- 1Microbiology, Immunology, and Parasitology Department, Institute of Basic Health Sciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil
- 2Gram-Positive Cocci Laboratory, Federal University of Health Sciences of Porto Alegre, Porto Alegre, Brazil
- 3Brazilian Institute of Environment and Renewable Natural Resources, IBAMA, Brasília, Brazil
- 4Institute of Food Science and Technology, Federal University of Rio Grande do Sul, Porto Alegre, Brazil
The environment, human, and animals play an important role in the spread of antibiotic-resistant bacteria. Enterococci are members of the gastrointestinal tracts of humans and animals and represent important reservoirs of antibiotic resistance genes. Until today, few studies have examined antibiotic susceptibility in enterococci isolated from primates. Therefore, the present study investigated species distribution, antibiotic susceptibility, and resistance genes in enterococci isolated from wild and captive black capuchins monkeys (Sapajus nigritus) in Rio Grande do Sul, South Brazil. A total of 24 swabs/fecal samples were collected, including 19 from wild monkeys living in two forest fragments [São Sebastião do Caí (SSC) and Santa Cruz do Sul (SCS)], and five in captive [Parque Zoológico da Fundação Zoobotânica (ZOO)], between August 2016 and November 2017. Fifteen colonies were randomly selected from each sample. Enterococci were identified by MALDI-TOF, tested for susceptibility to 12 antibiotics; and screened for tet(S), tet(M), tet(L), msrC, and erm(B) genes by PCR. Two-hundred ninety-six enterococci were isolated (SSC n = 137; SCS n = 86; ZOO n = 73) and differences in Enterococcus species distribution were detected on three monkey groups, with low abundance in SCS (1 - D = 0.2), followed by ZOO (1 - D = 0.68), and SSC (1 - D = 0.73). The enterococci frequently recovered include the following: Enterococcus faecalis (42.6%), E. hirae (29.1%), and E. faecium (15.9%). Antibiotic-nonsusceptible was observed in 202 (67.9%) strains. The rate of non-susceptibility to rifampicin, tetracycline, erythromycin, nitrofurantoin, chloramphenicol, and ampicillin was 46%, 26%, 22% and 19%, 13%, 0.3%, and 0.3%, respectively. All strains were susceptible to vancomycin, streptomycin, gentamycin, and linezolid. Forty-three (14.52%) isolates were identified as multidrug resistant (MDR), and the highest number of MDR enterococci were E. faecium recovered from wild monkeys living close to a hospital and water treatment plant. Elevated rates of antibiotic resistance genes msrC and tet(L) were isolates from ZOO. In conclusion, differences in the frequency of enterococci species, antibiotic-nonsusceptible and antibiotic resistance genes in all groups of monkeys were identified. These data suggest that anthropogenic activities could have an impact in the resistome of primate gut enterococci communities.
Introduction
Brazil has the greatest biodiversity on the planet, comprising approximately 103,870 different animal species and the highest diversity of Primates, around 77 species, including the howler monkey, the capuchin monkey, the marmoset, and the tamarin (Brazilian Society of Primatology [SBP], 2016). Sapajus nigritus (black-horned capuchin or black capuchin monkeys) are part of the Cebidae family, characterized as robust capuchin monkeys with adornments or tufts on the head (Rylands et al., 2012). They are considered the largest omnivorous Neotropical primate, which is able to adapt its diet according to food availability, thus bringing them into contact with a wide diversity of microorganisms. Their diet is composed of approximately 55% fruits, 33% insects, 8% seeds, 8% leaves (mainly young), and 2% flowers (National Research Council of the National Academies [NRC], 2003). Currently, this species occurs in Minas Gerais, Rio de Janeiro, São Paulo, Paraná, Santa Catarina, and Rio Grande do Sul states, extending to the Argentinean province of Misiones (International Union for Conservation of Nature [IUCN], 2017).
The black capuchin monkeys (S. nigritus) live in different habitats, from large remnants or continuous to small forests fragments. Outside of their natural environment, they can be found in zoological, rehabilitation, or research centers, and even in urban and rural environments. Additionally, these animals exhibit a niche overlap with humans in the case of semi-wild areas (Muehlenbein, 2017). Since the natural habitats of primates are forests, most interactions between humans and primates occur in this high-risk interface. In many regions of the world, omnivorous primate species are adapting to human activities. Furthermore, the frequency of such interactions has increased due to ecotourism and/or increasing forest invasion, and these interactions could lead bacteria exchanges by multiple routes, namely through the offering of food (Mikich and Liebsch, 2014). Glover (2014) compared the enteric bacteria of monkeys with three levels of human contact and determined that the closer the animals were to humans, the more resistant was the enteric bacteria to antibiotics. Importantly, Rolland et al. (1985) observed that wild baboons (Papio cynocephalus) that fed on human debris, maintained a high proportion of antibiotic-resistant enteric bacteria than those without human contact.
The environment, humans, and animals play an important role in the emergence and spread of antibiotic-resistant bacteria. Singer et al. (2016) described three well-characterized classes of chemicals – antimicrobials, heavy metals, and biocides – related to the selection of antibiotic resistance genes. Biological fluids (e.g., urine and feces) contaminated with antimicrobials or resistant bacteria from human and animal origins are released into the environment – especially in soil, sewage, water, and wastewater – thereby contributing to the spread of resistance (Baquero et al., 2008; Gothwal and Shashidhar, 2014). The proximity to human activity has showed to increase the number of resistant bacteria in wild animals, with animals living near waste or agricultural water harboring more antibiotic-resistant bacteria than animals living close by unpolluted water (Allen et al., 2010). Recently, it was demonstrated that exposure to human antibiotics was associated with changes in the microbiota composition of baboons (Tsukayama et al., 2018).
Enterococci are a large genus of bacteria widely distributed on plants, soil, water, humans, and animals. In humans and other species, inhabit various sites including the oral cavity, genitourinary and gastrointestinal tracts (Lebreton et al., 2014). The genus Enterococcus consists of over 50 diverse species, and Enterococcus faecalis, E. faecium, E. hirae, E. durans, E. casseliflavus, E. gallinarum, and E. mundtii are the most frequently encountered in the gastrointestinal tracts of animals (Poeta et al., 2005; Cassenego et al., 2011; Lozano et al., 2016; Medeiros et al., 2017). However, the species evaluation in the gastrointestinal tract of primates remains little known (Xavier et al., 2010; Glover, 2014). The species distribution, as well as their proportions in the different niche can change according to the host and its age, diet, underlying diseases, and prior antimicrobial therapy (Lebreton et al., 2014).
Otherwise, enterococci are considered an opportunistic pathogen, associated with serious infection, such as endocarditis, urinary, and bloodstream infections, intra-abdominal end intra-pelvic abscesses, which has been attributed, in part; to the increasing resistance to a wide range of antimicrobial agents. The presence of resistant and multidrug-resistant enterococci in patients has a clinical relevance because of limited therapeutic options (Higuita and Huycke, 2014). Antimicrobial resistance to several classes of agents is a remarkable characteristic of enterococcal isolates. These microorganisms are intrinsically resistant to some antimicrobial agents commonly prescribed for Gram-positive cocci, and exhibit resistance to a wide variety of other antimicrobials by mutation and/or acquisition of genes through the plasmids and transposons. In fact, many species are recognized for their ability to acquire and transfer resistance and virulence genes, which give a selective advantage to Enterococcus spp. survival and dispersion in the environment (Lebreton et al., 2014; Miller et al., 2014). The occurrence of antimicrobial resistance among enterococci is not restricted to the nosocomial setting, and therefore, resistant strains has been investigated and monitored in different habitats, providing important information regarding about environmental disturbances (Poeta et al., 2005; Frazzon et al., 2010; Barros et al., 2011; Cassenego et al., 2011; Santos et al., 2013; Santestevan et al., 2015; Prichula et al., 2016).
