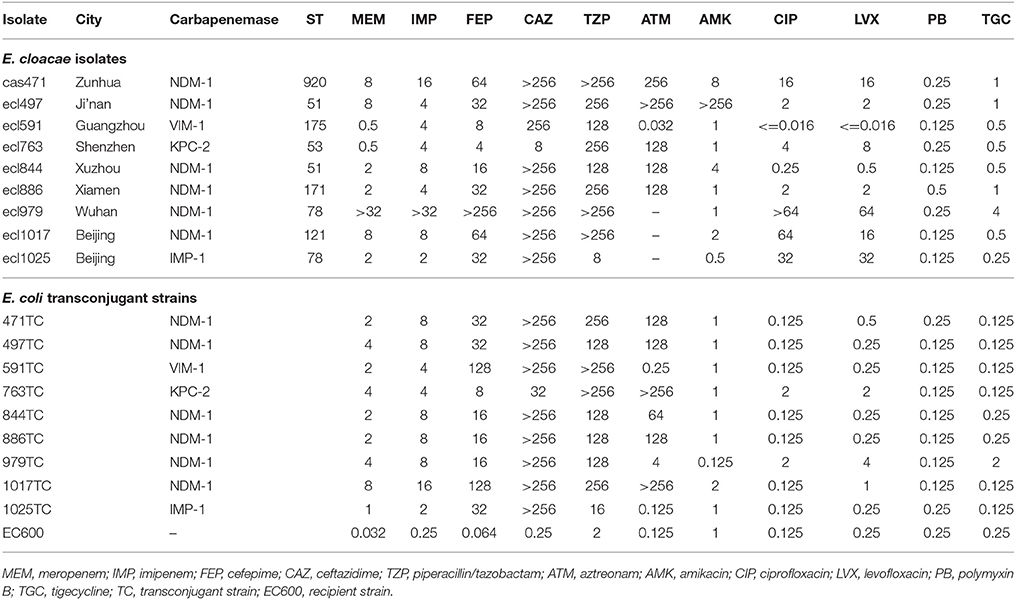
- 1Department of Clinical Laboratory, Yanbian University Hospital, Yanji, China
- 2Department of Clinical Laboratory, Peking University People's Hospital, Beijing, China
Carbapenem-resistant Enterobacteriaceae (CRE) are usually resistant to most of antibiotics. Infections caused by such bacteria have a high mortality and pose a serious threat to clinical management and public health. Enterobacter cloacae ranks third among Enterobacteriaceae that cause nosocomial infections. In this study, the molecular characteristics of carbapenem-resistant E. cloacae in China were investigated. From November 2012 to August 2016, 55 non-repetitive strains of carbapenem-resistant E. cloacae were collected from 12 hospitals in 11 Chinese cities. The bacteria were identified with matrix-assisted laser desorption/ionization time of flight mass spectrometry. Antimicrobial susceptibility tests were determined by agar dilution method. Carbapenemase and other β-lactamase genes were detected with PCR and sequencing. Multilocus sequence typing and plasmid conjugation tests were performed. Among the 55 E. cloacae strains, 50 strains were detected to produce 8 types of carbapenemase including NDM-1, NDM-5, IMP-4, IMP-26, IMP-1, KPC-2, and VIM-1. NDM-1 accounted for 68.0% (34/50) among the carbapenemase-producing E. cloacae. A total of 24 sequence types were identified and ST418 was the most common, accounting for 20% (11/55). For further investigation, a pulsed-field gel electrophoresis (PFGE) assay was conducted to identify the PFGE patterns of the strains. These 23 isolates yielded 13 PFGE patterns, which were designated as type A–M. Eight isolates obtained from Shenzhen had the same PFGE pattern (type A) and the remaining 15 isolates belonged to the other 12 PFGE patterns (type B–M). The observation that 8 of the 15 blaNDM−1-positive E. cloacae isolates obtained from Shenzhen with the same PFGE pattern (type A) suggested a transmission outbreak of a common strain. S1-nuclease PFGE and Southern blotting were also conducted to estimate the size of plasmids harbored by blaNDM−1-positive strains. The results showed that the plasmids harboring the blaNDM−1 gene ranged in size from approximately 52–58 kilobases. Our study indicates that carbapenem-resistant E. cloacae strains that produce NDM carbapenemase have strong resistance. Early detection and monitoring of the prevalence of these strains are urgent.
