recurrence and perineural invasion is low too (47). Choi et al. evaluated 240 patients with diffuse-type gastric cancer. The study revealed that the patients with higher CMTM2 expression had a better overall survival (50). The Human Protein Atlas showed that CMTM2 is not a prognostic index in HCC (Figure 1), but several studies have shown CMTM2 had clinical significance in HCC. By using bioinformatics and immunohistochemistry (IHC) methods, Guo et al. found that the CMTM2 expression in HCC tissues is lower than the expression level in paired parocancerous tissues (29). In addition, its expression correlates with HCC pathological grades as well. They further confirmed that knockdown of CMTM2 in vitro significantly decreased the expression of E-cadherin and β-catenin and increased the expression of N-cadherin, Vimentin, ZEB1 and ZEB2. CMTM2 inhibited invasion and migration in HCC cells (Huh-7 and SMMC7721) by suppressing the epithelial-mesenchymal transition (EMT) process (30). In summary, CMTM2 plays tumor suppressive role and the expression level in tumor tissues could be a predictor of HCC, which could further guide postoperative treatment. Furthermore, the regulatory mechanisms of CMTM2 in HCC require further study which could provide a new direction for HCC treatment.
CMTM3
CMTM3 and CMTM4 genes are closely located at the tumor suppressor locus 16q22.1. CMTM3 promoter contains a typical CpG island. Different from other members in the CMTM family, alternatively spliced transcript variants of CMTM3 contain different 5′ UTRs, but encode the same protein. The protein only owns three transmembrane regions, in which one form is a secretory protein released via exosomes (2, 51). CMTM3 showed tumor-suppressive function while the DNA methylation of CMTM3 promoter inhibited CMTM3 expression in multiple tumors, such as gastric, breast, nasopharyngeal (52), male laryngeal (53), esophageal (54), and colon carcinomas (55). Li et al. revealed that CMTM3 could potentially induce a G2 cell cycle arrest and apoptosis in a p53-independent manner, thereby suppressing the proliferation and migration capabilities of testicular cancer cell line (NCCIT) (56). High expression of CMTM3 might correlate with favorable prognosis in gastric cancer, which not only inhibits the EGF-mediated tumorigenicity by promoting Rab5 activity, but also suppresses metastasis of gastric cancer via the STAT3/Twist1/EMT pathway (57–59). High expression of CMTM3 in prostate cancer cell also showed anti-tumor effects but the specific molecular mechanism remains unclear (60). In contrast with the anti-tumor effect, CMTM3 showed cancer-promoting effects in glioblastomas. Delic et al. found that higher CMTM3 expression was significantly correlated with shorter overall survival in glioblastomas by analyzing a large clinically annotated dataset. Knockdown of CMTM3 significantly reduced proliferation and invasiveness in U251MG cells (46). In HCC research, Li et al. confirmed that the expression of CMTM3 in HCC cell lines (HepG2, 97H, Hep3B, and HCCLM3) was low, and CMTM3 inhibits the proliferation and metastasis of HCC cells partially via suppressing the JAK2/STAT3 signaling pathway (31). Moreover, Zhao et al. identified the different expression of chemokines and chemokine-related genes between subsets of myeloid-derived suppressor, and put forward that CMTM3 may participate in regulating the tumor microenvironment of HCC (61). However, the results of the UALCAN and the Human Protein Atlas showed that higher CMTM3 expression was detected in liver tumor tissues and associated with poor prognosis, which is not consistent with the current researches (Figures 1 and 2). The relationship between CMTM3 expression and clinicopathological features of HCC patients is not clear yet. Therefore, more studies are needed to elucidate the clinical significance of CMTM3 in HCC diagnosis and prognosis from clinical samples.
CMTM4
CMTM4 is the most conserved member in CMTM family, and consists of three transcript variants including CMTM4-v1, CMTM4-v2, and CMTM4-v3. The CMTM4-v1 and -v2 are the main forms and widely expressed in multiple human tissue while on the other hand, the CMTM4-v3 only exists in certain renal and placental tissues (2). Kittler et al. successfully identified that CMTM4 was one of the 37 genes required for cell division in HeLa cells by using an endoribonuclease-prepared short interfering RNAs technique (62). The knockdown of CMTM4 caused the defect on cellular cleavage and resulted in bi-nucleated cells after mitosis. Plate et al. further studied the characteristics and functions of CMTM4 in HeLa cells. To the contrary, they found that CMTM4-v1 and -v2 inhibited HeLa cell growth by inducing G2/M phase accumulation without apoptosis (63). After that, many studies explored the expression and function of CMTM4 in a plenty of tumors, such as clear cell renal cell carcinoma (64), colorectal cancer (65) and pancreatic cancer (66). CMTM4 is frequently downregulated and act as a tumor-suppressor. The potential pathway mainly involved PI3K/AKT, ERK1/2, and STAT3 signaling pathways. Interestingly, Mezzadra et al. demonstrated that CMTM4 was a back-up regulator of programmed cell death ligand 1 (PD-L1) expression, which had the ability to protect PD-L1 protein from ubiquitination (67). Another research analyzed genes consistently overexpressed in 23 solid tumor types, and CMTM4 was identified as one of CD8+ T cell low tumor associated genes (68).
CMTM4 also showed clinical importance in liver cancer. Rong et al. found that CMTM4 was an oncogene in intrahepatic cholangiocarcinoma, and could be upregulated after knocking-down of FTO (69). In HCC, Bei et al. compared the expression of CMTM4 in cancer and parocancerous tissues using IHC (32). They concluded that there was a lower CMTM4 protein expression level in HCC tissues and further suggested that it might be a risk factor for poor prognosis in HCC patients. However, this result wasn’t consistent with the data obtained from the Human Protein Atlas (Figure 1) and UALCAN (Figure 2). Recently, by analyzing the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database, Zhou et al. found the expression of CMTM4 mRNA copies were significantly upregulated in HCC tissues and correlated with poor prognosis (33). Furthermore, they discovered CMTM4 had negative correlations with immune cells in HCC, and proposed CMTM4 might play an important role in HCC immune microenvironment. To date, the difference of CMTM4 in protein and mRNA expression levels has not been well explored. Whether there is mutation, promoter methylation, posttranscriptional regulation, or microRNAs regulation of CMTM4 expression in HCC is unknown. In addition, the expression of CMTM4 in HCC cell lines has not been well studied, whether existing of difference between mRNA and protein expression is unclear. Considering that CMTM4 may be involved in cell cycle regulation and related to PD-L1 protein re-expression, the role of CMTM4 in immunotherapy for HCC worthy further investigation.