To date, few studies have examined the presence of enterococci in monkeys, and these studies have focused primarily on captive animals, perhaps due to the inherent difficulty in obtaining samples from free-living wild animals (Xavier et al., 2010; Glover, 2014; Woods et al., 2017). The investigation of the persistence of enterococci in these animals highlights the impact of human activities on the environment. Moreover, antibiotic-resistant enterococci in monkeys are an important point that must be addressed in the host–microorganism–environment interactions. Therefore, the objective of the present study was to evaluate the distribution of enterococci in fecal samples of free-living and captive black capuchin monkeys from South Brazil. In addition, the prevalence of antibiotic susceptibility and antibiotic resistance genes in enterococci isolated from these primate populations were determinates.
Materials and Methods
Sample Collection
Twenty-four samples collected from black capuchin monkeys between August 2016 and November 2017 were used in the present study, including samples from animals with free lifestyle (n = 19) and animals living in captivity (n = 5). Samples were obtained in Rio Grande do Sul, South Brazil (Supplementary Data 1).
Samples were taken from three groups of black capuchin monkeys. Two groups include wild animals from two forest fragments in Rio Grande do Sul State (Figure 1). In the first forest fragment located in São Sebastião do Caí (SSC) (29° 35′ 13″ S; 51° 22′ 17″ W), samples were obtained from 11 animals, corresponding to 30% of overall group composition. This forest fragment is located near to a hospital and water treatment plant. The area comprises 2% of vegetation, totalizing 9611 hectares of forest (SOS Mata Atlântica, 2016). In the second forest fragment, located in Parque Municipal da Gruta dos Índios (Indian Grotto Municipal Park) in Santa Cruz do Sul (SCS) (29° 43′ 03″ S; 52° 25′ 33″ W), samples were obtained from eight animals, corresponding to 27% of the overall group. This forest fragment is located inside of the park, and the animals come without indirect contact with any park visitor, but maintain contact with garbage and other food sources. The area comprises 13% of vegetation, totalizing 539.8 hectares of forest (SOS Mata Atlântica, 2016). The third group was in captive condition at the Zoological Park of the Zoobotânica Foundation of Rio Grande do Sul (ZOO) in Sapucaia do Sul (29° 49′ 29″ S; 51° 08′ 54″ W), and five samples were collected. The animals were isolated in quarantine at ZOO since they were rescued from illegal or abusive situations by the Wild Animals Triage Center (CETAS – IBAMA). The diet of captive monkeys was composed of extruded ration for primates (Nuvital Primatas Neotropicais, Nuvital Nutrientes S/A, Colombo, Brazil) complemented with fruits and vegetables.
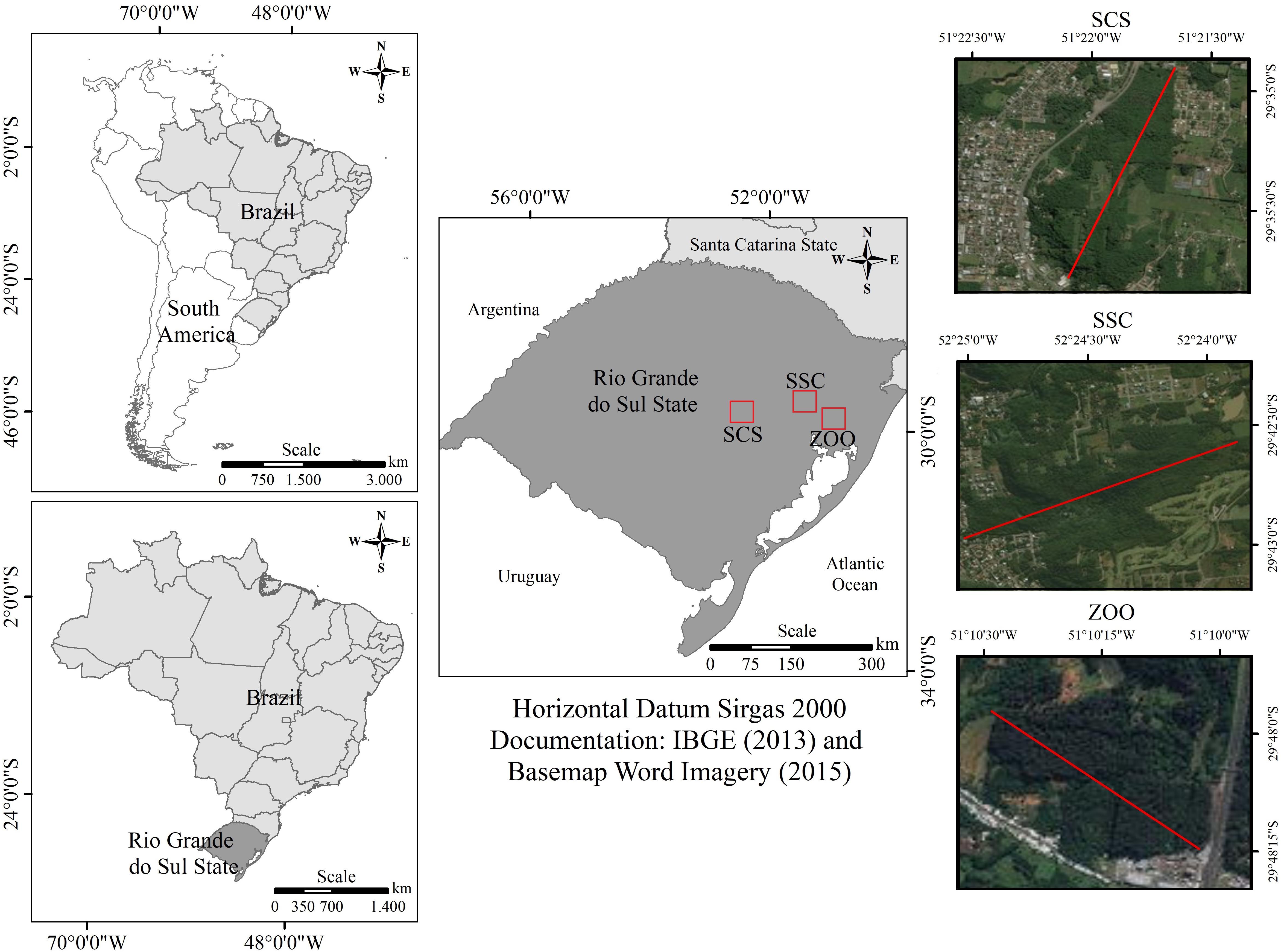
FIGURE 1. On the left Southern American (top) and Brazil (bottom) maps. On the center, Rio Grande do Sul map showing locations of collected samples site. On the right-detailed map of the Rio Grande do Sul showing collected samples locations: Top; SSC, São Sebastião do Caí – a fragmented forest coverage of 2% vegetation of Atlantic Forest, totaling 9611 hectares of forest: Center; SCS, Santa Cruz do Sul – a fragmented forest comprised of 13% vegetation, totaling 539.8 hectares of forest; Botton; ZOO, Zoological (ZOO) at Zoobotanical Foundation of Rio Grande do Sul, in Sapucaia do Sul city.
Wild capuchin monkeys were captured and manipulated using conventional methods according to the protocol for sample collection described by Instituto Chico Mendes de Conservação da Biodiversidade [ICMBio] (2012) using Tomahawk-type cages. The ketamine (100 mg/mL) and xylazine (20 mg/mL) were used intramuscularly for wildlife immobilization (Miranda et al., 2011).