Introduction
In recent years, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) has become a serious issue both on community-acquired infections and healthcare-associated infections (van Duin and Doi, 2017). As well as other Enterobacteriaceae, Enterobacter cloacae (E. cloacae) is a conditional pathogen found in the intestine. Healthcare-associated infections caused by E. cloacae ranked third among all the Enterobacteriaceae (Dai et al., 2013). Enterobacter cloacae can produce chromosome mediated AmpC β-lactamase and has resistance to ampicillin, amoxicillin/clavulanic, cephamycin and first and second generation cephalosporin. A wide spectrum of antibacterial drugs such as carbapenems may be used in treatment more often. Thus, multidrug resistance has emerged rapidly under antibiotic selection pressure. Carbapenem-resistant E. cloacae infections have been reported in many countries such as Spain, Australia, the United States, India, and China (Kiedrowski et al., 2014; Fernández et al., 2015; Liu et al., 2015; Sidjabat et al., 2015). The emergence of carbapenem-resistant E. cloacae is an enormous challenge to clinical treatment. It is well known that the main mechanism for reduced susceptibility to carbapenems in E. cloacae is the deregulation of ACT (the natural cephalosporinase of E. cloacae), which is associated with a decrease in membrane permeability. In addition to this, producing carbapenemases is another important mechanism of Enterobacteriaceae in carbapenem resistance (Walsh et al., 2005; Nordmann et al., 2009; Tzouvelekis et al., 2012). Also, the mechanism of combinations of either ESBL or AmpC and mutation of porins may hold a certain proportion (Yang et al., 2010).
Up until now, there was a lack of multicenter research on carbapenem-resistant E. cloacae in China. So, we conducted this molecular epidemiological study on carbapenem-resistant E. cloacae to further understand the prevalence of the bacteria in China.
Materials and Methods
Sample Collection
From November 2012 to August 2016, we collected 55 unrepeated strains of carbapenem-resistant [any carbapenem (imipenem, meropenem, or ertapenem) as determined by standard methods] E. cloacae from 12 hospitals in 11 Chinese cities (Beijing, Chengde, Zunhua, Ji'nan, Xuzhou, Xi'an, Wuhan, Xiamen, Guangzhou, Dongguan, and Shenzhen; Figure 1). The participating hospitals include Peking University People's Hospital, Peking Union Medical College Hospital, Affiliated Hospital of Chengde Medical University, People's Hospital of Zunhua, Qilu Hospital of Shandong University, Affiliated Hospital of Xuzhou Medical University, Xijing Hospital, Tongji Hospital, The First Affiliated Hospital of Xiamen University, The First Affiliated Hospital Sun Yat-sen University, Donghua Hospital Sun Yat-sen University, and Shenzhen Second People's Hospital.
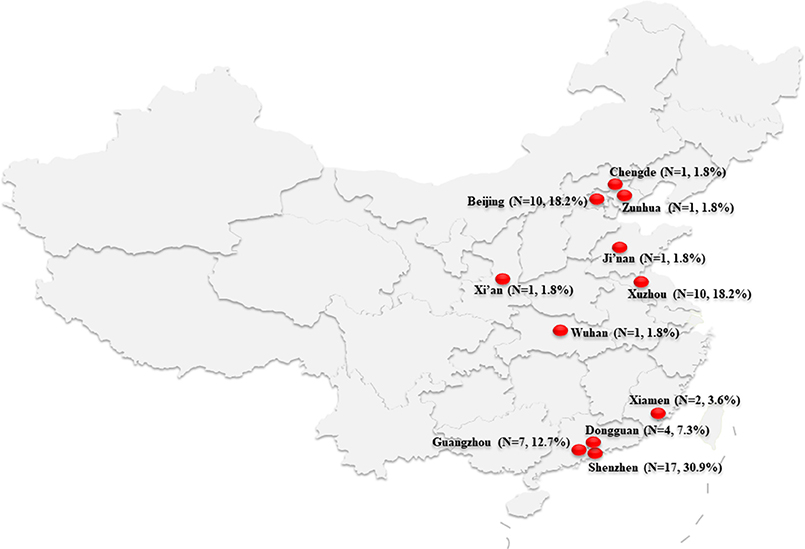
Figure 1. Map of China showing the location of the 11 cities where the carbapenem-resistant Enterobacteriaceae cloacae isolates were collected.
