- Division of Cell and Molecular Biology, Centre for Molecular Microbiology and Infection, Imperial College London, London, UK
Protein secretion systems are molecular nanomachines used by Gram-negative bacteria to thrive within their environment. They are used to release enzymes that hydrolyze complex carbon sources into usable compounds, or to release proteins that capture essential ions such as iron. They are also used to colonize and survive within eukaryotic hosts, causing acute or chronic infections, subverting the host cell response and escaping the immune system. In this article, the opportunistic human pathogen Pseudomonas aeruginosa is used as a model to review the diversity of secretion systems that bacteria have evolved to achieve these goals. This diversity may result from a progressive transformation of cell envelope complexes that initially may not have been dedicated to secretion. The striking similarities between secretion systems and type IV pili, flagella, bacteriophage tail, or efflux pumps is a nice illustration of this evolution. Differences are also needed since various secretion configurations call for diversity. For example, some proteins are released in the extracellular medium while others are directly injected into the cytosol of eukaryotic cells. Some proteins are folded before being released and transit into the periplasm. Other proteins cross the whole cell envelope at once in an unfolded state. However, the secretion system requires conserved basic elements or features. For example, there is a need for an energy source or for an outer membrane channel. The structure of this review is thus quite unconventional. Instead of listing secretion types one after each other, it presents a melting pot of concepts indicating that secretion types are in constant evolution and use basic principles. In other words, emergence of new secretion systems could be predicted the way Mendeleïev had anticipated characteristics of yet unknown elements.
As for any other living cells, a hydrophobic membrane that guarantees their insulation against toxic substances present in the environment surrounds bacteria. Yet it is crucial for bacteria to establish a communication with the external milieu and paths and channels should be created across these membranes to allow the passage of molecules. Most bacteria need to release enzymes such as proteases or lipases to transform complex macromolecules in nutrients usable by the cell. Pathogenic bacteria developed strategies to efficiently infect their hosts. They deliver virulence factors into host cells which then act to minimize efficiency of host defense, subvert host cell signaling to their own benefit and/or utilize the host as a niche for replication (Cossart and Sansonetti, 2004; Merrell and Falkow, 2004). In most cases, the virulence factors produced by the pathogen need to be transported not only across the bacterial cell envelope but also across the host cell plasma membrane. In order to achieve these processes bacteria have evolved a wide range of protein secretion systems (Desvaux et al., 2009; Bleves et al., 2010). Studying protein secretion and identifying new secretion systems rely on evidences which demonstrate that a protein produced by a bacterium can reach the extracellular compartment in a specific manner (Filloux and Hardie, 1998). This means that appearance of exoproteins in the extracellular milieu should be independent of any cell leakage, cell lysis or even release of outer membrane vesicles.
There are many ways for describing and discussing protein secretion systems. To avoid a shopping list-style presentation, the review is formatted on the basis of two criteria. The first one was to choose Pseudomonas aeruginosa as a model organism. The versatility of this microorganism and the large size of its genome provide a rich playground for protein secretion specialists (Bleves et al., 2010). The second criterion was not to list protein secretion systems one after each other but rather consider basic molecular concepts.
Protein Secretion Systems in Gram-Negative Bacteria
In Gram-negative bacteria, such as P. aeruginosa, the cell envelope is composed of two hydrophobic membranes. The inner and outer membranes are separated by a hydrophilic space, the periplasm. The secreted proteins, enzymes, or toxins, do have to travel through the hydrophobic environment of the membranes. These proteins are usually large hydrophilic molecules and therefore need to be accommodated into aqueous channels or other type of conduits that are spanning the cell envelope. These paths to the external medium are built via the assembly of more or less sophisticated macromolecular complexes which we may call secretion machines. The composition and nature of these complexes may vary, but they are broadly conserved across Gram-negative bacteria (Filloux et al., 1990). Nowadays, six different types have been identified which are recognizable by the characteristics of proteins/components forming the secretion machine. These are called type I to type VI secretion systems (T1SS–T6SS; Desvaux et al., 2009; Durand et al., 2009). The secretion systems could also be split into two categories. Those that transport proteins at once across the bacterial cell envelope, thus directly from the cytoplasm to the cell surface, are known as one-step secretion mechanism. Alternatively, the secreted proteins could transit through the periplasm before crossing the outer membrane. These systems are known as two-step secretion mechanism.
Two-Step Secretion Mechanism
A two-step mechanism involves a stopover of the secreted proteins into the periplasm. It is likely that the periplasmic stay is very brief and the protein is trapped there only when the secretion machine is faulty. In principle, the secreted protein reaches the periplasm independently of the secretion system. Instead, it uses a general export pathway that is also involved in the transport of periplasmic and outer membrane proteins. The most essential general export pathway is known as the Sec pathway and is made with the Sec proteins, among which SecA (ATPase) and SecYEG (translocon; Papanikou et al., 2007). The proteins using the Sec pathway have a N-terminal signal peptide and are released in the periplasm after cleavage of this peptide (Choo et al., 2009). Another pathway for protein translocation across the inner membrane, though not essential, was later on identified for the transport of folded protein. This system called Tat, for Twin Arginine Translocation, involves three membrane components, TatABC (Sargent, 2007). The Tat-dependent proteins have a cleavable signal peptide that is recognizable from the Sec signal by its length and the presence of tandem arginine residues.