Rectal swabs and fecal samples were collected by veterinarians, all animals were clinically healthy and were classified according to gender and age group. Rectal swabs were collected from the perirectal area, stored in Stuart transport medium (Kasvi, Paraná, Brazil), and transported to our laboratory for microbiological analyses. Fecal samples were collected, individually or in groups, directly from cages using sterilized wooden sticks. Fecal samples were placed in sterile tubes, kept on ice, and sent to our laboratory for storage at -80°C.
This study was carried out in accordance with the recommendations of Brazilian Institute of Environment and Renewable Natural Resources (IBAMA) and Chico Mendes Institute for Biodiversity Conservation (ICMBio). The protocol was approved by Information Authorization System in Biodiversity (SISBIO) number 56640.
Isolation and Identification of Enterococci
Isolation, enumeration, and characterization of enterococci in fecal/rectal swabs were performed as previously described by Prichula et al. (2016) and Santestevan et al. (2015). Swabs or fecal samples (0.1 g) were inoculated in 9 mL of azide dextrose broth (Himedia, Mumbai, India) and incubated for 24 h at 37°C. Aliquots of 1 mL were placed in 9 mL of saline water, and initial samples were further diluted 10-fold to obtain a final dilution factor of 1/1000. From each dilution, 100 μL was inoculated in brain heart infusion (BHI) agar plates (Himedia, Mumbai, India) supplemented with 6.5% NaCl, before being incubated as previously described (Santestevan et al., 2015; Prichula et al., 2016). Fifteen colonies were randomly selected from each sample. Phenotypic criteria, such as size/volume, shape, color, gram staining, catalase production, growth capacity at 45°C, and bile aesculin reaction were used to separate the enterococci group and the non-enterococcal strains. Selected pure colonies were stored at -20°C in a 10% (w/v) solution of skim milk (Difco, Sparks, MD, United States) and 10% (v/v) glycerol (Neon Comercial Ltda, São Paulo, SP, BR).
The isolates collected were identified using matrix-assisted laser desorption and ionization time-of-flight technique (MALDI-TOF) applied to Enterococcus spp. according to the protocol previously described by Sauget et al. (2017).
Antimicrobial Susceptibility Testing
Susceptibility to antimicrobial agents was performed using the Kirby–Bauer disk diffusion method recommended by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2016). Twelve antibiotics commonly used in clinical and veterinary medicine were evaluated: ampicillin 10 μg (AMP), ciprofloxacin 5 μg (CIP), chloramphenicol 30 μg (CHL), erythromycin 15 μg (ERY), streptomycin 300 μg (STR), gentamicin 120 μg (GEN), linezolid 30 μg (LNZ), nitrofurantoin 300 μg (NIT), norfloxacin 10 μg (NOR), rifampicin 5 μg (RIF), tetracycline 30 μg (TET), and vancomycin 30 μg (VAN). Minimum inhibitory concentration (MIC) of linezolid was determined by broth microdilution and interpretation of the results was performed following CLSI guidelines.
E. faecalis ATCC 51299 and E. faecium ATCC 53519 were included as control strains.
Strains resistant to three or more unrelated antibiotics were considered as multidrug-resistant (MDR). Intermediate and resistant strains were considered in a single category and classified as antibiotic-nonsusceptible.
Detection of Resistance-Related Genes in Enterococcus sp.
DNA extraction was performed as described by Depardieu et al. (2004). PCR was carried out for the detection of six different resistance-related genes commonly observed in clinical and environmental enterococci, namely, erm(B), msrC, tet(M), tet(S), and tet(L) (Sutcliffe et al., 1996; Aarestrup et al., 2000; Werner et al., 2001; Frazzon et al., 2010; Rathnayake et al., 2011). erm(B) encodes a ribosomal methylase that mediates macrolides, lincosamides, and type B streptogramins resistance; msrC encodes for a macrolide and streptogramin B efflux pump; tet(M) and tet(S) encodes for tetracycline resistance via a ribosomal protection protein mechanism; and tet(L) encodes for tetracycline resistance via efflux pumps proteins.
Statistical Analysis
The correlation between antimicrobial susceptibility presented by Enterococcus spp. and monkey collection origins were analyzed using a cross-table with Pearson’s chi-square test (χ2) (p ≤ 0.05) and Fisher’s exact test for small samples (≤5). Simpson’s index of diversity (D) was calculated to assess the differentiation of enterococci species among the monkeys from the different locations (Hunter and Gaston, 1988).
Results
Enterococcus spp. Isolation and Identification in Fecal Samples
The distribution of Enterococcus species recovered from fecal/rectal samples of wild and captive black capuchins monkeys is provided in Table 1. A total of 296 enterococci were isolated, of those 223 (75%) were recovered from wild (SSC n = 137; SCS n = 86), and 73 (25%) from captive monkeys (ZOO). Among enterococci isolated, E. faecalis (42.6%; n = 126), E. hirae (29.1%; n = 86), and E. faecium (15.9%; n = 47) were detected in all groups of monkeys; and E. durans (6.8%; n = 20), E. casseliflavus (4.4%; n = 13), E. raffinosus (0.3%; n = 1), E. avium (0.3%; n = 1), E. gallinarum (0.3%; n = 1), and Enterococcus sp. (0.3%; n = 1) were occasionally detected in the animals.
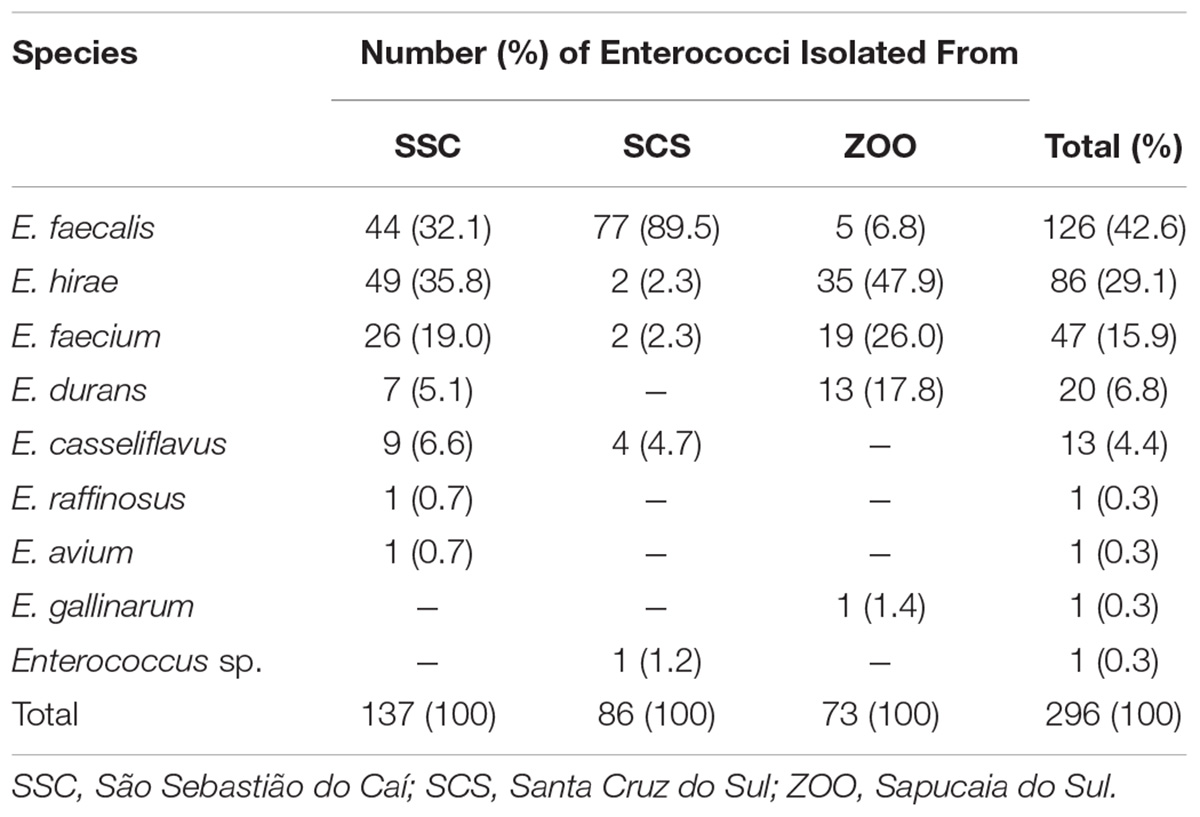
TABLE 1. Species distribution of enterococci in fecal samples of wild and captive black capuchin monkeys (Sapajus nigritus).