Identification of the Bacterial Strains and Antimicrobial Susceptibility Tests
All isolates were identified with matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany). Minimum inhibitory concentrations (MICs) were determined by the agar dilution method according to CLSI guidelines (M100-S27). The tested drugs included ceftriaxone (Roche China, Shanghai, China), cefotaxime, ceftazidime, cefepime, aztreonam, amikacin, levofloxacin, minocycline, fosfomycin (National Institute for Food and Drug Control of China, Beijing, China), piperacillin/tazobactam, tigecycline (Pfizer, NY, USA), imipenem (Merck Sharp & Dohme, Hangzhou, China), meropenem (Sumitomo Pharmaceuticals, Suzhou, China), ciprofloxacin (Bayer, Leverkusen, Germany), and polymyxin B (Amresco, Solon, USA). Strains used in quality control were Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853. The results were interpreted according to 2017 CLSI standards (M100-S27). The tigecycline test was performed according to the Food and Drug Administration standards.
Detection of Antimicrobial Resistance Genes
Phenotypic screening for the resistance genes of carbapenem-resistant E. cloacae strains was based on the 2017 CLSI guidelines. Modified Hodge test (MHT), imipenem-EDTA double-disk synergy test (DDST) (Lee et al., 2001), and modified carbapenem inactivation method (mCIM) were used to test carbapenemase production. Polymerase chain reaction (PCR) was used to detect carbapenemase genes (blaNDM, blaKPC, blaIMP, blaIMI, blaNMC, blaGES, blaSME, blaSIM, blaVIM, and blaOXA−48) and other β-lactamase genes (blaCTX−M, blaTEM, blaSHV, blaDHA, and blaCMY) (Lewis et al., 2007; Yang et al., 2010). The products were submitted for sequencing.
Multilocus Sequence Typing (MLST)
MLST was performed according to a previously described method (https://pubmlst.org/ecloacae/). New alleles and sequence types were submitted to the MLST website and approved. Sequence Type Analysis and Recombinational Tests 2 (START2) (http://pubmlst.org/software/analysis/start2/) software was used to generate the phylogenetic tree (Jolley et al., 2001).
Plasmid Conjugation Test
The plasmid conjugation test was used to test carbapenem-resistant gene transfer. Ten strains were selected for the test. Escherichia coli EC600 (rifampicin resistant) was used as the recipient. Conjugants were screened using China blue lactose agar plates containing rifampicin (300 μg/ml) and imipenem (1 μg/ml). The donor and the recipient were mixed at a ratio of 1:1 for 24 h. Transconjugants were selected on China blue lactose agar plates (OXOID, Basingstoke Hampshire, UK), supplemented with rifampicin (100 μg/ml) and imipenem (1 μg/ml). PCR was used to screen for blaNDM−1, blaVIM−1, blaKPC−2, and blaIMP−1 as previously described (Wang et al., 2014).
Pulsed-Field Gel Electrophoresis (PFGE)
Enterobacter cloacae isolates were characterized by PFGE according to the previously published protocol by Ribot et al., with modifications (Ribot et al., 2002). We selected 23 blaNDM−1-positive isolates (including the 15 isolates from Shenzhen, 6 isolates involved in the conjugation experiments, and another 2 representative isolates).
Electrophoresis conditions were altered to have an initial switch time of 2.16 s and a final switch time of 54.17 s, and gels were run for 18 h. The resulting PFGE patterns were analyzed in BioNumerics software (Applied Maths, Austin, TX, USA) with dendrograms based on the Dice coefficient with a band position tolerance of 1%. Patterns with no discernible differences were considered indistinguishable and given the same PFGE pattern designation.
S1-Nuclease PFGE and Southern Blotting
S1-nuclease PFGE and Southern blotting were performed to estimate the size of plasmids harbored by blaNDM−1-positive strains as described previously. We selected 23 blaNDM−1-positive isolates as mentioned above.
The blaNDM−1 gene was detected by digoxigenin-labeled specific probes (DIG High Prime DNA Labeling and Detection Starter Kit II, Roche Diagnostics, Mannheim, Germany). Salmonella enterica H9812 was used as a size marker.
Statistical Analyses
WHONET (version 5.6) software (http://www.whonet.org/software.html) and SPSS (version 22.0) software (SPSS Inc., Chicago, IL, USA) were used for statistical analyses.