The type II secretion system
Pseudomonas aeruginosa studies contributed to the pioneering work on the discovery of the type II secretion system (T2SS). Mutations in genes, called xcp, resulted in the periplasmic accumulation of otherwise secreted enzymes such as elastase (LasB; Bever and Iglewski, 1988), lipase (LipA; Wohlfarth et al., 1992), alkaline phosphatase (PhoA; Filloux et al., 1988), or phospholipase C (PlcH; Wretlind and Pavlovskis, 1984; Filloux et al., 1987, 1989; Ostroff and Vasil, 1987). The xcp genes encode proteins with similarity to the Pul proteins that were shown to be involved in the secretion of pullulanase in Klebsiella species (d’Enfert et al., 1987; Filloux et al., 1990; Bally et al., 1992; Akrim et al., 1993). This observation pointed out that each bacterium do not have its own specific secretion system. Instead secretion systems are conserved across Gram-negative bacteria (Filloux et al., 1990). However, the T2SS remains specific and heterologous secretion rarely occurs (de Groot et al., 1991, 1996; Michel et al., 1995).
The Xcp system is encoded in a cluster containing 11 genes, xcpP–Z (Filloux, 2004). A twelfth gene is essential to Xcp function, namely xcpA, which is located elsewhere on the chromosome (Bally et al., 1991, 1992). In fact XcpA is also known as PilD (Nunn and Lory, 1991) and is also involved in type IV piliation. Data mining in the genome sequence of the PAO1 strain (Stover et al., 2000) showed that it exists a second T2SS gene cluster which is called hxc (homologous to xcp; Ball et al., 2002). The genetic organization of the hxc and xcp clusters is different but the hxc cluster contains the 11 xcp homologs, hxcP-Z. The Hxc system also requires the function of the XcpA/PilD protein.
There are as much as 14 characterized Xcp substrates whose genes are scattered all over the chromosome (Table 1). In contrast, there is only one Hxc-dependent substrate, namely LapA, known as low-molecular weight alkaline phosphatase (Ball et al., 2002). In this case, the lapA gene is clustered with the hxc genes indicating that the Hxc system may exclusively be assigned to LapA secretion.
Most Xcp and Hxc substrates carry a cleavable N-terminal signal peptide and are routed to the periplasm in a Sec-dependent manner. Once in the periplasm the proteins adopt a 3D conformation that is a prerequisite for recognition by the T2SS machinery. For example, in the case of the Xcp-dependent elastase (LasB), it was shown that the N-terminal propeptide acts as an intra-molecular chaperone to promote the folding of the LasB catalytic domain (Kessler and Safrin, 1994; McIver et al., 1995, 2004; Braun et al., 1996). In the absence of a functional propeptide, LasB does not fold, is not secreted and accumulates in the periplasm. The lack of secretion is proposed to result from the lack of a conformational signal thus preventing specific recognition by the Xcp machinery (Voulhoux et al., 2000). What exactly is the nature of the secretion signal in Xcp- or Hxc-dependent proteins is unknown. Only in a few cases T2SS secretion motifs have been highlighted, e. g. with the pullulanase from Klebsiella species in which a series of motifs have been proposed to be essential for recognition by the Pul system (Francetic and Pugsley, 2005). Another issue about targeting of substrates to the secretion machine is the identification of the T2SS component involved in the specific recognition. In P. aeruginosa, there is no study indicating which Xcp component could directly bind to a T2SS substrate. However, it was shown that exchange of xcp genes between two related Pseudomonas species, P. aeruginosa and Pseudomonas alcaligenes, results in the assembly of a hybrid T2SS machine which remains functional (de Groot et al., 2001). Whereas this is true for most xcp genes, it is not for the xcpP and xcpQ genes. From this observation two possibilities were considered, i.e., (i) the XcpP and XcpQ component have complementary interface and their interaction is species-specific; (ii) XcpP and/or XcpQ directly interacts with the secreted substrate in a species-specific manner. XcpP is a bitopic inner membrane protein with a large domain protruding in the periplasm and which interacts with the XcpQ outer membrane protein called secretin (Bleves et al., 1996; Gerard-Vincent et al., 2002; Robert et al., 2005b). It is proposed that the interaction between these two components is crucial to control the gating of the secretory channel formed by XcpQ (Tommassen et al., 1992). Whether XcpP and/or XcpQ directly interact with a T2SS substrate is not documented in Pseudomonas species. Nevertheless, a specific interaction between a T2SS substrate and the secretin is described in at least two cases. OutD of Erwinia chrysanthemi binds specifically to the PelB pectate lyase (Shevchik et al., 1997) and an interaction has been demonstrated between the cholera toxin and the secretin EpsD of Vibrio cholerae (Reichow et al., 2010). Interestingly, XphA and XqhA are two other XcpP and XcpQ homologs, respectively. The xphA/xqhA genetic locus does not contain any other xcp-like genes. However, it was shown that XphA and XqhA could replace the XcpP and XcpQ protein in the T2SS/Xcp system. The hybrid Xcp machine containing XphA/XqhA is functional and achieves the secretion of all Xcp substrates but the aminopeptidase PaAP (Michel et al., 2007). This is another indication of a specific interaction between the XcpP/XcpQ-like subcomplex and T2SS substrates.