CMTM5
CMTM5 is independently located at chromosome 14q11.2, and contains at least 6 alternatively spliced isoforms, ranging from CMTM5-v1 to -v6 (2). CMTM5-v1, as an evolutionarily conserved protein with 42% homology to CMTM3, is the major form in human CMTM5 and could be secreted via a vesicle mediated secretory pathway (51, 70). CMTM5 is highly expressed in human brain and blood (basophil, neutrophil), but frequently downregulated or silenced in pancreatic (71), cervical (72), oral (73), ovarian (74), prostate (75), renal (76), liver (34), and breast cancers (77). Similarity to the CMTM3 gene, the promoter methylation of CMTM5 gene is the main mechanism of tumor evading anticancer effect (78). By far, CMTM5 exhibits tumor suppressor activity in different types of cancer. For example, CMTM5-v1 induces pancreatic cancer cell lines (AsPC-1, BxPC-3, PANC-1, and MIA PaCa-2) apoptosis with activation of caspase 3, 8, and 9, and has synergistic effects with TNF-α (71). In renal cancer, restoration of CMTM5 could significantly inhibit ACHN cells growth by inducing cell cycle arrest in G0/G1 phase and apoptosis, and obviously suppress the cells migration and invasion (76). Another team confirmed that CMTM5-v1 is significantly downregulated in all the prostate cancer cell lines (PC3, DU 145, 22Rv1, and LNCaP). Ectopic expression of CMTM5-v1 could inhibit prostate cancer cell proliferation and migration by downregulating oncogenic EGFR signaling. Furthermore, CMTM5-v1 can improve the sensitivity of PC3 cells to Gefetinib, an EGFR tyrosine kinase inhibitor (79).
A case-control study indicated a strong correlation between rs3811178 in CMTM5 and risk of HCC in the southern Chinese population (35). Xu and Dang found CMTM5 was significantly reduced in 77.6% (59/76) of HCC tissues compared with the paired adjacent nontumor tissues, as well as in Huh7, Hep3B, HepG2, and SMMC-7721 cell lines. Overexpression of CMTM5 significantly inhibited Huh7 cell growth and metastasis in vitro and inhibited xenograft tumor growth in vivo. They further observed that CMTM5 is negatively correlate with the expression of PI3K, pAKT, Bcl2, cyclinD1, cyclinE, MMP2 and MMP9, and is positive correlation with the expression of p21, Bax, Bad, and cleaved caspase3 in HCC. The results suggested that CMTM5 may suppress HCC growth and metastasis through inhibiting PI3K-AKT signaling (34). In another study, Guan et al. proved that miR-10b-3p, acting as an oncogenic role, was dramatically up-regulation in HCC cell lines (HepG2) and the expression of CMTM5 was significantly suppressed (80). In addition, previous study indicated that the expression of CMTM5 in HCC could be restored by the treatment with PXD101, a histone deacetylase inhibitor (81). Taken together, CMTM5 may be expected to become a prognostic biomarker and valuable therapeutic target for HCC.
CMTM6
CMTM6 is another member of CMTM family located at chromosome 3p22.3 where it shares 55% amino acid identity with CMTM4. The protein is broadly expressed in many normal tissues, mainly in the plasma membrane, but the exact function is still unknown. Recent studies identified that CMTM6 was co-localization with PD-L1 and acted as a critical regulator for the maintenance of PD-L1 expression in various cancer types (67, 82). By using a CRISPR–Cas9 deletion library screen, Burr et al. discovered that CMTM6 could protect internalized PD-L1 from lysosomal degradation and effectively recycled PD-L1 back to the cell surface (82). Through a haploid genetic screen, Mezzadra et al. demonstrated that CMTM6 could stabilize PD-L1 in the membrane by preventing ubiquitination, which then induced the T-cell suppression (67). Blocking the interaction of the two proteins by H1A, a PD-L1 antibody, results PD-L1 degradation (83). Both high expression of CMTM6 and PD-L1 were associated with better survival rate in breast (84) and lung cancer (85). CMTM6 overexpression could enhance the therapeutic effect of immune checkpoint inhibitors in advanced-stage non-small-cell lung cancer (86). In contrast, high CMTM6 expression was associated with poor prognosis for gliomas (87), pancreatic adenocarcinomas (84) and squamous cell carcinoma (88, 89). Interestingly, Zhao et al. found the effect of PD-L1 in renal cancer was bidirectional regulation by the CMTM6 level (90). High expression of PD-L1 will promote cancer progression when CMTM6 was overexpressed, but the effect will be reversed when CMTM6 was down-regulated.
In HCC, CMTM6 expression of mRNA in tumor samples was significantly lower than in normal samples (Figure 2). Zhu et al. compared the expression of CMTM6 in 75 paired HCC and adjacent nontumor tissues through IHC (36). They found that CMTM6 was in a lower expression level in HCC, and which correlated with tumor metastasis and low survival of patients. The confusing part is that the results of other studies are the opposite. Yafune et al. found that the protein level of CMTM6 in HCC was higher than that in non-tumor tissues (91). CMTM6 was co-expressed with CK8/18+, and may become a detection marker of hepatocyte proliferative lesions. Liu et al. demonstrated that CMTM6/PD-L1 coexpression was associated with poorer survival rate in HCC patients, especially macrotrabecular−massive HCC patients (38). The results of the Human Protein Atlas also showed that higher CMTM6 expression was associated with a poorer survival probability (Figure 1). Additionally, Bei et al. evaluated the genetic variant in CMTM6 with HCC risks, and found the individuals with rs164207 AA genotype have a higher risk of HCC than with CC genotype (35). Yamamoto et al. found that CMTM6 could be upregulated by anti-HBV drug Entecavir, then induced PD-L1 on the hepatocyte surface (37). The reasons for the discrepancy in the CMTM6 mRNA level and results of IHC between these studies are still not clear. Post-translation and posttranscriptional regulations are the possible factors. Small scale populations and observational bias may also influence the results. Given the participation of CMTM6 in the carcinogenesis of HCC and biological process of PD-L1 stabilization, combined treatment of anti-CMTM6 and anti-PD-L1 may be a new method to enhance the therapeutic benefits of immune checkpoint inhibitors in HCC.