Differences in the distribution of Enterococcus spp. was detected amongst the three groups of black capuchin monkeys, as shown in Table 1. Samples from SSC presented the higher difference and relative abundance of enterococci, when compared to SCS and ZOO. The Simpsons diversity indexes showed differences between the three groups, with low abundance to SCS (1 - D = 0.2), followed by ZOO (1 - D = 0.68) and SSC (1 - D = 0.73). E. faecalis was the predominant species recovered from wild monkeys from SCS (89.5%; n = 77). On the other hand, in fecal samples of wild monkeys from SSC, the more commonly observed species were E. faecalis (32.1%; n = 44), E. hirae (35.8%; n = 49), and E. faecium (19.0%; n = 26). Whereas E. hirae (47.9%; n = 35), E. faecium (26.0%; n = 19), and E. durans (17.8%; n = 13) were the most abundant species isolated in fecal samples of captive monkeys.
Antimicrobial Susceptibility Profile
Among the 296 Enterococcus spp. obtained from fecal samples of black capuchin monkeys, 201 (67.90%) were nonsusceptible to at least one antibiotic evaluated (Figure 2). Nonsusceptible to rifampicin (46%), tetracycline (26%), erythromycin (22%), and quinolones (ciprofloxacin/norfloxacin) (19%) was commonly observed, whereas nonsusceptible to nitrofurantoin, chloramphenicol, and ampicillin was observed only in 13%, 0.3%, and 0.3% of the strains, respectively. Further, all isolates were susceptible to vancomycin, streptomycin, gentamycin, and linezolid (Table 2). Chi-squared testing showed significant differences (p ≤ 0.05) in tetracycline-nonsusceptible strains isolated from wild black capuchin monkeys from SSC when compared to the other groups.
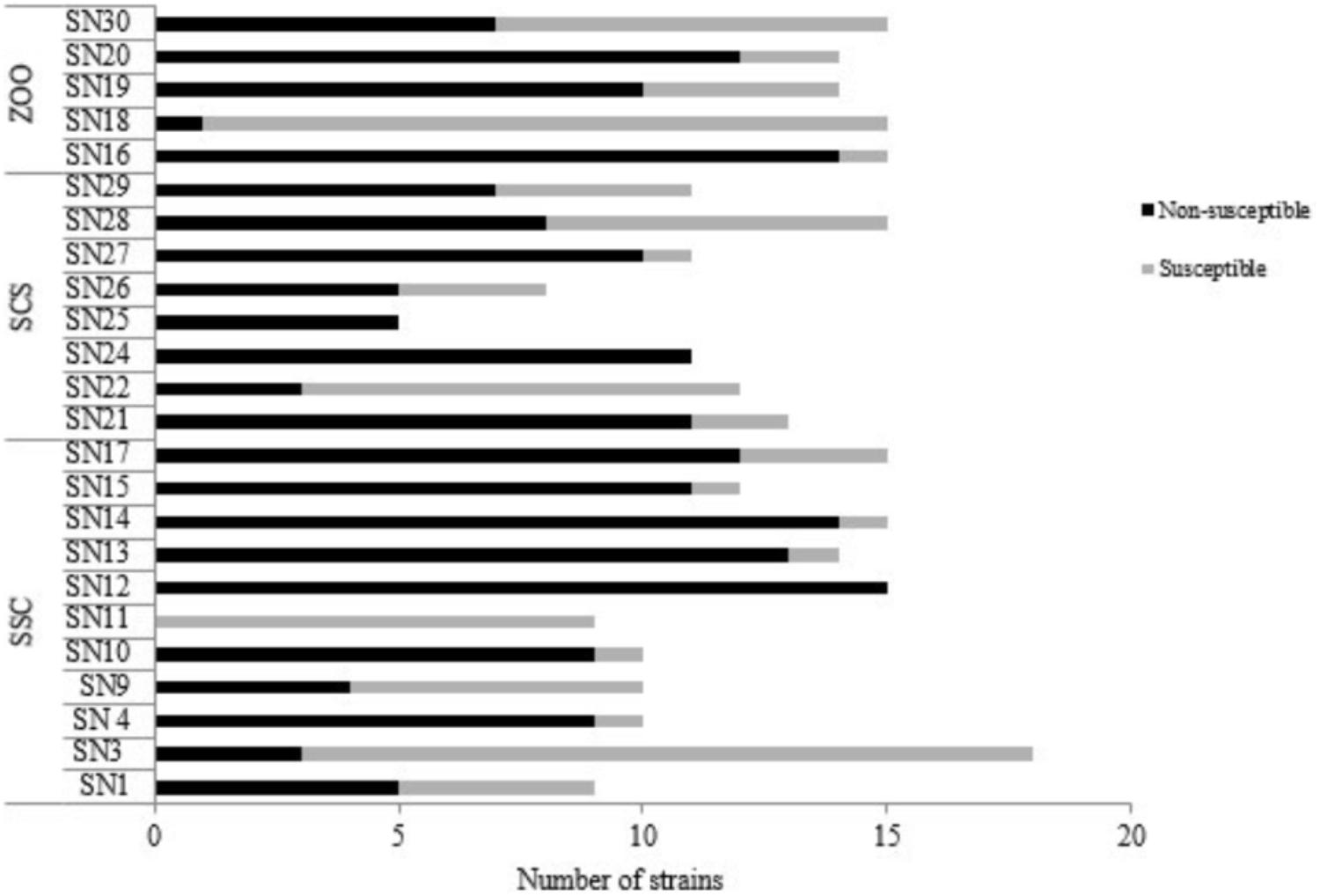
FIGURE 2. Numbers of susceptible and nonsusceptible enterococci species isolated from fecal samples of wild (SSC and SCS) and captive (ZOO) black capuchin monkeys (Sapajus nigritus). SN, identification sample of black capuchin monkeys. SSC, São Sebastião do Caí; SCS, Santa Cruz do Sul; ZOO, Sapucaia do Sul.