From previous studies it is clear that T2SS substrates need to be folded in the periplasm for recognition by the secretion machine. This is in contrast with what is observed in other secretion systems such as with the T3SS (Feldman et al., 2002). Whereas Sec-dependent substrates are translocated across the inner membrane in an unfolded conformation, the Tat-dependent substrates are transported through the membrane in a folded conformation. The majority of the Tat-dependent substrates are periplasmic proteins that need to acquire a co-factor in the cytoplasm in order to fulfill their function. This is for example the case of the multicopper enzyme nitrous oxide reductase in Pseudomonas stutzeri (Heikkila et al., 2001). However, it was shown that P. aeruginosa T2SS substrate, such as the phospholipases PlcH and PlcN are transported to the periplasm in a Tat-dependent manner (Voulhoux et al., 2001). This observation clearly shows that in the two-step secretion mechanism, such as with the T2SS, the two successive steps of translocation across the membranes are disconnected. In the first step of translocation across the inner membrane the T2SS substrates can use either the Sec or Tat machinery. However, in the second step both Tat and Sec-dependent substrates converge to the T2SS, Xcp, or Hxc in the case of P. aeruginosa, for translocation across the outer membrane and release in the extracellular medium.
The type V secretion system
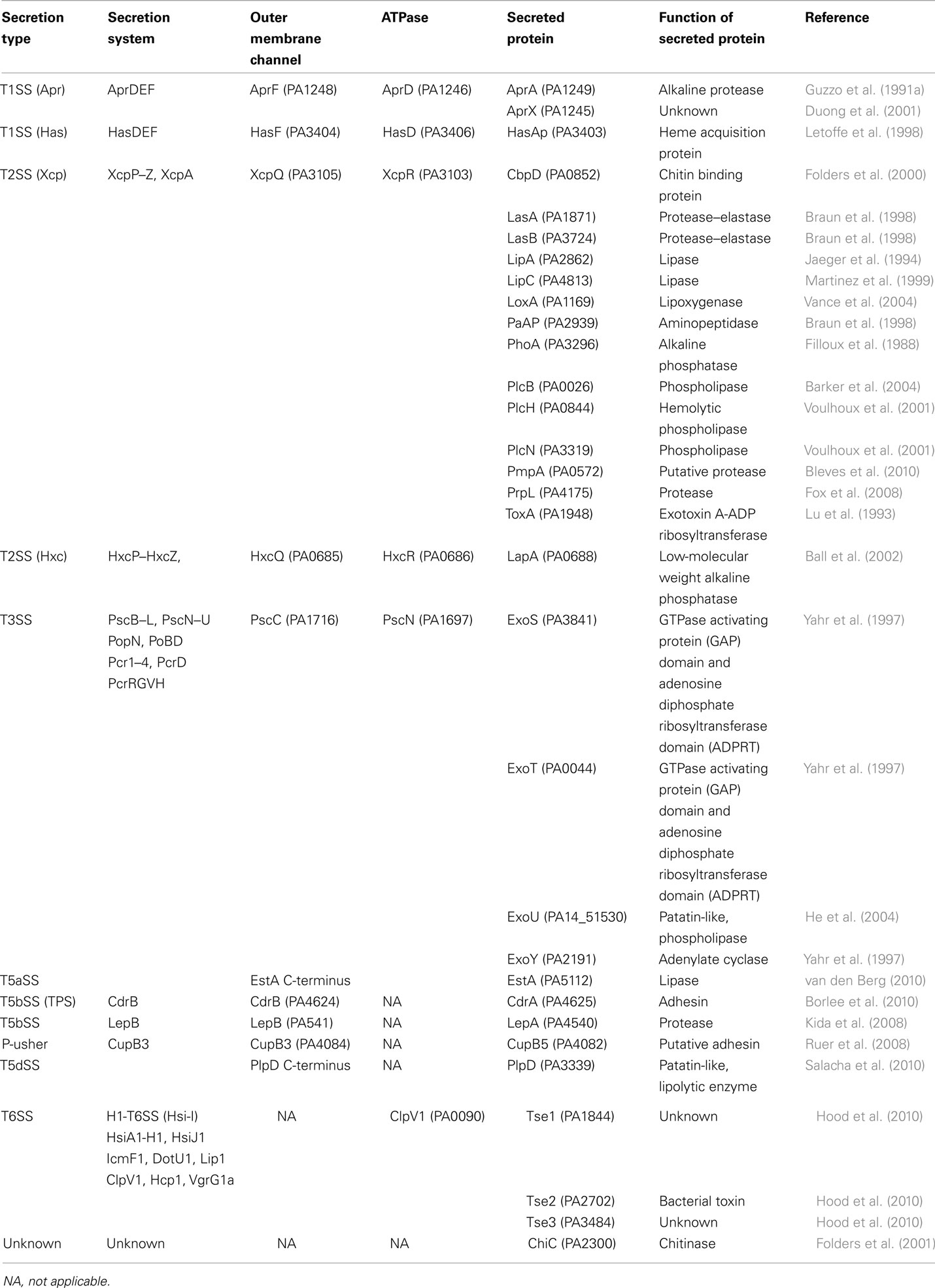
The type V secretion system is the most simple of all. It was originally described for the IgA protease in Neisseria gonorrhoeae (Pohlner et al., 1987). Basically, all the secretion information is contained within one single polypeptide. The protein has a cleavable signal peptide for targeting to the periplasm by the Sec system and a C-terminal domain that forms a β-barrel structure into the outer membrane. The passenger, or protease domain, could travel through the channel formed by theβ-barrel. This is still under debate since other studies indicate that the β-barrel of the autotransporter hemoglobin protease might not be fully folded and inserted in the outer membrane while the passenger domain is already accessible from the surface (Sauri et al., 2009). An alternative transport pathway in this case may involve the Bam/Omp85 complex. Once exposed to the cell surface the protease is released from the β-barrel via autocatalytic cleavage. Because there is no need for accessory proteins to help the secretion process, the mechanism was called autotransporter and coined Type Va secretion system (T5aSS; Dautin and Bernstein, 2007; Yen et al., 2008). The P. aeruginosa PAO1 genome contains three genes encoding proteins with a typical autotransporter C-terminal β-barrel attached to a N-terminal passenger domain. Two, PA3535 ad PA0328, have not been functionally characterized but are predicted to carry serine protease and metallopeptidase activity, respectively. In contrast, EstA has been extensively characterized. It has lipolytic activity (Wilhelm et al., 1999) and its 3D structure has been solved (Figure 1; van den Berg, 2010). The passenger domain is a member of the GDSL family of lipases (Akoh et al., 2004). The structure shows that the size of the barrel is too narrow to accommodate a folded protein and therefore translocation across the outer membrane should occur in an unfolded state. It is suggested that the EstA passenger may fold sequentially starting from the C-terminus. The vectorial passenger folding provides the driving force for the translocation. The catalytic triad is located on the apical surface of the passenger protein. Finally, there is a central α-helix that connects the extracellular passenger to the periplasmic side of the β-barrel that further suggests that the passenger is traveling through the channel. In this case the so-called secreted protein remains anchored at the bacterial surface and is not released into the supernatant.