CMTM7
CMTM7 is located on the chromosome 3p22 which is rich in tumor-suppressor genes and the promoter contains a typical CpG island. The gene is evolutionarily conserved and encodes two isoforms, CMTM7-v1 and CMTM7-v2. CMTM7-v1 is the main form of expression. CMTM7 is widely expressed in human normal tissues, especially in immune cells while its protein is mainly located in cytoplasm and cell membrane. Previous studies have found that CMTM7 is frequently downregulated or absent in some cancers, partly because of the aberrant promoter CpG methylation and loss of heterozygosity. Overexpression of CMTM7 could inhibit cancer cells (KYSE410 and KYSE180) growth by inducing G1/S phase arrest and repressing EGFR-PI3K/AKT signaling in KYSE180 cells (92). The expression of CMTM7 could be dynamically adjusted by transcription factor FLI1 and SOX10 during tumor pathogenesis (93, 94). Another study indicated that the silence of CMTM7 could decrease Rab5 activation, promoting tumor growth and migration in non-small cell lung cancer (95). Huang et al. investigated the expression, function, and mechanism of CMTM7 in HCC through IHC and in vitro cell experiments (39). They found CMTM7 was significantly downregulated in HCC tissues and cell lines (Hep3B, SK-HEP-1, Huh7, and HepG2), and exhibited tumor-suppressor activities. CMTM7 is negatively correlated with the TNM staging of HCC and tumor metastasis. They further revealed that overexpression of CMTM7 could induce cell cycle arrest in G0/G1 phase by downregulation of cyclin D1 and cyclin-dependent kinase 4/6 (CDK4/6) expressions and upregulation of p27 expression, then inhibit the HCC cells growth and migration. Therefore, CMTM7 may serve as a potential biomarker to predict the possible of HCC invasion and metastasis. But similar with CMTM4, the results of the Human Protein Atlas are contrary, which showed that high CMTM7 expression is associated with poor prognosis (Figure 1). The causes of this phenomenon have yet to be elucidated, and should be explored in the future studies.
CMTM8
CMTM8 has high similarity to TM4SF in the CMTM family, with 39.3% homology (2). Bioinformatics analysis reveals that CMTM8 is conserved during evolution, the similarities between human and mouse CMTM8 is 95.4% (2). CMTM8, the full-length cDNA product, is the predominant isoform and expresses in many cell lines and normal tissues. Jin et al. revealed that CMTM8 inhibited tumor cells (HEK293, HeLa and PC3) growth by accelerating the internalization of EGFR to attenuate EGFR-mediated signaling pathway (96). Further study demonstrated that CMTM8 induced cell apoptosis through both caspase-dependent and -independent pathway (97). Li et al. reported CMTM8-v2 as an alternative spliced isoform of CMTM8 to maintain the ability to induce cell apoptosis (98). But it doesn’t affect EGFR internalization because of lacking second exon which codes MARVEL domain and cytosolic YXXø motif. Previous studies have demonstrated that CMTM8 was frequently downregulated or silenced in multiple solid tumors (liver, lung, colon, rectum, esophagus, stomach, bladder, bone) (99–101). Overexpression of CMTM8 in bladder cancer can inhibit cell growth, migration and invasion both in vivo and in vitro (100). Downregulation of CMTM8 induced EMT-like processes via HGF/c-MET/ERK signaling in HCC cells (HepG2) and other epithelial cells, indicating CMTM8 was a key in regulating cell motility and invasion (40). Fewer study on the relationship between CMTM8 and HCC patient survival have been reported to date, but the Human Protein Atlas showed that CMTM8 is not a prognostic index in HCC (Figure 1). Therefore, more studies are needed to examine the real clinical significance of CMTM8 in HCC.
Conclusions and Prospects
In conclusion, CMTMs have different expression profiles in HCC and normal tissues (Figure 2). Individual members of the CMTM family play different roles in the development and progression of HCC, mainly involved in the cellular proliferation, apoptosis, metastasis and invasion (Figure 3). CKLFs promoted carcinogenesis and enhanced the ability to resist chemotherapy in HCC. Current studies showed that CMTM2-8 act as tumor suppressors in HCC and the negative expression are risk factors for poor prognosis of HCC. These findings truly exist, but the interesting distinctions found between these findings and the results of database make them become controversial. The anti-tumor effect mainly relate with the regulation of cell cycle, inhibition of the EGFR-induced cell growth and the EMT process. DNA methylation and/or microRNAs regulation of these CMTMs is the dominant mechanism of HCC to evade anticancer effect. Small molecular agonist/inhibitor of CMTMs might be a kind of treatment method in the future. Additionally, CMTM4 and CMTM6, as PD-L1 protein regulators, are expected to be potential immunotherapy targets of HCC. It’s still unknown whether CMTM1 displays positive or negative effect on HCC. Hence, further exploration of CMTMs expression, molecular mechanisms and related signaling pathways in HCC is necessary. The CMTM family could provide new ideas and targets for HCC diagnosis and treatment.