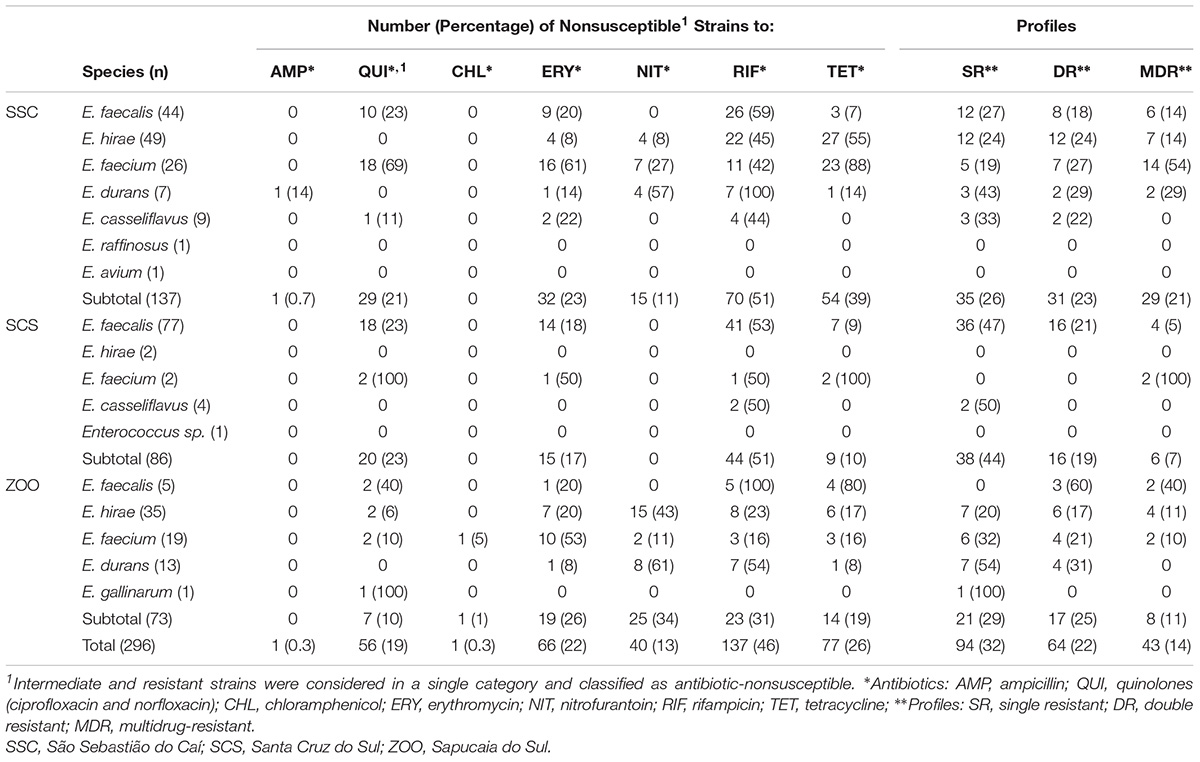
TABLE 2. Antibiotic resistance patterns among enterococci recovered from fecal samples and rectal of wild and captive black capuchin monkeys (Sapajus nigritus).
In relation to species isolated from black capuchin monkeys, E. durans (90%) and E. faecium (85%), showed elevated frequency of antibiotic non-susceptibility, followed by E. faecalis (69%), E. hirae (56%), and E. casseliflavus (54%). Enterococcus gallinarum strain was only nonsusceptible to quinolones. Unlike the other species, E. raffinosus, E. avium, and Enterococcus spp. were susceptible to all antimicrobials tested. Regarding the source of samples, the occurrence of antibiotic non-susceptible strains was observed more frequently in isolates from SSC (Figure 2).
Single, double, and MDR profiles were observed in 32% (n = 94), 22% (n = 64), and 14.52% (n = 43) of strains, respectively. The percentages of double and MDR strains isolated from wild monkeys from SCS (10%; n = 9 and 7%; n = 6) and the captive (16%; n = 12 and 11%; n = 8) were lower compared to wild monkeys from SSC (39%; n = 54 and 21%; n = 29). Among the 29 MDR strains from SSC, E. faecium was the species with higher prevalence (54%; n = 14) (Supplementary Data 2).
Frequency of Antibiotic Resistance Genes
Among the 66 erythromycin-nonsusceptible strains (11 were resistance and 56 were intermediate resistance), 24 (36%) contained the msrC, and none the erm(B) gene. Of the 77 tetracycline-nonsusceptible strains, 43 (56%) harbored only the tet(M), and 24 (31%) have both tet(M) and tet(L) genes. The tet(S) gene was not found in this study (Table 3).
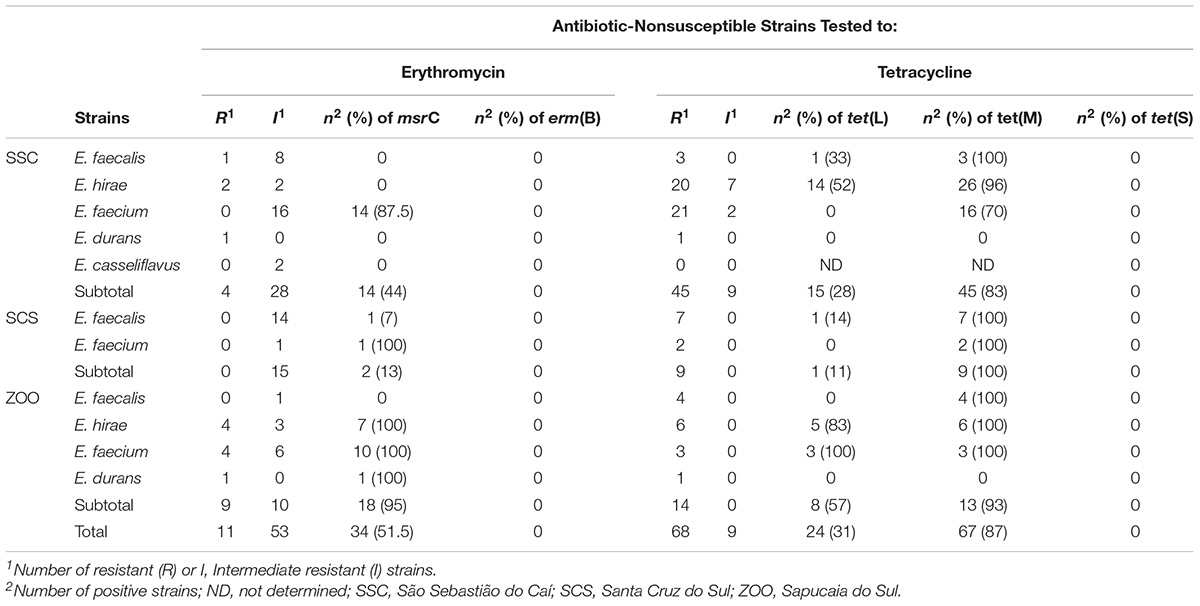
TABLE 3. Resistance-related genes among antibiotic-nonsusceptible enterococci isolated from fecal samples of wild and captive black capuchin monkeys (Sapajus nigritus).
In relation to species, the results showed that 92.5% E. faecium, 64% E. hirae, and 4% E. faecalis strains harbored msrC gene. The tet(M) was present in all E. faecalis, E. faecium, and E. hirae tetracycline-nonsusceptible strains, and tet(L) was detected in 14% E. faecalis, 57.5% E. hirae, and in 11% E. faecium tetracycline-nonsusceptible strains.
We investigated the association between resistance-related genes and the sample sources where enterococci species isolated from captive monkeys presented a higher frequency of msrC (95%) and tet(L) (57%) genes when compared to wild monkeys (Table 3). In addition, seven (21%) erythromycin and tetracycline-nonsusceptible strains from the ZOO harbored both msrC, tet(M), and tet(L) genes.
Discussion
In this study using fecal samples collected of wild and captive black capuchin monkeys (S. nigritus) from South Brazil, we were able to detected different Enterococcus species. To date, only a few studies have investigated the distribution of enterococci species in the fecal samples/rectal swabs of wild and captive black capuchin monkeys. The genus Enterococcus was first reported in fecal samples from captive capuchin monkeys (Cebus apella) and common marmoset (Callithrix penicillata) in the Primate Center of the University of Brasília, Brazil (Xavier et al., 2010). Thereafter, Glover (2014) identified the genus Enterococcus in the fecal samples from the baboons (Papio) and vervet monkeys (Chlorocebus pygerythrus) in two rehabilitation centers in South Africa.