Figure 1. Structure of the P. aeruginosa EstA autotransporter (T5aSS), reproduced from van den Berg (2010). (A) Backbone representation viewed from the side, with the protein colored by a gradient from blue at the N-terminus to red at the C-terminus. (B) Backbone view 90° rotated within the plane of the membrane relative to (A), with helices colored red, β-strands colored green, and loops colored gray. The catalytic triad residues are shown as blue stick models. Horizontal lines indicate the approximate location of the outer membrane core. (C) Surface view of EstA from the side with the β-barrel domain colored green and the passenger domain colored red. (D) Stereo view of the EstA passenger from the extracellular side, colored as a rainbow from dark blue at the N-terminus to dark red at the C-terminus. The numbers are those for the central residue of the α-helix.
Slightly more sophisticated as compared to autotransporters are the two partner secretion systems, coined TPS or T5bSS (Jacob-Dubuisson et al., 2001; Mazar and Cotter, 2007). In this case the β-barrel domain and the passenger domain are two distinct polypeptides, TpsB and TspsA, respectively. The β-barrel structure in T5aSS involved 12 β-strands (Oomen et al., 2004), whereas it is 16 β-strands that are found in T5bSS (Clantin et al., 2007). One extensively characterized TPS system is involved in the secretion of the filamentous hemagglutinin (FHA) in Bordetella pertussis (Hodak et al., 2006). In addition to a classical Sec-dependent signal peptide, the mature FHA passenger displays a N-terminal secretion motif known as TPS motif that is required for targeting/interaction with the β-barrel domain formed by the TpsB component, in this case FhaC. The interaction is specific and occurs between the TPS motif and a polypeptide-transport-associated (POTRA) domain found at the N-terminus of the β-barrel (Clantin et al., 2007). The POTRA domains are connected to the barrel but protrude into the periplasm where they can interact with the passenger protein. Mining the PAO1 genome identified at least five putative TPSs, Tps1: PA2462–PA2463; Tps2: PA0040–PA0041; Tps3: PA4624–PA4625; Tps4: PA4540–PA4541 and Tps5: PA0690–PA0692. Among these, only few have been functionally characterized. In the case of LepA/LepB (Tps4-PA4541/PA4540; Kida et al., 2008) the passenger protein, LepA, contains a RGD motif that may contribute to eukaryotic cell attachment. However, the most striking feature is the identification of a trypsin-like serine protease motif and the demonstration that LepA has proteolytic activity. In the case of CdrA/CdrB (Tps3-PA4625/PA4624), CdrA is a putative c-diGMP regulated adhesin with a β-helix structural motif (Borlee et al., 2010). The CdrA adhesin was shown to promote P. aeruginosa biofilm formation and bacterial auto-aggregation in liquid culture. Finally, from the PAO1 genome, a sixth putative TPS could be considered, PA2542/PA2543, since PA2542 displays some similarities with the C-terminal domain of TpsA1 and PA2543 is predicted to be an outer membrane protein.
In addition to the 5–6 pairs of TpsA–TpsB, which are canonical examples of the T5bSS, P. aeruginosa contains an additional TpsA-encoding gene which is located within a gene cluster encoding cup fimbrial genes (Vallet et al., 2001). The secretion of this protein, CupB5 (Figure 5B), does not involve a cognate TpsB partner. Instead, it involves an outer membrane protein, CupB3, which is a typical usher protein involved in the assembly of fimbriae at the bacterial cell surface (Ruer et al., 2008). Furthermore, the CupB3 usher contains an N-terminal extension that shares similarities with POTRA domains found in TpsB proteins (Clantin et al., 2007). It is proposed that CupB3 is a chimera between an usher and a TpsB protein and that the POTRA domain is required for proper secretion of the putative CupB5 adhesin, but dispensable for the assembly of the CupB1 fimbriae (Ruer et al., 2008). CupB3 was called a P-usher for POTRA-containing Usher.