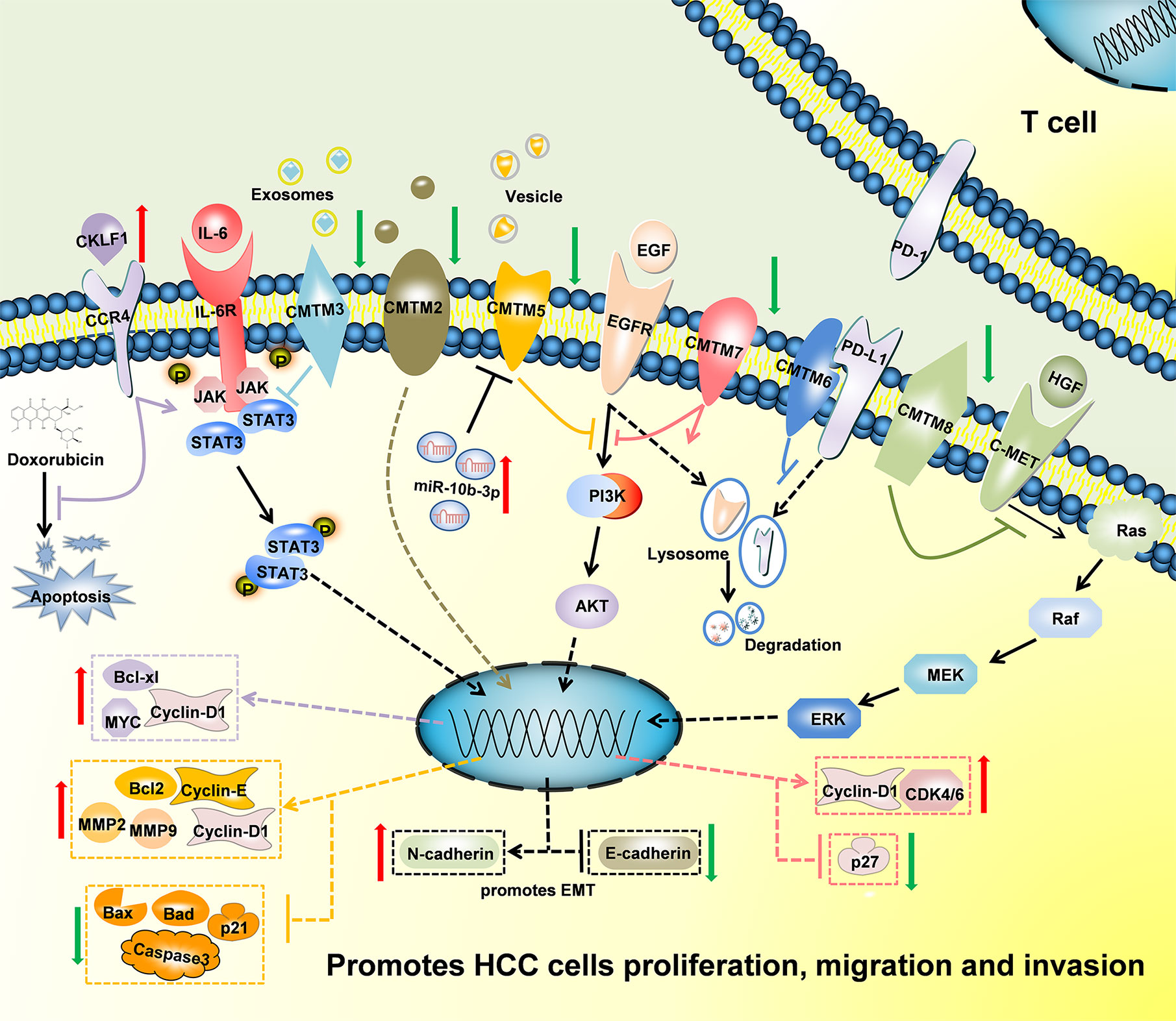
Figure 3 Schematic of the mechanisms regulated by CMTMs in Hepatocellular Carcinoma. CKLF1 enhanced hepatocarcinogenesis and prevented Doxorubicin-induced apoptosis via IL-6/STAT3 signaling pathway. Down-Regulated CMTM2, CMTM3, CMTM5, CMTM7 and CMTM8 promoted cells proliferation, migration, invasion, and induced epithelial-mesenchymal transition in Hepatocellular Carcinoma via JAK2/STAT3, PI3K/AKT, and c-MET/ERK signaling pathways. CMTM6 suppressed T-cell by stabilizing PD-L1 in the membrane. The mechanism of CMTM1 and CMTM4 is not clear yet.
Author Contributions
ML and FL collected related papers and drafted the manuscript. ML drafted the figures. XT and SY participated in the design of the review. LZ and SZ were responsible for the supervision of the work. All authors contributed to the article and approved the submitted version.
Funding
Innovative Research Groups of National Natural Science Foundation of China (No. 81721091), National S&T Major Project (No. 2017ZX10203205), Zhejiang International Science and Technology Cooperation Project (No. 2016C04003), Research Unit Project of Chinese Academy of Medical Sciences (2019-I2M-5-030), and Grant from Health Commission of Zhejiang Province (JBZX-202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Han W, Lou Y, Tang J, Zhang Y, Chen Y, Li Y, et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J (2001) 357(Pt 1):127–35. doi: 10.1042/0264-6021:3570127
2. Han W, Ding P, Xu M, Wang L, Rui M, Shi S, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics (2003) 81(6):609–17. doi: 10.1016/S0888-7543(03)00095-8
3. Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci (2002) 27(12):599–601. doi: 10.1016/S0968-0004(02)02229-6
4. Wu K, Li X, Gu H, Yang Q, Liu Y, Wang L. Research Advances in CKLF-like MARVEL Transmembrane Domain-containing Family in Non-small Cell Lung Cancer. Int J Biol Sci (2019) 15(12):2576–83. doi: 10.7150/ijbs.33733
5. Duan HJ, Li XY, Liu C, Deng XL. Chemokine-like factor-like MARVEL transmembrane domain-containing family in autoimmune diseases. Chin Med J (Engl) (2020) 133(8):951–8. doi: 10.1097/CM9.0000000000000747
6. Tian L, Li W, Wang J, Zhang Y, Zheng Y, Qi H, et al. The CKLF1-C19 peptide attenuates allergic lung inflammation by inhibiting CCR3- and CCR4-mediated chemotaxis in a mouse model of asthma. Allergy (2011) 66(2):287–97. doi: 10.1111/j.1398-9995.2010.02478.x
7. van Doorn R, Slieker RC, Boonk SE, Zoutman WH, Goeman JJ, Bagot M, et al. Epigenomic Analysis of Sézary Syndrome Defines Patterns of Aberrant DNA Methylation and Identifies Diagnostic Markers. J Invest Dermatol (2016) 136(9):1876–84. doi: 10.1016/j.jid.2016.03.042
8. Wang X, Lei D, Ding J, Liu S, Tao L, Zhang F, et al. A DNA-Methylated Sight on Autoimmune Inflammation Network across RA, pSS, and SLE. J Immunol Res (2018) 2018:4390789. doi: 10.1155/2018/4390789
9. Hu S, Vissink A, Arellano M, Roozendaal C, Zhou H, Kallenberg CGM, et al. Identification of autoantibody biomarkers for primary Sjögren’s syndrome using protein microarrays. Proteomics (2011) 11(8):1499–507. doi: 10.1002/pmic.201000206
10. Fujihara Y, Oji A, Kojima-Kita K, Larasati T, Ikawa M. Co-expression of sperm membrane proteins CMTM2A and CMTM2B is essential for ADAM3 localization and male fertility in mice. J Cell Sci (2018) 131(19):jcs221481. doi: 10.1242/jcs.221481
11. Liu F, Liu X, Liu X, Li T, Zhu P, Liu Z, et al. Integrated Analyses of Phenotype and Quantitative Proteome of CMTM4 Deficient Mice Reveal Its Association with Male Fertility. Mol Cell Proteomics (2019) 18(6):1070–84. doi: 10.1074/mcp.RA119.001416
12. Wang Y, Li T, Qiu X, Mo X, Zhang Y, Song Q, et al. CMTM3 can affect the transcription activity of androgen receptor and inhibit the expression level of PSA in LNCaP cells. Biochem Biophys Res Commun (2008) 371(1):54–8. doi: 10.1016/j.bbrc.2008.03.143