The enterococci species identified here from both wild and captive black capuchin monkeys have been reported to be predominant in fecal samples of different animals. Studies evaluating enterococci species in fecal samples of domestic and wild animals revealed presence of similar species (Layton et al., 2010; Cassenego et al., 2011; Franz et al., 2011; Silva et al., 2012; Nowakiewicz et al., 2014; Santestevan et al., 2015; Prichula et al., 2016; Medeiros et al., 2017). Among the species identified in the present study, E. faecalis was predominant. This species was also the most prevalent species in fecal samples of captive capuchin monkeys, common marmoset, domesticated mammals, birds, and wildlife feces, described in previous studies (Lanthier et al., 2010; Xavier et al., 2010). Nevertheless, it is important to highlight that some species could be underestimated in the present study due to the limitation of the method on used for enterococci isolation based on culturable methods. Although this method is widely used to isolate enterococci from different samples; we know that methods evaluating bacterial species in biological samples based on cultivation could limit the ability to recover some species occurring in small proportion.
Differences in the frequency of enterococci species in fecal samples among the three groups of monkeys were observed. Confinement, diet, and human contact are factors that may be responsible for this difference (Lebreton et al., 2014). In fecal samples of wild monkeys from SCS, the E. faecalis was the dominant species. In contrast, the species distribution of enterococci in samples of wild monkeys from SSC was more heterogeneous. These differences in the frequency of enterococci could be explained by the environmental conditions. In spite of the fact that both monkeys live in a free-living condition, monkeys from SCS are in a forest fragment surrounded by an urban area. Urban forest fragments are considered the most fragile area, which suffers directly the negative impacts of the anthropic action (Pereira et al., 2018). The urbanization also affects the insect species composition, as recently demonstrated by Melliger et al. (2018), whereas changes in the composition of ants and spiders were associated with increasing degree of urbanization. The anthropic action on the forest fragment in SCS may have reduced the contact of monkeys with diverse routes transmitting variable enterococci, including insect that comprised approximately 33% of the diet of these animals. Besides, the monkeys from SCS are feeding by human and have access to the garbage left by visitors on the park.
Contrasting with wild monkeys from SCS, the fecal samples from wild monkeys of SSC showed more dissimilar Enterococcus species, including E. faecalis, E. hirae, E. faecium, E. durans, E. casseliflavus, E. raffinosus, E. avium, and E. gallinarum. These monkeys live in a less urbanized forest fragment with a general diet, composed by insects, fruits, stems, flowers, and leaves, and consequently exposed to several Enterococcus species. In captive monkeys from ZOO, which are feeding with nonhuman dry food – composed by proteins, crude fiber and fat – supplemented with fruits and vegetables, the E. hirae, E. faecium, and E. durans were the most prevalent species. The presence of these Enterococcus species might be associated with the food source since enterococci were detected in the feed and feed ingredients samples as described by da Costa et al. (2007) and Ge et al. (2013).
Antibiotic-nonsusceptible enterococci species were found in captive and black capuchin monkeys. Similar studies, detected resistant bacteria in captive and wild animal from different environments (Xavier et al., 2010; Santos et al., 2013; Glover, 2014; Smith et al., 2014; Santestevan et al., 2015; Bondarczuk et al., 2016; Prichula et al., 2016; Furness et al., 2017; Bengtsson-Palme et al., 2018). In addition, samples from wild black capuchin monkeys from SSC presented a high number of antibiotic-nonsusceptible strains. The antibiotic-nonsusceptible strains isolated from wild monkeys are a matter of concern since these animals did not have a history of therapeutic antibiotic exposure. The analysis of resistant enterococci in these animals emphasizes the role of human activities on the environment. However, we cannot forget to mention that wild black capuchin monkeys from SSC live in a forest fragment near a public hospital and water treatment plant, and this proximity with these environments should represent a source of antibiotic-nonsusceptible strains in these animals. The presence of bacteria antibiotic-nonsusceptible and antibiotic resistance genes in hospital effluents has been observed and related to dissemination of resistance in the environment (Brown et al., 2006; Rodríguez-Mozaz et al., 2014; Xu et al., 2014). For example, tetracycline and erythromycin prescribed in human and animal medicine are excreted as active metabolites and remain stable in the environment (Rahardjo et al., 2011; Rudra et al., 2018; Schafhauser et al., 2018) to be considered modern pollutants in soils and aquatic environment (Gothwal and Shashidhar, 2014; Dizavandi et al., 2016). Another aspect to be considered is the antibiotic resistome (Wright, 2007; Stewart et al., 2014). Previous reports have noted the occurrence of resistant bacteria in soil independent of human activity (Allen et al., 2010). As such, we cannot exclude the possibility that the resistance found in monkeys is derived from the gut microbial communities. Tsukayama et al. (2018) showed that antibiotic resistance is an ancient feature of gut microbial communities of primate and that sharing habitats with humans may have an important impact on the structure and function of this microbiota.
In our study, 14% of the isolated strains were resistant at least to three or more drugs. The MDR enterococci species have been isolated from wild and captive animals (Nowakiewicz et al., 2014; Prichula et al., 2016). It is important to note that an elevate number of MDR E. faecium isolated from wild monkey that lives near to the hospital was detected. In the last years, the emergence of MDR bacteria has become a hospital-acquired infection problem and, a high number of MDR enterococcal infections are caused by E. faecium (Kristich et al., 2014).
The resistance-related genes commonly observed in this environment, msrC, tet(M), and tet(L) was detected in our samples. Those resistance genes were found in higher frequency in samples from captive monkeys when compared to wild monkeys. Perhaps, the captive condition of animals might be contributing to the acquisition/dispersion and persistence of these genes in this environmental. Up until now, only two studies have evaluated resistant genes in enterococci-resistant isolated from monkeys (Xavier et al., 2010; Woods et al., 2017). Our data demonstrated the tet(M) gene is widely distributed among our isolates followed by tet(L). Furthermore, when studying enterococci from wild marine animals, Prichula et al. (2016) identified a high prevalence (73.07%) of the tet(M) gene and a low prevalence (23.07%) of tet(L), which corroborates with the findings in the present study. Notably, Poeta et al. (2005) determined that the tet(M) gene is the more prevalent in enterococci from wild animals in Portugal, other than monkeys. Moreover, 50% of samples from Santestevan et al. (2015), isolated from wild sea lions presented tet(M) gene. Despite erm(B) gene is frequently observed in macrolide-resistant strains isolated from animals (Poeta et al., 2005; Cassenego et al., 2011), this gene was not detected in our samples. In addition, the msrC gene was detected at low frequency in wild monkeys. Additionally, Prichula et al. (2016) tested the erm(B) and msrC in enterococci strains isolated from wild marine animals and reported only the presence of the msrC. It is possible that other genes could be associated with erythromycin-nonsusceptible strains isolated from monkeys, like erm(A), erm(C), erm(D), erm(E), erm(F), erm(G), erm(Q), and the macrolide efflux pump (msrA).
In conclusion, the enterococci isolated in this study from monkeys living in three distinct areas, showed differences in the species, in the frequency of antibiotic-nonsusceptible and antibiotic resistance genes. These differences could be related to food web interactions, environmental pollutants, and/or antibiotic resistome. High frequency of MDR strains was observed in fecal samples of wild monkeys, which live in a forest fragment near a public hospital. The data presented in this study suggest that anthropogenic action might be affecting primate-gut enterococci community.
Finally, further research is necessary to better understand the evolution of resistance mechanisms presented by enterococci. Therefore, this study contributes in part to the comprehension of black capuchin monkey’s microbiota, and to the elucidation of resistant bacterial strains and spread in wild and captive environments.