As indicated earlier, three autotransporters (T5aSS) have been identified in P. aeruginosa. Recently, a fourth autotransporter was described, PlpD (Salacha et al., 2010). PlpD is a member of the lipolytic enzyme family of patatin-like proteins (PLPs). Patatins are major proteins in potato tubers. They display lipid acyl hydrolase activity and have been proposed to be protective against plant pathogens (Banerji and Flieger, 2004). Interestingly, one of the P. aeruginosa T3SS effectors, ExoU, is also classified as a patatin-like protein and carries phospholipase A2 activity (Sato and Frank, 2004; Lee et al., 2007). PlpD is considered an autotransporter since it has a two-domain organization. This includes the passenger lipase domain and a domain predicted to form the β-barrel in the outer membrane. It is predicted that the β-barrel of PlpD has 16 and not 12 strands which suggests that the protein does not belong to the T5aSS family but is closer from the T5bSS. Sequence analysis also revealed that a POTRA like domain could be identified within the PlpD protein, also a characteristic of TpsB and T5bSS family. PlpD thus represents a novel secretion type that mixes T5aSS and T5bSS and was given the name T5dSS (Salacha et al., 2010). Of note, T5cSS was already coined for a particular category of autotransporter whose β-barrel is formed by the assembly of a trimer (Kajava and Steven, 2006; Meng et al., 2006). Such trimeric assembly is somehow similar to what was described for the TolC protein (Koronakis et al., 2000).
One-Step Secretion Mechanism
By contrast with the two-step mechanism, the one-step secretion mechanism is based on the concept that no pool of secreted protein should be identified in the periplasm. In other words, when the secretion system is defective the secreted protein is blocked in the cytosol and possibly degraded. A continuous channel crossing the bacterial cell envelope, channel consisting of one or a combination of proteins, could make the transition from the bacterial cytosol to the extracellular environment.
The type I secretion system
The Escherichia coli α-hemolysin (HlyA) represents the T1SS archetype (Mackman et al., 1985). The T1SS consists of three components of which an ATP-binding cassette (ABC) protein (HlyB), an outer membrane protein (TolC) and an inner membrane protein (HlyD) largely protruding in the periplasm (Membrane fusion protein, MFP; Holland et al., 2005). HlyB and HlyD are plasmid-encoded together with HlyA, whereas TolC is chromosome-encoded. Studies conducted with P. aeruginosa have also pioneered the field with the alkaline protease, AprA (Guzzo et al., 1990, 1991a,b; Baumann et al., 1993). AprA has long remained the only type I-dependent substrate in P. aeruginosa until AprX, a protein with unknown function, was shown to use the same secretion system (Duong et al., 2001). The genes encoding the secretion system, AprDEF that are homologous to HlyBC and TolC, respectively, and the genes encoding AprA and AprX, are clustered together (Figure 2). This observation suggests that the Apr system is specific for these two substrates. This is in contrast with the Xcp system that is used to secrete proteins encoded from genes scattered all over the chromosome. Another type I secretion system was discovered later which is involved in iron uptake (Letoffe et al., 1998). This system called HasDEF (Figure 2) is very similar to the Serratia marcescens system (Ghigo et al., 1997; Arnoux et al., 1999; Wandersman and Delepelaire, 2004) and involves a heme-binding protein, HasAp. The protein is secreted in a T1SS-dependent manner, binds heme from hemoglobin and is then taken up via the cell surface receptor HasR (Letoffe et al., 1999). The NMR structure of HasAp has been solved and gave insight on the kinetic parameter for heme-binding and uptake (Jepkorir et al., 2010; Yukl et al., 2010). Another P. aeruginosa T1SS could be identified by genome mining. The system is coded by PA1875–PA1877 while a gene adjacent to the secretion cluster is encoding a large protein (PA1874) reminiscent of proteins secreted by the T5bSS (van Ulsen et al., 2008). This organization recalls what was reported with Pseudomonas putida in which LapA, a large adhesin involved in biofilm formation, is transported to the cell surface by a T1SS made with the LapBCD components (Hinsa et al., 2003). A fourth complete T1SS is found encoded in the PAO1 genome, which is PA4142–PA4144. No functional data are available regarding this system but PA4142 and PA4143 are homologous to CvaA and CvaB, a T1SS involved in the secretion of colicin V (ColV) in E. coli (Zhang et al., 1995; Zhong et al., 1996). ColV is a 109 amino-acids-long toxin and it is striking to observe that the gene located downstream the putative P. aeruginosa T1SS cluster, PA4141, encodes a small protein of 99 amino acids.