13. Chrifi I, Louzao-Martinez L, Brandt M, van Dijk CGM, Burgisser P, Zhu C, et al. CMTM3 (CKLF-Like Marvel Transmembrane Domain 3) Mediates Angiogenesis by Regulating Cell Surface Availability of VE-Cadherin in Endothelial Adherens Junctions. Arteriosclerosis Thromb Vasc Biol (2017) 37(6):1098–114. doi: 10.1161/ATVBAHA.116.308792
14. Chrifi I, Louzao-Martinez L, Brandt MM, van Dijk CGM, Burgisser PE, Zhu C, et al. CMTM4 regulates angiogenesis by promoting cell surface recycling of VE-cadherin to endothelial adherens junctions. Angiogenesis (2019) 22(1):75–93. doi: 10.1007/s10456-018-9638-1
15. Morrison AC, Felix JF, Cupples LA, Glazer NL, Loehr LR, Dehghan A, et al. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet (2010) 3(3):248–55. doi: 10.1161/CIRCGENETICS.109.895995
16. Kong LL, Wang ZY, Han N, Zhuang XM, Wang ZZ, Li H, et al. Neutralization of chemokine-like factor 1, a novel C-C chemokine, protects against focal cerebral ischemia by inhibiting neutrophil infiltration via MAPK pathways in rats. J Neuroinflam (2014) 11:112. doi: 10.1186/1742-2094-11-112
17. Zhang T, Zhang X, Yu W, Chen J, Li Q, Jiao Y, et al. Effects of chemokine-like factor 1 on vascular smooth muscle cell migration and proliferation in vascular inflammation. Atherosclerosis (2013) 226(1):49–57. doi: 10.1016/j.atherosclerosis.2012.09.023
18. Tao K, Tang X, Wang B, Li R-J, Zhang B-Q, Lin J-H, et al. Distinct expression of chemokine-like factor 1 in synovium of osteoarthritis, rheumatoid arthritis and ankylosing spondylitis. J Huazhong Univ Sci Technol Med Sci (2016) 36(1):70–6. doi: 10.1007/s11596-016-1544-4
19. Yang GY, Chen X, Sun YC, Ma CL, Qian G. Chemokine-like factor 1 (CLFK1) is over-expressed in patients with atopic dermatitis. Int J Biol Sci (2013) 9(8):759–65. doi: 10.7150/ijbs.6291
20. Ai Q, Chen C, Chu S, Luo Y, Zhang Z, Zhang S, et al. IMM-H004 Protects against Cerebral Ischemia Injury and Cardiopulmonary Complications via CKLF1 Mediated Inflammation Pathway in Adult and Aged Rats. Int J Mol Sci (2019) 20(7):1661. doi: 10.3390/ijms20071661
21. Wang L, Wu C, Zheng Y, Qiu X, Wang L, Fan H, et al. Molecular cloning and characterization of chemokine-like factor super family member 1 (CKLFSF1), a novel human gene with at least 23 alternative splicing isoforms in testis tissue. Int J Biochem Cell Biol (2004) 36(8):1492–501. doi: 10.1016/j.biocel.2003.11.017
22. Song HS, Shi S, Lu XZ, Gao F, Yan L, Wang Y, et al. Intracellular CMTM2 negatively regulates human immunodeficiency virus type-1 transcription through targeting the transcription factors AP-1 and CREB. Chin Med J (Engl) (2010) 123(17):2440–5.
23. Zhang Y, Wang J-Y, Han W. A role for CMTM7 in BCR expression and survival in B-1a but not B-2 cells. Int Immunol (2014) 26(1):47–57. doi: 10.1093/intimm/dxt042
24. Zhu Q, Xue K, Guo HW, Deng FF, Yang YH. Interaction of the CMTM7 rs347134 Polymorphism with Dietary Patterns and the Risk of Obesity in Han Chinese Male Children. Int J Environ Res Public Health (2020) 17(5):1515. doi: 10.3390/ijerph17051515
25. H’Ng CH, Camp E, Anderson PJ, Zannettino ACW, Gronthos S. CMTM8 is a suppressor of human mesenchymal stem cell osteogenic differentiation and promoter of proliferation via EGFR signalling. Stem Cells Dev (2020) 29(13):823–34. doi: 10.1089/scd.2020.0007
26. Wu J, Li L, Wu S, Xu B. CMTM family proteins 1-8: roles in cancer biological processes and potential clinical value. Cancer Biol Med (2020) 17(3):528–42. doi: 10.20892/j.issn.2095-3941.2020.0032
27. Liu Y, Liu L, Zhou Y, Zhou P, Yan Q, Chen X, et al. CKLF1 Enhances Inflammation-Mediated Carcinogenesis and Prevents Doxorubicin-Induced Apoptosis via IL6/STAT3 Signaling in HCC. Clin Cancer Res Off J Am Assoc Cancer Res (2019) 25(13):4141–54. doi: 10.1158/1078-0432.CCR-18-3510
28. Wang J, Zhang G, Zhang Y, Luo Y, Song Q, Qiu X, et al. CMTM1_v17 is a novel potential therapeutic target in breast cancer. Oncol Rep (2014) 32(5):1829–36. doi: 10.3892/or.2014.3429
29. Guo X, Zhang S, Tan S, Bei C, Zhang H, Zhu X, et al. Downregulated CMTM2 Poses Potential Clinical Significance in Hepatocellular Carcinoma. DNA Cell Biol (2020) 39(4):683–9. doi: 10.1089/dna.2019.5237