Ethical Approval
This study was approved by the research ethics board at Peking University People's Hospital. Informed consent was not needed as this study was retrospective and participants were anonymized. Medical records and patient's information were retrospectively reviewed and collected.
Results
Characteristics of Collected Samples
The most common specimens were respiratory tract (20 cases, 36.4%), followed by urine (13 cases, 23.6%), blood (11 cases, 20%), ascitic fluid (7 cases, 12.7%), bile (2 cases, 3.6%), catheter (1 case, 1.8%), and wound (1 case, 1.8%).
Antimicrobial Susceptibility Tests
Of all the antimicrobials tested, the most susceptible antimicrobial was polymyxin B (100%, 55/55), followed by amikacin (89.1%, 49/55), fosfomycin (80%, 44/55), tigecycline (78.2%, 43/55), minocycline (52.7%, 29/55), levofloxacin (30.9%, 17/55), and ciprofloxacin (25.5%, 14/55). All of the 36 blaNDM-positive strains were resistant to piperacillin-tazobactam, ceftazidime, cefotaxime, ceftriaxone, cefepime, imipenem, and meropenem. It is worth noting that the most susceptible antimicrobials to blaNDM-positive strains was polymyxin B (100%, 36/36), followed by amikacin (86.1%, 31/36), and fosfomycin (86.1%, 31/36). There was some differences between blaNDM-positive strains and strains with other carbapenemases (Table 1).
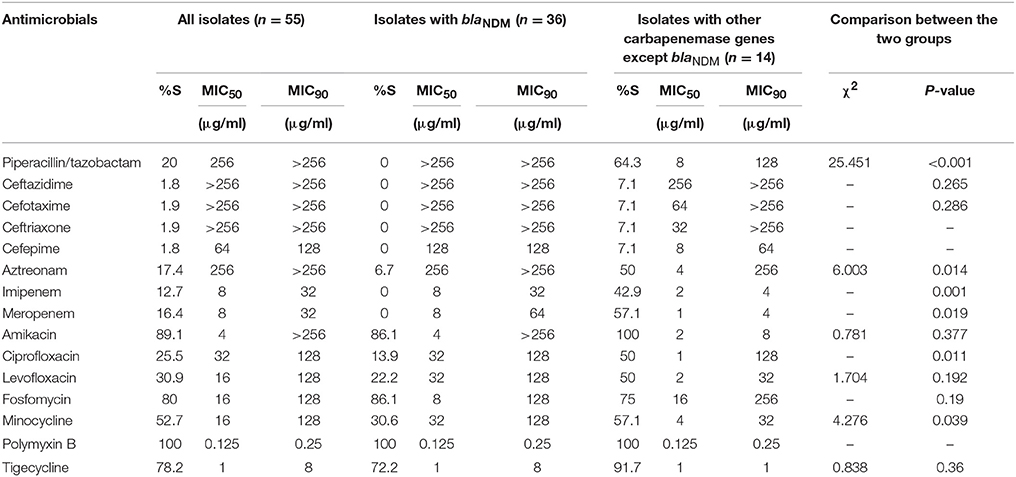
Table 1. In vitro activities of antimicrobial agents against carbapenemase-producing Enterobacteriaceae.
Genotype Analysis
Among the 55 strains, 50 were confirmed to produce 8 types of carbapenemases including NDM-1, NDM-5, IMP-4, IMP-26, IMP-1, KPC-2, and VIM-1. The corresponding numbers of the strains that produced the foregoing types of carbapenemases were 34, 2, 6, 3, 2, 2, and 1. Other carbapenemase genes were not detected. No strains contained two or more carbapenemase genes. NDM-1-producing carbapenem-resistant E. cloacae was primarily distributed in Shenzhen (Table 2). Carbapenemase genes were not detected in the other 5 strains.

Table 2. Microbiological and molecular characteristics of 34 blaNDM−1-positive Enterobacter cloacae strains.
NDM-1-producing E. cloacae isolates were mainly collected from Shenzhen (44.1%, 15/34), followed by Xuzhou (20.6%, 7/34), Beijing, Dongguan, Guangzhou, Ji'nan, Xi'an, Xiamen, Wuhan, and Zunhua. These samples were primarily collected from the Intensive Care Unit and the Emergency Intensive Care Unit (41.2%, 14/34), followed by the department of neurosurgery (23.5%, 8/34). NDM-1-producing E. cloacae isolates were most commonly identified in sputum samples (35.3%, 12/34), followed by urine samples (32.4%, 11/34). All strains were positive for MHT, imipenem-EDTA-DDST, and mCIM. Results of the three tests were consistent. In addition, 6 strains also produced the AmpC enzyme DHA-1, 8 strains produced CTX-M-3, and 2 strains produced both CTX-M and DHA-1.