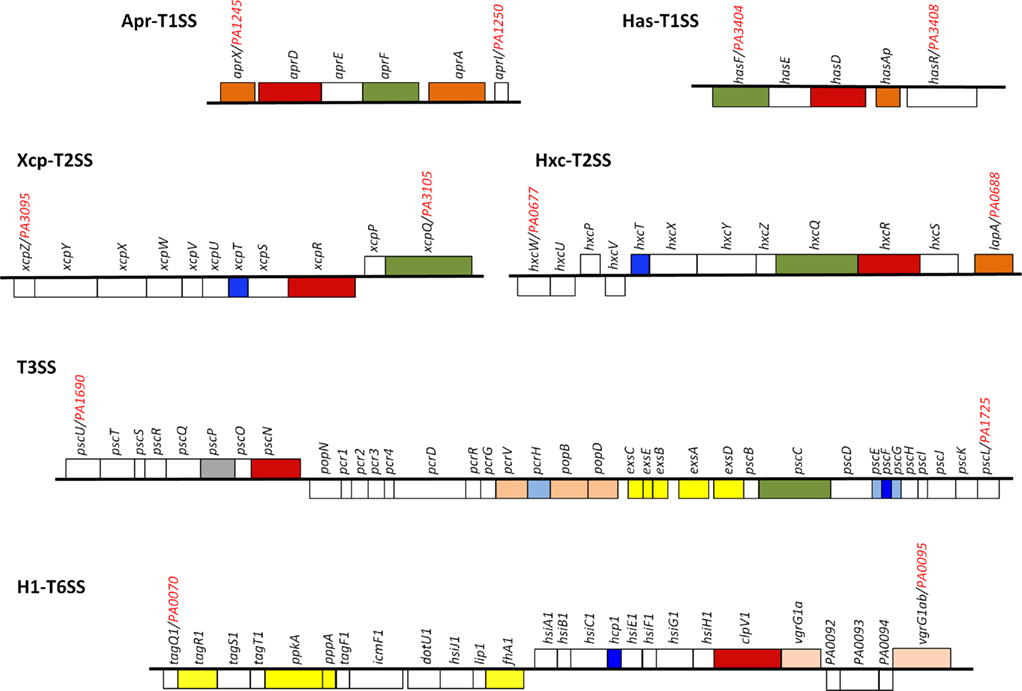
Figure 2. Pseudomonas aeruginosa secretion gene clusters. Within the clusters shown, the gene encoding the ATPase is colored red, the gene encoding the outer membrane channel is colored green. The genes encoding secreted proteins are colored dark orange. Genes encoding components of the T2SS pseudopilus, the T3SS needle and the T6SS tail tube are colored dark blue. The genes in light orange encode either translocators of the T3SS or puncturing device of the T6SS. Shown in light blue are T3SS chaperones and in gray the T3SS ruler. Shown in yellow are genes encoding proteins involved in regulation. For more details see text.
The type III secretion system
The T3SS is a fascinating secretion system. In Yersinia enterocolitica, it was proposed to secrete proteins called Yersinia outer membrane protein (Yop; Michiels et al., 1990). These Yop proteins were recovered with outer membrane proteins during fractionation procedures. This unusual localization was due to aggregation of the proteins once released in the extracellular medium. In these experiments, extracellular secretion was obtained upon artificial induction of the T3SS using Ca2+ chelation. It is now established that T3SS activity is cell contact-dependent and therefore the T3SS substrates are injected directly from the bacterial cytoplasm into the eukaryotic cells cytosol (Cornelis, 2010).
In P. aeruginosa the T3SS was discovered in 1996 (Yahr et al., 1996) and since 36 genes have been associated with T3SS function (Frank, 1997; Brutinel et al., 2008). The genes encoding elements of the secretion machine and genes encoding regulatory proteins are clustered but organized in several operons (Figure 2). In contrast, the genes encoding the effector proteins are scattered all over the chromosome (Table 1). The secretion machine is made of over 20 different proteins that in P. aeruginosa are called the Psc, Pop, or Pcr components. The T3SS machine resembles a needle whose first observation in Salmonella Typhimurium (Figure 3; Kubori et al., 1998) came as a real flash of inspiration for the scientific community. It materialized the idea of injection that led to name the T3SS nanomachine, injectisome. High-resolution images are now available for S. Typhimurium and Shigella flexneri injectisomes (Hodgkinson et al., 2009; Schraidt and Marlovits, 2011). Whereas the needle-like structure is central to the injection process (Blocker et al., 2008), a set of three proteins, PopBD and PcrV in P. aeruginosa, are crucial to establish a pore into the membrane of eukaryotic cells (Dacheux et al., 2001; Schoehn et al., 2003; Goure et al., 2004). In fact these proteins are the first proteins to be transported through the needle but they are not injected into the cytosol. Instead, they remain inserted into the membrane to form a pore 2.8–6.0 nm in diameter that is called the translocon. Genuine effectors are subsequently transported through the needle and injected in the eukaryotic cell cytosol by passing through the pore formed by the translocon. PopB and PopD are the proteins that insert into the eukaryotic cell membrane. The role of PcrV is unclear but it seems to form a scaffold at the tip of the needle and antibodies against PcrV inhibit injection of T3SS effectors (Sawa et al., 1999; Goure et al., 2005; Broz et al., 2007).
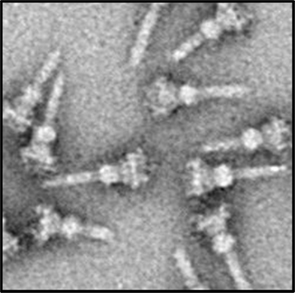
Figure 3. A transmission electron-microscope image of isolated T3SS needle complexes from S. typhimurium reproduced from Schraidt et al. (2010).