30. Zhang S, Tian R, Bei C, Zhang H, Kong J, Zheng C, et al. Down-Regulated CMTM2 Promotes Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Onco Targets Ther (2020) 13:5731–41. doi: 10.2147/OTT.S250370
31. Li W, Zhang S. CKLF-Like MARVEL Transmembrane Domain-Containing Member 3 (CMTM3) Inhibits the Proliferation and Tumorigenisis in Hepatocellular Carcinoma Cells. Oncol Res (2017) 25(2):285–93. doi: 10.3727/096504016X14732523471442
32. Bei C, Zhang Y, Wei R, Zhu X, Wang Z, Zeng W, et al. Clinical significance of CMTM4 expression in hepatocellular carcinoma. OncoTargets Ther (2017) 10:5439–43. doi: 10.2147/OTT.S149786
33. Zhou H-Q, Li J-H, Liu L-W, Lou J-M, Ren Z-G. Increased CMTM4 mRNA expression predicts a poor prognosis in patients with hepatocellular carcinoma. HBPD Int (2020) S1499–3872(20):30119–3. doi: 10.1016/j.hbpd.2020.06.004
34. Xu G, Dang C. CMTM5 is downregulated and suppresses tumour growth in hepatocellular carcinoma through regulating PI3K-AKT signalling. Cancer Cell Int (2017) 17:113. doi: 10.1186/s12935-017-0485-8
35. Bei C, Tan C, Zhu X, Wang Z, Tan S. Association Between Polymorphisms in CMTM Family Genes and Hepatocellular Carcinoma in Guangxi of China. DNA Cell Biol (2018) 37(8):691–6. doi: 10.1089/dna.2018.4274
36. Zhu X, Qi G, Li C, Bei C, Tan C, Zhang Y, et al. Expression and Clinical Significance of CMTM6 in Hepatocellular Carcinoma. DNA Cell Biol (2019) 38(2):193–7. doi: 10.1089/dna.2018.4513
37. Yamamoto Y, Kakizaki M, Shimizu T, Carreras J, Chiba T, Chamoto K, et al. PD-L1 is induced on hepatocyte surface via CKLF-like MARVEL transmembrane domain-containing protein 6 upregulation by the anti-HBV drug Entecavir. Int Immunol (2020) 32(8):519–31. doi: 10.1093/intimm/dxaa018
38. Liu L-L, Zhang S-W, Chao X, Wang C-H, Yang X, Zhang X-K, et al. Coexpression of CMTM6 and PD-L1 as a predictor of poor prognosis in macrotrabecular-massive hepatocellular carcinoma. Cancer Immunol Immunother CII (2020) Aug 7. doi: 10.1007/s00262-020-02691-9
39. Huang ZM, Li PL, Yang P, Hou XD, Yang YL, Xu X, et al. Overexpression of CMTM7 inhibits cell growth and migration in liver cancer. Kaohsiung J Med Sci (2019) 35(6):332–40. doi: 10.1002/kjm2.12058
40. Zhang W, Mendoza MC, Pei X, Ilter D, Mahoney SJ, Zhang Y, et al. Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem (2012) 287(15):11850–8. doi: 10.1074/jbc.M111.258236
41. Wang Y, Zhang Y, Han W, Li D, Tian L, Yin C, et al. Two C-terminal peptides of human CKLF1 interact with the chemokine receptor CCR4. Int J Biochem Cell Biol (2008) 40(5):909–19. doi: 10.1016/j.biocel.2007.10.028
42. Cai X, Deng J, Ming Q, Cai H, Chen Z. Chemokine-like factor 1: A promising therapeutic target in human diseases. Exp Biol Med (Maywood NJ) (2020) 245(16):1518–28. doi: 10.1177/1535370220945225
43. Li J, Bao X, Li Y, Wang Y, Zhao Z, Jin X. Study of the functional mechanisms of osteopontin and chemokine-like factor 1 in the development and progression of abdominal aortic aneurysms in rats. Exp Ther Med (2016) 12(6):4007–11. doi: 10.3892/etm.2016.3891
44. Zhang M, Xu Y, Liu Y, Cheng Y, Zhao P, Liu H, et al. Chemokine-Like Factor 1 (CKLF-1) is Overexpressed in Keloid Patients: A Potential Indicating Factor for Keloid-Predisposed Individuals. Medicine (2016) 95(11):e3082. doi: 10.1097/MD.0000000000003082
45. Si J, Zhang P, Tian D, Wang X, Ma Y, Zhang J, et al. CMTM1_v17 is associated with chemotherapy resistance and poor prognosis in non-small cell lung cancer. World J Surg Oncol (2017) 15(1):34. doi: 10.1186/s12957-016-1094-z
46. Delic S, Thuy A, Schulze M, Proescholdt MA, Dietrich P, Bosserhoff AK, et al. Systematic investigation of CMTM family genes suggests relevance to glioblastoma pathogenesis and CMTM1 and CMTM3 as priority targets. Genes Chromosomes Cancer (2015) 54(7):433–43. doi: 10.1002/gcc.22255
47. Mays AC, Feng X, Browne JD, Sullivan CA. Chemokine and Chemokine Receptor Profiles in Metastatic Salivary Adenoid Cystic Carcinoma. Anticancer Res (2016) 36(8):4013–8.