MLST
The results of the MLST are shown in Figure 2. A total of 24 sequence types were detected in the 55 E. cloacae strains. ST418 was the most common (20%, 11/55), followed by ST93 (14.5%, 8/55).

Figure 2. Multilocus sequence typing (MLST) phylogenetic tree of the 55 carbapenem-resistant E. cloacae strains. ab, abdominal fluid; ba, broncho-alveolar lavage.
Plasmid Conjugation Test
Plasmids from 9 carbapenemase-producing E. cloacae strains were successfully transferred to E. coli EC600. Drug sensitivity tests showed the MICs of meropenem increased by 5- to 8-fold in the 9 conjugators; for imipenem, cefepime, ceftazidime, and piperacillin/tazobactam, the MICs increased by 3~6-, 7~11-, 7~10-, and 3~7-fold, respectively (Table 3).
PFGE, S1-Nuclease PFGE and Southern Blotting
When typed by PFGE to determine if they were related, the 23 isolates yielded 13 PFGE patterns, which were designated as type A–M. Eight isolates (ecl768, ecl771, ecl774, ecl776, ecl779, ecl782, ecl784, and ecl786) obtained from Shenzhen had the same PFGE pattern (type A) and the remaining 15 isolates belonged to the other 12 PFGE patterns (type B–M) (Table 2). The observation that 8 of the 15 blaNDM−1-positive E. cloacae isolates with the same PFGE pattern (type A) and the same sequence type (ST418) suggested a transmission outbreak of a common strain.
The results of S1-nuclease PFGE and Southern blotting showed that the plasmids harboring the blaNDM−1 gene ranged in size from approximately 52–58 kilobases, respectively (Table 2). The plasmids harboring the blaNDM−1 gene of the 8 isolates obtained from Shenzhen were the same size (approximately 52 kilobases).
Discussion
Carbapenemase-producing E. cloacae has been reported in many countries, such as strains producing OXA-48 and VIM-1 have been reported in Spain. In Brazil, Australia and America, strains producing NDM-1, IMP-4, and KPC-3, respectively, have been reported (Kiedrowski et al., 2014; Rozales et al., 2014; Villa et al., 2014; Fernández et al., 2015; Sidjabat et al., 2015), while in Chongqing, Henan and Ningxia of China, strains producing NDM-1 have been identified (Dai et al., 2013; Liu et al., 2015; Shi et al., 2017). Strains that produced other carbapenemases have also been reported in the Sichuan province of China (Huang et al., 2015). In the present study, we found that the E. cloacae prevalent in China mainly produced NDM-1 (68.0%, 34/50) and IMP-4 (12.0%, 6/50). NDM-1 was found in the highest proportion and may represent a significant drug-resistant mechanism of carbapenem-producing Enterobacteriaceae in China.
The plasmid conjugation test was completed with 6 blaNDM−1-positive strains. Conjugants were all detected to have the blaNDM−1. Susceptibility results showed that compared with the receptor bacteria EC600, the conjugants have a higher MIC value on cephalosporins and carbapenems. There were no MIC promotions on polymyxin B and tigecycline between conjugants and EC600. Many studies have demonstrated that the plasmid owned blaNDM also have other resistant genes, such as blaTEM−1, blaCMY, qnrA6, and qnrB1 for quinolone resistance, armA, rmtA, and rmtC for aminoglycoside resistance (Poirel et al., 2011a,b; Kocsis et al., 2016). But all strains in this study have no blaTEM−1 and blaCMY. Five of the conjugants have no MIC difference on quinolone with the EC600. Maybe the relative plasmid did not harbor the quinolone resistant gene.