In P. aeruginosa, the T3SS is a major virulence weapon that contributes to cytotoxicity and acute infections (Hauser et al., 2002; Hauser, 2009). Cytotoxicity is associated with the activity of effector proteins that are secreted in a T3SS-dependent manner. In P. aeruginosa only four effectors, named ExoS, ExoT, ExoU, and ExoY, have been identified. ExoS and ExoT have a GTPase activating protein (GAP) domain and an adenosine diphosphate ribosyl transferase domain (ADPRT; Fleiszig et al., 1997). ExoU is the most potent cytotoxin, it has a phospholipase activity and can be classified within the PLP family (Sato and Frank, 2004). ExoY has an adenylate cyclase activity. The presence of the genes encoding these effectors may vary from one isolate to the other but it is generally observed that exoS and exoU genes are mutually exclusive (Finck-Barbancon et al., 1997; Berthelot et al., 2003; Wolfgang et al., 2003; Kulasekara et al., 2006). For example when looking at the sequence of available P. aeruginosa genomes, exoS is found only in PAO1 and LESB58. In contrast, exoU is found exclusively in PA14 and is located within a well-characterized pathogenicity island, PAPI-I (Qiu et al., 2006). Finally, exoT and exoY are found in all three strains, PAO1, PA14, and LESB58. The fourth sequenced P. aeruginosa strain, PA7(Roy et al., 2010), is a non-respiratory human isolate that is highly resistant to antibiotics. Strikingly, not only this strain lacks genes encoding all four effectors but it also lacks all the genes encoding the components of the T3SS machinery. The effector proteins are transported through the needle in an unfolded manner and are targeted to the T3SS by a cognate chaperone which they bind to prior secretion (Parsot et al., 2003). The chaperone for ExoS and ExoT is called SpsC, whereas the chaperone for ExoU is SpcU (Finck-Barbancon et al., 1998; Shen et al., 2008). The spsC gene is clustered with the exoS gene. Interestingly, in the PA14 strain the exoS gene is lacking whereas the spsC gene is kept, likely for ExoT secretion. No cognate chaperone has been characterized to date for ExoY (Yahr et al., 1998). The PopBD translocon proteins that are also transported by the needle-like structure have an identified cognate chaperone, namely PcrH (Broms et al., 2003).
The Type VI Secretion System
It is yet unclear whether the T6SS is definitely a one-step mechanism since proteins secreted by the T6SS have not been clearly identified, or their fate incompletely characterized. In P. aeruginosa there are three different gene clusters encoding a set of 15–20 genes involved in T6SS function (Filloux et al., 2008). These clusters were initially identified through the characterization of a gene pair encoding proteins similar to IcmF and DotU (Das and Chaudhuri, 2003). These proteins are required in several bacteria for the function of the T4SS (Sexton and Vogel, 2002). Genomic analyses revealed that in many cases the pair icmF/dotU is clustered with a conserved set of genes unrelated to T4SS genes and the name T6SS was coined. The three P. aeruginosa T6SS clusters were initially called HSI, for Hcp Secretion Island (Mougous et al., 2006). Later on, the three clusters were renamed H1-T6SS to H3-T6SS (Hood et al., 2010). Though the three clusters displayed a set of core genes, there is a level of variability in their composition and genetic organization. The most complex and best studied is the H1-T6SS cluster (Figure 2) since it is highly expressed in a strain mutated for a gene encoding the RetS kinase sensor (Mougous et al., 2006; Goodman et al., 2009).
Historically, the T6SS substrates were considered to be proteins named Hcp and VgrG (Pukatzki et al., 2006). On the P. aeruginosa PAO1 genome, a BLAST search identified 10 vgrG genes and 5 hcp genes (Hachani et al., 2011). Some of these genes are clustered with T6SS genes but others are scattered around the chromosome. One hcp gene, hcp1, and two vgrG genes, vgrG1a and vgrG1b, are associated with the H1-T6SS cluster (Figure 2). Hcp1 and VgrG1a are secreted in a T6SS-dependent manner (Mougous et al., 2006; Hood et al., 2010; Hachani et al., 2011). However, the structure of Hcp1(Mougous et al., 2006), and later VgrG1a (Leiman et al., 2009; Hachani et al., 2011), showed that these proteins form multimeric complexes and are likely to be part of the secretion machine rather than being genuine effectors (Figure 4). The occurrence of these proteins in the extracellular medium could be incidental. They may constitute a cell surface exposed portion of the T6SS machine that may be sheared off during sub-cellular fractionation procedures. Recently, three genuine effectors for the P. aeruginosa H1-T6SS machine have been identified and given the name Tse1–3, for type six exported (Hood et al., 2010). Importantly, it was shown that the Tse2 protein is a toxin able to kill competing bacteria. The tse2 gene is clustered with a gene called tsi2. The tsi2 gene encodes a protein that protects the producing strain from the effect of the Tse2 toxin.
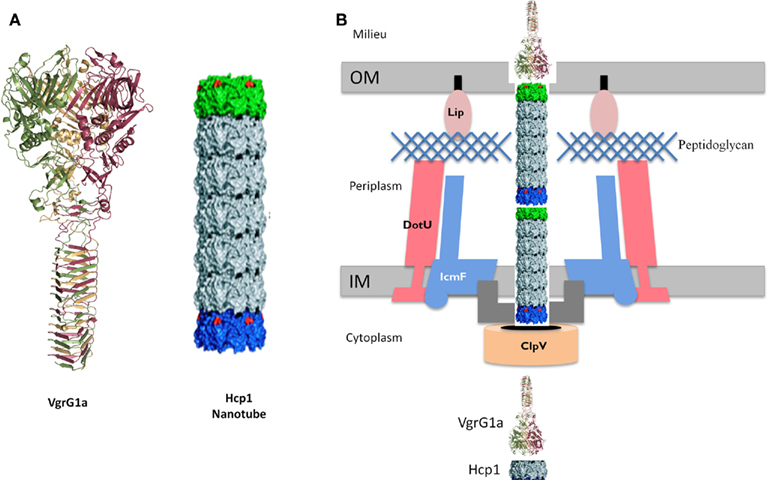
Figure 4. Schematic model for the T6SS. (A) Structural-based homology prediction for the VgrG1a protein of P. aeruginosa reproduced from Hachani et al. (2011). The Hcp1 nanotube reproduced from Ballister et al. (2008). (B) Schematic representation of the T6SS, in which VgrG1a (puncturing device) and Hcp1 nanotube have been included as depicted in (A). Few other T6SS components are shown and the system punctures the bacterial cell envelope from inside to outside. See text for more details.