Author Contributions
TG, JF, PW, and AF designed the study. RP and RS performed the MIC. TG, DZ, PW, and AA carried out the sampling work. TG, LC, JF, and AF analyzed the data and drafted the manuscript. All authors have read and approved the final manuscript.
Funding
We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico do Brasil (CNPq – Grant Nos. 302574/2017-4, 401714/2016-0, and 303603/2015-1) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) of the Brazilian government partially supported this work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02366/full#supplementary-material
References
Aarestrup, F. M., Agerso, Y., Gerner-Smidt, P., Madsen, M., and Jensen, L. B. (2000). Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37, 127–137.
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Barros, J., Igrejas, G., Andrade, M., Radhouani, H., López, M., Torres, C., et al. (2011). Gilthead seabream (Sparus aurata) carrying antibiotic-resistant enterococci. A potential bioindicator of marine contamination?. Mar. Pollut. Bull. 62, 1245–1248. doi: 10.1016/j.marpolbul.2011.03.021
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux053. doi: 10.1093/femsre/fux053
Bondarczuk, K., Markowicz, A., and Piotrowska-Seget, Z. (2016). The urgent need for risk assessment on the antibiotic resistance spread via sewage sludge land application. Environ. Int. 87, 49–55. doi: 10.1016/j.envint.2015.11.011
Brazilian Society of Primatology [SBP] (2016). Brasil: SBP. Available at: http://sbprimatologia.org.br/os-primatas/
Brown, K. D., Kulis, J., Thomson, B., Chapman, T. H., and Mawhinney, D. B. (2006). Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 366, 772–783. doi: 10.1016/j.scitotenv.2005.10.007
Cassenego, A. P. V., d’Azevedo, P. A., Ribeiro, A. M. L., Frazzon, J., Van Der Sand, S. T., and Frazzon, A. P. G. (2011). Species distribution and antimicrobial susceptibility of enterococci isolated from broilers infected experimentally with Eimeria spp and fed with diets containing different supplements. Braz. J. Microbiol. 42, 480–488. doi: 10.1590/S1517-83822011000200012
Clinical and Laboratory Standards Institute [CLSI] (2016). Performance Standards for Antimicrobial Susceptibility Testing – Twelve-Six Edition 100S. Wayne, PA: CLSI.
da Costa, P. M., Oliveira, M., Bica, A., Vaz-Pires, P., and Bernardo, F. (2007). Antimicrobial resistance in Enterococcus spp. and Escherichia coli isolated from poultry feed and feed ingredients. Vet. Microbiol. 120, 122–131. doi: 10.1016/j.vetmic.2006.10.005
Depardieu, F., Perichon, B., and Courvalin, P. (2004). Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42, 5857–5860 doi: 10.1128/JCM.42.12.5857-5860.2004
Dizavandi, Z. R., Aliakbar, A., and Sheykhan, M. (2016). A novel Pb-poly aminophenol glassy carbon electrode for determination of tetracycline by adsorptive differential pulse cathodic stripping voltammetry. Electrochim. Acta 227, 345–356. doi: 10.1016/j.electacta.2016.12.167
Franz, C. M., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Frazzon, A. P. G., Gama, B. A., Hermes, V., Bierhals, C. G., Pereira, R. I., Guedes, A. G., et al. (2010). Prevalence of antimicrobial resistance and molecular characterization of tetracycline resistance mediated by tet(M) and tet(L) genes in Enterococcus spp. isolated from food in Southern Brazil. World J. Microbiol. Biotechnol. 26, 365–370. doi: 10.1007/s11274-009-0160-x
Furness, L. E., Campbell, A., Zhang, L., Gaze, W. H., and McDonald, R. A. (2017). Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ. Res. 154, 28–34. doi: 10.1016/j.envres.2016.12.014
Ge, B., LaFon, P. C., Carter, P. J., McDermott, S. D., Abbott, J., Glenn, A., et al. (2013). Retrospective analysis of Salmonella, Campylobacter, Escherichia coli, and Enterococcus in animal feed ingredients. Foodborne Pathog. Dis. 10, 684–691. doi: 10.1089/fpd.2012.1470
Glover, B. A. (2014). Characterization and Resistance Profiles of Selected Enteric Bacteria Isolated From Non-Human Primates at a Wildlife-Human Interface. Doctoral dissertation, Universidade de Pretoria, Pretoria.
Gothwal, R., and Shashidhar, T. (2014). Antibiotic pollution in the environment: a review. Clean 43, 479–489. doi: 10.1002/clen.201300989
Higuita, N. I. A., and Huycke, M. M. (2014). “Enterococcal disease, epidemiology, and implications for treatment”, in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Eye and Ear Infirmary), 1–34.
Hunter, P. R., and Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microb. 26, 2465–2466.
Instituto Chico Mendes de Conservação da Biodiversidade [ICMBio] (2012). Protocolo Para Coleta de Dados Sobre Primatas em Unidades de Conservação da Amazônia. Brasília: ICMBio.
International Union for Conservation of Nature [IUCN] (2017). Sapajus nigritus. Available at: http://www.iucnredlist.org/details/136717/0
Kristich, C. J., Rice, L. B., and Arias, C. A. (2014). “Enterococcal infection—treatment and antibiotic resistance,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (New York, NY: Eye and Ear Infirmary), 1–62.
Lanthier, M., Scott, A., Lapen, D. R., Zhang, Y., and Topp, E. (2010). Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can. J. Microbiol. 56, 715–729. doi: 10.1139/w10-046
Layton, B. A., Walters, S. P., Lam, L. H., and Boehm, A. B. (2010). Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109, 539–547. doi: 10.1111/j.1365-2672.2010.04675.x
Lebreton, F., Willems, R. J. L., and Gilmore, M. S. (2014). “Enterococcus diversity, origins in nature, and gut colonization,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (New York, NY: Eye and Ear Infirmary), 1–82.
Lozano, C., Gonzalez-Barrio, D., Camacho, M. C., Lima-Barbero, J. F., de la Puente, J., Höfle, U., et al. (2016). Characterization of fecal vancomycin-resistant enterococci with acquired and intrinsic resistance mechanisms in wild animals, Spain. Microb. Ecol. 72, 813–820. doi: 10.1007/s00248-015-0648-x
Medeiros, A. W., Amorim, D. B., Tavares, M., Moura, T. M., Franco, A. C., d’Azevedo, P. A., et al. (2017). Enterococcus species diversity in fecal samples of wild marine species as determined by real-time PCR. Can. J. Microbiol. 63, 129–136. doi: 10.1139/cjm-2016-0427
Melliger, R. L., Braschler, B., Rusterholz, H.-P., and Baur, B. (2018). Diverse effects of degree of urbanisation and forest size on species richness and functional diversity of plants, and ground surface-active ants and spiders. PLoS One 13:e0199245. doi: 10.1371/journal.pone.0199245
Mikich, S. B., and Liebsch, D. (2014). Damage to forest plantations by tufted capuchins (Sapajus nigritus): too many monkeys or not enough fruits? For. Ecol. Manage. 314, 9–16. doi: 10.1016/j.foreco.2013.11.026
Miller, W. R., Munita, J. M., and Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert. Rev. Anti Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Miranda, C. B., Cabala, R. W., Atan, J. B. C. D., Müller, L. C. C. M., Silva, P. C., Vivas, L. A. M., et al. (2011). Drug retention in non-human primates with employment of ketamine and xylazine. Rev. Pubvet. 5:8.