MLST showed subtype diversity. A total of 24 sequence types were detected in 55 E. cloacae strains. ST418 was detected the most frequently (11/55, 20%), and the second was ST93 (14.5%, 8/55). Three new sequence types were found, namely ST916, ST919, and ST920. Our study reveals the diversity of carbapenem-resistant E. cloacae and the difference in genetic affinity, which is consistent with the study of Gomez-Simmonds et al. (2016). Our study showed that ST418 is the main epidemic strain in Shenzhen in China; while in America, Central de Asturias of Spain, and the Henan province of China, the main epidemic strains were ST171, ST74, and ST120, respectively (Fernández et al., 2015; Liu et al., 2015; Gomez-Simmonds et al., 2016). We found that ST418 was genetically closer to ST127 and ST755 with START2 analysis. Studies have found that all ST418 strains produced NDM-1 carbapenemase, indicating that there might be a small outbreak of NDM-1-ST418 carbapenem-resistant E. cloacae in Shenzhen and Dongguan City of Guangdong province of China. In this study, ST78-NDM-1-type carbapenem-resistant E. cloacae was also found in Xuzhou, Xi'an, Wuhan, and Xiamen, which should be taken seriously concern.
In conclusion, our study indicates that ST418, which produces NDM-1 carbapenemase, is the main epidemic strain of carbapenem-resistant E. cloacae in Shenzhen and Dongguan City of China. Early detection and monitoring are necessary to prevent the further spread of the bacteria.
Author Contributions
HW conceived and designed the study. CJ and JZ wrote this paper. CJ, QW, and JZ performed the experiments. QW and JZ analyzed the data. HC, XW, and YZ assisted CJ and JZ to finish the experiments. All authors approved the final version.
Funding
This study was supported by National Natural Science Foundation of China (grant No.81625014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the hospitals that provided carbapenem-resistant E. cloacae isolates (Peking University People's Hospital, Peking Union Medical College Hospital, Affiliated Hospital of Chengde Medical University, People's Hospital of Zunhua, Qilu Hospital of Shandong University, Affiliated Hospital of Xuzhou Medical University, Xijing Hospital, Tongji Hospital, The First Affiliated Hospital of Xiamen University, The First Affiliated Hospital Sun Yat-sen University, Donghua Hospital Sun Yat-sen University, and Shenzhen Second People's Hospital).
References
Dai, W., Sun, S., Yang, P., Huang, S., Zhang, X., and Zhang, L. (2013). Characterization of carbapenemases, extended spectrum β-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter Cloacae in a Chinese hospital in Chongqing. Infect. Genet. Evol. 14, 1–7. doi: 10.1016/j.meegid.2012.10.010
Fernández, J., Montero, I., Martínez, Ó., Fleites, A., Poirel, L., Nordmann, P., et al. (2015). Dissemination of multiresistant Enterobacter Cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int. J. Antimicrob. Agents 46, 469–474. doi: 10.1016/j.ijantimicag.2015.07.003
Gomez-Simmonds, A., Hu, Y., Sullivan, S. B., Wang, Z., Whittier, S., and Uhlemann, A.-C. (2016). Evidence from a New York City hospital of rising incidence of genetically diverse carbapenem-resistant Enterobacter Cloacae and dominance of ST171, 2007–14. J. Antimicrob. Chemother. 71, 2351–2353. doi: 10.1093/jac/dkw132
Huang, L., Wang, X., Feng, Y., Xie, Y., Xie, L., and Zong, Z. (2015). First identification of an IMI-1 carbapenemase-producing colistin-resistant Enterobacter Cloacae in China. Ann. Clin. Microbiol. Antimicrob. 14:51. doi: 10.1186/s12941-015-0112-2
Jolley, K. A., Feil, E., Chan, M.-S., and Maiden, M. C. J. (2001). Sequence type analysis and recombinational tests (START). Bioinformatics 17, 1230–1231. doi: 10.1093/bioinformatics/17.12.1230
Kiedrowski, L. M., Guerrero, D. M., Perez, F., Viau, R. A., Rojas, L. J., Mojica, M. F., et al. (2014). Carbapenem-resistant Enterobacter Cloacae isolates producing KPC-3, North Dakota, USA. Emerging Infect. Dis. 20, 1583–1585. doi: 10.3201/eid2009.140344
Kocsis, E., GuŽvinec, M., Butić, I., Krešić, S., Crnek, S. Š., Tambić, A., et al. (2016). bla NDM-1 carriage on incR plasmid in Enterobacteriaceae strains. Microbial. Drug Resist. 22, 123–128. doi: 10.1089/mdr.2015.0083