48. Cao L, Yang C, Zhu B, Zhang G, Zhao N, Liu X, et al. A novel protein CMTM1-v5 specifically induced human lymphoma cells apoptosis in vitro and in vivo. Exp Cell Res (2019) 385(1):111623. doi: 10.1016/j.yexcr.2019.111623
49. Shi S, Rui M, Han W, Wang Y, Qiu X, Ding P, et al. CKLFSF2 is highly expressed in testis and can be secreted into the seminiferous tubules. Int J Biochem Cell Biol (2005) 37(8):1633–40. doi: 10.1016/j.biocel.2004.04.028
50. Choi JH, Kim YB, Ahn JM, Kim MJ, Bae WJ, Han SU, et al. Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp Mol Med (2018) 50(4):6. doi: 10.1038/s12276-017-0009-6
51. Liu B, Li H, Fu W, Cheng Y, Yuan W, Liu W, et al. CMTM3 presents a secreted form released via exosomes. Acta Biochim Biophys Sin (2016) 48(6):584–6. doi: 10.1093/abbs/gmw029
52. Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H, et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res (2009) 69(12):5194–201. doi: 10.1158/0008-5472.CAN-08-3694
53. Shen Z, Chen X, Li Q, Zhou C, Xu Y, Yu R, et al. Elevated methylation of CMTM3 promoter in the male laryngeal squamous cell carcinoma patients. Clin Biochem (2016) 49(16-17):1278–82. doi: 10.1016/j.clinbiochem.2016.08.002
54. Han T, Shu T, Dong S, Li P, Li W, Liu D, et al. Chemokine-like factor-like MARVEL transmembrane domain-containing 3 expression is associated with a favorable prognosis in esophageal squamous cell carcinoma. Oncol Lett (2017) 13(5):2982–8. doi: 10.3892/ol.2017.5837
55. Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene (2017) 630:1–7. doi: 10.1016/j.gene.2017.07.082
56. Li Z, Xie J, Wu J, Li W, Nie L, Sun X, et al. CMTM3 inhibits human testicular cancer cell growth through inducing cell-cycle arrest and apoptosis. PloS One (2014) 9(2):e88965. doi: 10.1371/journal.pone.0088965
57. Su Y, Lin Y, Zhang L, Liu B, Yuan W, Mo X, et al. CMTM3 inhibits cell migration and invasion and correlates with favorable prognosis in gastric cancer. Cancer Sci (2014) 105(1):26–34. doi: 10.1111/cas.12304
58. Yuan W, Li T, Mo X, Wang X, Liu B, Wang W, et al. Knockdown of CMTM3 promotes metastasis of gastric cancer via the STAT3/Twist1/EMT signaling pathway. Oncotarget (2016) 7(20):29507–19. doi: 10.18632/oncotarget.8789
59. Yuan W, Liu B, Wang X, Li T, Xue H, Mo X, et al. CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett (2017) 386:77–86. doi: 10.1016/j.canlet.2016.11.015
60. Hu F, Yuan W, Wang X, Sheng Z, Yuan Y, Qin C, et al. CMTM3 is reduced in prostate cancer and inhibits migration, invasion and growth of LNCaP cells. Clin Trans Oncol Off Publ Fed Spanish Oncol Soc Natl Cancer Inst Mexico (2015) 17(8):632–9. doi: 10.1007/s12094-015-1288-9
61. Zhao W, Xu Y, Xu J, Wu D, Zhao B, Yin Z, et al. Subsets of myeloid-derived suppressor cells in hepatocellular carcinoma express chemokines and chemokine receptors differentially. Int Immunopharmacol (2015) 26(2):314–21. doi: 10.1016/j.intimp.2015.04.010
62. Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature (2004) 432(7020):1036–40. doi: 10.1038/nature03159
63. Plate M, Li T, Wang Y, Mo X, Zhang Y, Ma D, et al. Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol Cells (2010) 29(4):355–61. doi: 10.1007/s10059-010-0038-7
64. Li T, Cheng Y, Wang P, Wang W, Hu F, Mo X, et al. CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cell carcinoma. J Exp Clin Cancer Res CR (2015) 34:122. doi: 10.1186/s13046-015-0236-4
65. Xue H, Li T, Wang P, Mo X, Zhang H, Ding S, et al. CMTM4 inhibits cell proliferation and migration via AKT, ERK1/2, and STAT3 pathway in colorectal cancer. Acta Biochim Biophys Sin (2019) 51(9):915–24. doi: 10.1093/abbs/gmz084
66. Li M, Guo H, Wang Q, Chen K, Marko K, Tian X, et al. Pancreatic stellate cells derived exosomal miR-5703 promotes pancreatic cancer by downregulating CMTM4 and activating PI3K/Akt pathway. Cancer Lett (2020) 490:20–30. doi: 10.1016/j.canlet.2020.06.009
67. Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature (2017) 549(7670):106–10. doi: 10.1038/nature23669
68. Routh ED, Pullikuth AK, Jin G, Chifman J, Chou JW, D’Agostino RB, et al. Transcriptomic Features of T Cell-Barren Tumors Are Conserved Across Diverse Tumor Types. Front Immunol (2020) 11:57. doi: 10.3389/fimmu.2020.00057
69. Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao J, et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front Oncol (2019) 9:369. doi: 10.3389/fonc.2019.00369
70. Li H, Guo X, Shao L, Plate M, Mo X, Wang Y, et al. CMTM5-v1, a four-transmembrane protein, presents a secreted form released via a vesicle-mediated secretory pathway. BMB Rep (2010) 43(3):182–7. doi: 10.5483/BMBRep.2010.43.3.182
71. Guo X, Li T, Wang Y, Shao L, Zhang Y, Ma D, et al. CMTM5 induces apoptosis of pancreatic cancer cells and has synergistic effects with TNF-alpha. Biochem Biophys Res Commun (2009) 387(1):139–42. doi: 10.1016/j.bbrc.2009.06.148
72. Shao L, Guo X, Plate M, Li T, Wang Y, Ma D, et al. CMTM5-v1 induces apoptosis in cervical carcinoma cells. Biochem Biophys Res Commun (2009) 379(4):866–71. doi: 10.1016/j.bbrc.2008.12.126
73. Zhang H, Nan X, Li X, Chen Y, Zhang J, Sun L, et al. CMTM5 exhibits tumor suppressor activity through promoter methylation in oral squamous cell carcinoma. Biochem Biophys Res Commun (2014) 447(2):304–10. doi: 10.1016/j.bbrc.2014.03.158
74. Li P, Liu K, Li L, Yang M, Gao W, Feng J, et al. Reduced CMTM5 expression correlates with carcinogenesis in human epithelial ovarian cancer. Int J Gynecol Cancer (2011) 21(7):1248–55. doi: 10.1097/IGC.0b013e3182259c31
75. Xiao Y, Yuan Y, Zhang Y, Li J, Liu Z, Zhang X, et al. CMTM5 is reduced in prostate cancer and inhibits cancer cell growth in vitro and in vivo. Clin Trans Oncol Off Publ Fed Spanish Oncol Soc Natl Cancer Inst Mexico (2015) 17(6):431–7. doi: 10.1007/s12094-014-1253-z
76. Cai B, Xiao Y, Li Y, Zheng S. CMTM5 inhibits renal cancer cell growth through inducing cell-cycle arrest and apoptosis. Oncol Lett (2017) 14(2):1536–42. doi: 10.3892/ol.2017.6350