Muehlenbein, M. P. (2017). Primates on display: potential disease consequences beyond bushmeat. Am. J. Phys. Anthropol. 162(Suppl. 63), 32–43. doi: 10.1002/ajpa.23145
National Research Council of the National Academies [NRC] (2003). Nutrient Requirements of Nonhuman Primates. Available at: https://www.nap.edu/read/9826/chapter/1
Nowakiewicz, A., Ziółkowska, G., Zięba, P., and Kostruba, A. (2014). Undomesticated animals as a reservoir of multidrug-resistant Enterococcus in eastern Poland. J. Wildl. Dis. 50, 645–650. doi: 10.7589/2013-09-240
Pereira, H. S., Kudo, S. A., and Silva, S. C. P. (2018). Topophilia and environmental valuation of urban forest fragments in an Amazonian city. Ambient. Soc. 21:e01590. doi: 10.1590/1809-4422asoc170159vu18l1ao
Poeta, P., Costa, D., Saenz, Y., Klib, N., Ruiz-Larrea, F., Rodrigues, J., et al. (2005). Characterization of antibiotic resistance genes and virulence factors in faecal enterococci of wild animals in Portugal. J. Vet. Med. B Infect. Dis. Vet. Public Health 52, 396–402. doi: 10.1111/j.1439-0450.2005.00881.x
Prichula, J., Pereira, R. I., Wachholz, G. R., Cardoso, L. A., Tolfo, N. C. C., Santestevan, N. A., et al. (2016). Resistance to antimicrobial agents among enterococci isolated from fecal samples of wild marine species in the southern coast of Brazil. Mar. Pollut. Bull. 105, 51–57. doi: 10.1016/j.marpolbul.2016. 02.071
Rahardjo, A. K., Susanto, M. J. J., Kurniawan, A., and Ismadji, N. I. S. (2011). Modified ponorogo bentonite for the removal of ampicillin from wastewater. J. Hazard. Mater. 190, 1001–1008. doi: 10.1016/j.jhazmat.2011.04.052
Rathnayake, I., Hargreaves, M., and Huygens, F. (2011). SNP diversity of Enterococcus faecalis and Enterococcus faecium in a South East Queensland waterway, Australia, and associated antibiotic resistance gene profiles. BMC Microbiol. 11:201. doi: 10.1186/1471-2180-11-201
Rodríguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sànchez-Melsió, A., et al. (2014). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 69, 234–242. doi: 10.1016/j.watres.2014.11.021
Rolland, R. M., Hausfater, G., Marshall, B., and Levy, S. B. (1985). Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 49, 791–794.
Rudra, P., Hurst-Hess, K., Lappierre, P., and Ghosh, P. (2018). High levels of intrinsic tetracycline resistance in Mycobacterium abscessus is conferred by a tetracycline - modifying monooxygenase. Antimicrob. Agents Chemother. 62:e00119-18. doi: 10.1128/AAC.00119-18
Rylands, A. B., Mittermeier, R. A., and Silva, J. S. Jr. (2012). Neotropical primates: taxonomy and recently described species and subspecies. Int. Zoo Yearb. 46, 11–24. doi: 10.1111/j.1748-1090.2011.00152.x
Santestevan, N. A., Zvoboda, D. A., Prichula, J., Pereira, R. I., Wachholz, G. R., Cardoso, L. A., et al. (2015). Antimicrobial resistance and virulence factor gene profiles of Enterococcus spp. isolates from wild Arctocephalus australis (South America fur seal) and Arctocephalus tropicalis (Subantartic fur seal). World J. Microbiol. Biotechnol. 31, 1935–1946. doi: 10.1007/s11274-015-1938-7
Santos, T., Silva, N., Igrejas, G., Rodrigues, P., Micael, J., Rodrigues, T., et al. (2013). Dissemination of antibiotic-resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 24, 25–31. doi: 10.1016/j.anaerobe.2013.09.004
Sauget, M., Valot, B., Bertrand, X., and Hocquet, D. (2017). Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 25, 447–455. doi: 10.1016/j.tim.2016.12.006
Schafhauser, B. H., Kristofco, L. A., de Oliveira, C. M. R., and Brooks, B. W. (2018). Global review and analysis of erythromycin in the environment: occurrence, bioaccumulation and antibiotic resistance hazards. Environ. Pollut. 238, 440–451. doi: 10.1016/j.envpol.2018.03.052
Silva, N., Igrejas, G., Gonçalves, A., and Poeta, P. (2012). Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 62, 449–459. doi: 10.1007/s13213-011-0308-4
Singer, A. C., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Smith, S., Wang, J., Fanning, S., and McMahon, B. J. (2014). Antimicrobial resistant bacteria in wild mammals and birds: a coincidence or cause for concern? Ir. Vet. J. 67:8. doi: 10.1186/2046-0481-67-8
SOS Mata Atlântica (2016). Atlas Dos Remanescentes Florestais. Available at: http://mapas.sosma.org.br/
Stewart, J. R., Townsend, F. I., Lane, S. M., Dyar, E., Hohn, A. A., Rowles, T. K., et al. (2014). Survey of antibiotic-resistant bacteria isolated from bottlenose dolphins Tursiops truncates in the southeastern USA. Dis. Aquat. Organ. 108, 91–102. doi: 10.3354/dao02705
Sutcliffe, J., Grebe, T., Tait-Kamradt, A., and Wondrack, L. (1996). Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40, 2562–2566.
Tsukayama, P., Boolchandani, M., Patel, S., Pehrsson, E. C., Gibson, M. K., Chiou, K. L., et al. (2018). Characterization of wild and captive baboon gut microbiota and their antibiotic resistomes. mSystems 3:e00016-18. doi: 10.1128/mSystems.00016-18
Werner, G., Hildebrandt, B., and Witte, W. (2001). The newly described msrC gene is not equally distributed among all isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 45, 3672–3673. doi: 10.1128/AAC.45.12.3672-3673.2001
Woods, S. E., Lieberman, M. T., Lebreton, F., Trowel, E., Fuente-Núñez, C., Dzink-Fox, J., et al. (2017). Characterization of multi-drug resistant Enterococcus faecalis isolated from cephalic recording chambers in research macaques (Macaca spp.). PLoS One 12:e0169293. doi: 10.1371/journal.pone.0169293
Wright, G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186. doi: 10.1038/nrmicro1614
Xavier, D. B., Rosa, A. H., Sena, H. S., Teixeira, D. S., Tomaz, C., and Titze-de-Almeida, R. (2010). Absence of intestinal colonization by vancomycin resistant enterococci in nonhuman primates. Pesqui. Vet. Bras. 30, 491–496. doi: 10.1590/S0100-736X2010000600004
Keywords: Enterococcus, primates, wild and captive capuchin monkeys, Sapajus nigritus, antimicrobial resistance
Citation: Grassotti TT, de Angelis Zvoboda D, da Fontoura Xavier Costa L, de Araújo AJG, Pereira RI, Soares RO, Wagner PGC, Frazzon J and Frazzon APG (2018) Antimicrobial Resistance Profiles in Enterococcus spp. Isolates From Fecal Samples of Wild and Captive Black Capuchin Monkeys (Sapajus nigritus) in South Brazil. Front. Microbiol. 9:2366. doi: 10.3389/fmicb.2018.02366
Received: 10 June 2018; Accepted: 14 September 2018;
Published: 09 October 2018.
Edited by:
Gilberto Igrejas, Universidade de Trás-os-Montes e Alto Douro, PortugalCopyright © 2018 Grassotti, de Angelis Zvoboda, da Fontoura Xavier Costa, de Araújo, Pereira, Soares, Wagner, Frazzon and Frazzon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Paula Guedes Frazzon, ana.frazzon@ufrgs.br
 Tiela Trapp Grassotti
Tiela Trapp Grassotti Dejoara de Angelis Zvoboda1
Dejoara de Angelis Zvoboda1 Letícia da Fontoura Xavier Costa
Letícia da Fontoura Xavier Costa Jeverson Frazzon
Jeverson Frazzon