Lee, K., Chong, Y., Shin, H., Kim, Y., Yong, D., and Yum, J. (2001). Modified hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7, 88–91. doi: 10.1046/j.1469-0691.2001.00204.x
Lewis, J. S., Herrera, M., Wickes, B., Patterson, J. E., and Jorgensen, J. H. (2007). First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a US health care system. Antimicrob. Agents Chemother. 51, 4015–4021. doi: 10.1128/AAC.00576-07
Liu, C., Qin, S., Xu, H., Xu, L., Zhao, D., Liu, X., et al. (2015). New Delhi metallo-β-lactamase 1 (NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter Cloacae from Henan province, China. PLoS ONE 10:e0135044. doi: 10.1371/journal.pone.0135044
Nordmann, P., Cuzon, G., and Naas, T. (2009). The real threat of Klebsiella Pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9, 228–236. doi: 10.1016/S1473-3099(09)70054-4
Poirel, L., Bonnin, R. A., and Nordmann, P. (2011a). Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia Coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55, 4224–4229. doi: 10.1128/AAC.00165-11
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011b). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother.55, 5403–5407. doi: 10.1128/AAC.00585-11
Ribot, E. M., Wierzba, R. K., Angulo, F. J., and Barrett, T. J. (2002). Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1996. Emerg. Infect. Dis. 8, 387–391. doi: 10.3201/eid0804.010202
Rozales, F. P., Ribeiro, V. B., Magagnin, C. M., Pagano, M., Lutz, L., Falci, D. R., et al. (2014). Emergence of NDM-1-producing Enterobacteriaceae in Porto Alegre, Brazil. Int. J. Infect. Dis. 25, 79–81. doi: 10.1016/j.ijid.2014.01.005
Shi, Z., Zhao, H., Li, G., and Jia, W. (2017). Molecular characteristics of carbapenem-resistant Enterobacter Cloacae in Ningxia Province, China. Front. Microbiol. 8:94. doi: 10.3389/fmicb.2017.00094
Sidjabat, H. E., Townell, N., Nimmo, G. R., George, N. M., Robson, J., Vohra, R., et al. (2015). Dominance of IMP-4-producing Enterobacter Cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59, 4059–4066. doi: 10.1128/AAC.04378-14
Tzouvelekis, L., Markogiannakis, A., Psichogiou, M., Tassios, P., and Daikos, G. (2012). Carbapenemases in Klebsiella Pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Villa, J., Viedma, E., Brañas, P., Orellana, M. A., Otero, J. R., and Chaves, F. (2014). Multiclonal spread of VIM-1-producing Enterobacter Cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int. J. Antimicrob. Agents 43, 451–455. doi: 10.1016/j.ijantimicag.2014.02.006
Walsh, T. R., Toleman, M. A., Poirel, L., and Nordmann, P. (2005). Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18, 306–325. doi: 10.1128/CMR.18.2.306-325.2005
Wang, X., Xu, X., Li, Z., Chen, H., Wang, Q., Yang, P., et al. (2014). An outbreak of a nosocomial NDM-1-producing Klebsiella Pneumoniae ST147 at a teaching hospital in mainland China. Microbial. Drug Resist. 20, 144–149. doi: 10.1089/mdr.2013.0100
Yang, Q., Wang, H., Sun, H., Chen, H., Xu, Y., and Chen, M. (2010). Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob. Agents Chemother. 54, 573–577. doi: 10.1128/AAC.01099-09
Keywords: carbapenem-resistance, Enterobacter cloacae, carbapenemase, NDM-1, ST418
Citation: Jin C, Zhang J, Wang Q, Chen H, Wang X, Zhang Y and Wang H (2018) Molecular Characterization of Carbapenem-Resistant Enterobacter cloacae in 11 Chinese Cities. Front. Microbiol. 9:1597. doi: 10.3389/fmicb.2018.01597
Received: 27 February 2018; Accepted: 27 June 2018;
Published: 17 July 2018.
Edited by:
Yi-Wei Tang, Memorial Sloan Kettering Cancer Center, United StatesReviewed by:
Rong Zhang, Zhejiang University School of Medicine, ChinaRemy A. Bonnin, Université Paris-Saclay, France
Copyright © 2018 Jin, Zhang, Wang, Chen, Wang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Wang, wanghui@pkuph.edu.cn; whuibj@163.com
†These authors have contributed equally to this work and are co-first authors.
 Chunmei Jin1†
Chunmei Jin1† Jiangang Zhang
Jiangang Zhang Qi Wang
Qi Wang Yawei Zhang
Yawei Zhang Hui Wang
Hui Wang