77. Wu J. CMTM5/7 are biomarkers and prognostic factors in human breast carcinoma. Cancer Biomarkers section A Dis Markers (2020) 29(1):89–99. doi: 10.3233/CBM-191226
78. Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang Y, et al. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res (2007) 13(19):5756–62. doi: 10.1158/1078-0432.CCR-06-3082
79. Yuan Y, Sheng Z, Liu Z, Zhang X, Xiao Y, Xie J, et al. CMTM5-v1 inhibits cell proliferation and migration by downregulating oncogenic EGFR signaling in prostate cancer cells. J Cancer (2020) 11(13):3762–70. doi: 10.7150/jca.42314
80. Guan L, Ji D, Liang N, Li S, Sun B. Up-regulation of miR-10b-3p promotes the progression of hepatocellular carcinoma cells via targeting CMTM5. J Cell Mol Med (2018) 22(7):3434–41. doi: 10.1111/jcmm.13620
81. Ma BB, Sung F, Tao Q, Poon FF, Lui VW, Yeo W, et al. The preclinical activity of the histone deacetylase inhibitor PXD101 (belinostat) in hepatocellular carcinoma cell lines. Invest New Drugs (2010) 28(2):107–14. doi: 10.1007/s10637-009-9219-7
82. Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature (2017) 549(7670):101–5. doi: 10.1038/nature23643
83. Tu X, Qin B, Zhang Y, Zhang C, Kahila M, Nowsheen S, et al. PD-L1 (B7-H1) Competes with the RNA Exosome to Regulate the DNA Damage Response and Can Be Targeted to Sensitize to Radiation or Chemotherapy. Mol Cell (2019) 74(6):1215–26.e4. doi: 10.1016/j.molcel.2019.04.005
84. Mamessier E, Birnbaum DJ, Finetti P, Birnbaum D, Bertucci F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Trans Med (2018) 6(3):54. doi: 10.21037/atm.2017.11.26
85. Wang H, Gao J, Zhang R, Li M, Peng Z, Wang H. Molecular and immune characteristics for lung adenocarcinoma patients with CMTM6 overexpression. Int Immunopharmacol (2020) 83:106478. doi: 10.1016/j.intimp.2020.106478
86. Zugazagoitia J, Liu Y, Toki M, McGuire J, Ahmed FS, Henick BS, et al. Quantitative Assessment of CMTM6 in the Tumor Microenvironment and Association with Response to PD-1 Pathway Blockade in Advanced-Stage Non-Small Cell Lung Cancer. J Thorac Oncol (2019) 14(12):2084–96. doi: 10.1016/j.jtho.2019.09.014
87. Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine (2018) 35:233–43. doi: 10.1016/j.ebiom.2018.08.012
88. Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, et al. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res (2019) 8(2):179–91. doi: 10.1158/2326-6066.CIR-19-0394
89. Zheng Y, Wang C, Song A, Jiang F, Zhou J, Li G, et al. CMTM6 promotes cell proliferation and invasion in oral squamous cell carcinoma by interacting with NRP1. Am J Cancer Res (2020) 10(6):1691–709.
90. Zhao W, Zhao F, Yang K, Lu Y, Zhang Y, Wang W, et al. An immunophenotyping of renal clear cell carcinoma with characteristics and a potential therapeutic target for patients insensitive to immune checkpoint blockade. J Cell Biochem (2019) 120(8):13330–41. doi: 10.1002/jcb.28607
91. Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, et al. Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model. Toxicol Lett (2013) 222(3):295–302. doi: 10.1016/j.toxlet.2013.08.006
92. Li H, Li J, Su Y, Fan Y, Guo X, Li L, et al. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene (2014) 33(24):3109–18. doi: 10.1038/onc.2013.282
93. Miao B, Bauer AS, Hufnagel K, Wu Y, Trajkovic-Arsic M, Pirona AC, et al. The transcription factor FLI1 promotes cancer progression by affecting cell cycle regulation. Int J Cancer (2020) 147(1):189–201. doi: 10.1002/ijc.32831
94. Jin Y, Qin X, Jia G. SOX10-dependent CMTM7 expression inhibits cell proliferation and tumor growth in gastric carcinoma. Biochem Biophys Res Commun (2018) 507(1-4):91–9. doi: 10.1016/j.bbrc.2018.10.172
95. Liu B, Su Y, Li T, Yuan W, Mo X, Li H, et al. CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget (2015) 6(38):41092–107. doi: 10.18632/oncotarget.5732
96. Jin C, Ding P, Wang Y, Ma D. Regulation of EGF receptor signaling by the MARVEL domain-containing protein CKLFSF8. FEBS Lett (2005) 579(28):6375–82. doi: 10.1016/j.febslet.2005.10.021
97. Jin C, Wang Y, Han W, Zhang Y, He Q, Li D, et al. CMTM8 induces caspase-dependent and -independent apoptosis through a mitochondria-mediated pathway. J Cell Physiol (2007) 211(1):112–20. doi: 10.1002/jcp.20914
98. Li D, Jin C, Yin C, Zhang Y, Pang B, Tian L, et al. An alternative splice form of CMTM8 induces apoptosis. Int J Biochem Cell Biol (2007) 39(11):2107–19. doi: 10.1016/j.biocel.2007.06.002
99. Both J, Krijgsman O, Bras J, Schaap GR, Baas F, Ylstra B, et al. Focal chromosomal copy number aberrations identify CMTM8 and GPR177 as new candidate driver genes in osteosarcoma. PloS One (2014) 9(12):e115835. doi: 10.1371/journal.pone.0115835
100. Gao D, Hu H, Wang Y, Yu W, Zhou J, Wang X, et al. CMTM8 inhibits the carcinogenesis and progression of bladder cancer. Oncol Rep (2015) 34(6):2853–63. doi: 10.3892/or.2015.4310
Keywords: chemokine-like factor–like MARVEL transmembrane domain-containing family, chemokine-like factor, hepatocellular carcinoma, MARVEL, tumor suppressor gene
Citation: Li M, Luo F, Tian X, Yin S, Zhou L and Zheng S (2020) Chemokine-Like Factor-Like MARVEL Transmembrane Domain-Containing Family in Hepatocellular Carcinoma: Latest Advances. Front. Oncol. 10:595973. doi: 10.3389/fonc.2020.595973
Received: 26 August 2020; Accepted: 16 October 2020;
Published: 13 November 2020.
Edited by:
Davide Zella, University of Maryland, Baltimore, United StatesReviewed by:
Xiaonian Zhu, Guilin Medical University, ChinaFrancesca Benedetti, University of Maryland, United States
Copyright © 2020 Li, Luo, Tian, Yin, Zhou and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Zhou, zhoulin99@zju.edu.cn; Shusen Zheng, shusenzheng@zju.edu.cn
†These authors share first authorship
 Mengxia Li1,2,3,4,5†
Mengxia Li1,2,3,4,5† Shengyong Yin
Shengyong Yin Lin Zhou
Lin Zhou Shusen Zheng
Shusen Zheng