Molecular Needs for the Function of Secretion Machines
Most secretion machines are relatively complex and each secretion type is identified by the characteristics of the constituents involved. However, there are a number of features and rules that are hallmarks of all secretion systems. For example, these machines require a component providing energy to the process, usually an ATPase. It also requires an outer membrane protein that is the ultimate channel before the secreted protein can access the external milieu. Other components of the system are involved in the scaffolding and/or regulation of the macromolecular complex or in the specific recognition of secreted substrates.
Energizing the Secretion Process
Translocation of macromolecules across membranes is an active process that is powered by energy sources. The Sec machinery which is universally used by bacteria, requires an ATPase, SecA, which pushes substrates across the cytoplasmic membrane (Sardis and Economou, 2010). In contrast the Tat system that is involved in the translocation of folded proteins has been shown to use the proton motive force to move polypeptides across the cytoplasmic membrane (Palmer and Berks, 2003; Bageshwar and Musser, 2007). Most protein secretion systems use ATPases to promote movement of polypeptides across the whole bacterial cell envelope (Saier, 2006).
T1SS, T2SS, T3SS, and T6SS all contain at least one component that has a typical Walker motif characteristic of proteins able to bind and hydrolyze ATP. In P. aeruginosa, XcpR and HxcR are part of the T2SS and belong to the family of traffic ATPases (Planet et al., 2001). These proteins have a highly conserved Walker A, a less conserved Walker B and their own signature which is a highly conserved aspartate-rich region, or aspartate box, located between the Walker A and B (Possot and Pugsley, 1994). Additional conserved motifs in this family of ATPases are a histidine box with no assigned function and a tetracysteine motif whose role might be to coordinate divalent cations. The ATPase AprD (Duong et al., 1992), involved in the secretion of AprA, is a member of the ABC-transporter family (Higgins, 2001) which is found in all T1SS (Saier, 2006). This family of ATPases, in addition to the Walker motifs, displays a unique signature sequence (LSGGQ) in each nucleotide binding domain (NBD; Loo et al., 2002). In the case of T3SS a hydrophilic component homologous to F1/V1-ATPases (Pozidis et al., 2003) is involved in the secretion process, namely PscN in P. aeruginosa. Finally, for the T6SS a AAA ATPase, called ClpV1, ClpV2, or ClpV3 in P. aeruginosa, is involved. The AAA motif is a conserved sequence of about 250 residues that includes the Walker sequences and other domains unique to AAA proteins (Patel and Latterich, 1998). It is remarkable that each secretion system described previously involves an ATPase of a distinct family. This is not always the case since in the T4SS that does not exist in P. aeruginosa the energizing component is a traffic ATPase, such as the T2SS ATPases XcpR and HxcR (Sagulenko et al., 2001).
The 3D structures of some of these ATPases have been solved, although none were obtained from P. aeruginosa. These proteins show a hexameric structure and a central channel as with the T2SS traffic ATPase from Vibrio cholerae, EpsE, (Robien et al., 2003). The T6SS ATPases are also hexamers and the V. cholerae ClpV member has been proposed to modulate the formation of tubules which are made of two other T6SS components. These are VipA and VipB that are homologous to the HsiB and HsiC proteins in P. aeruginosa (Bonemann et al., 2009, 2010; Filloux, 2009). The T2SS and T6SS-associated ATPases are not integral membrane proteins. Instead, they could be peripherally associated with the inner face of the cytoplasmic membrane via interaction with another specific component. In the case of XcpR the integral membrane protein XcpY keeps XcpR associated with the membrane and with the rest of the T2SS machine (Ball et al., 1999). In the case of P. aeruginosa ClpV1, it was shown that the presence of other components of the T6SS is required to make a ClpV1–GFP fusion appeared as discrete foci, whereas ClpV1 distribution becomes diffuse in the absence of IcmF1 or Hcp1(Mougous et al., 2006).
Whereas the T2SS and T6SS-associated ATPases appeared to function as hexamers, the ABC-transporters involved in T1SS-dependent secretion are dimeric (Lin et al., 2009). The ABC-transporters from T1SS are usually integral membrane proteins with a transmembrane domain connected to the NBD domain that extends in the cytoplasm. The NBD domain structure of the canonical HlyB from E. coli has been solved and displays the common overall architecture of ABC-transporter NBDs (Schmitt et al., 2003). The direct interaction between the ABC-transporter and the secreted protein is largely documented (Letoffe et al., 1996; Thanabalu et al., 1998; Benabdelhak et al., 2003) which supports the idea that the ABC-transporter directly contributes to the translocation of the secreted protein across the cytoplasmic membrane.
In the case of T3SS, the ATPase is tightly associated but not inserted into the cytoplasmic membrane. It contains a NBD domain related to F1-ATPase. Biochemical and structural studies have been performed with HcrN from Pseudomonas syringae showing that the most active form of HcrN is an oligomer (Pozidis et al., 2003).
In the case of T5SS, the translocation of exoproteins across the outer membrane is disconnected from the cytoplasmic membrane. The one and only accessory component of the T5SS machine is an outer membrane protein. The dynamic of T5SS-dependent translocation across this outer membrane pore has been proposed to depend on energy resulting from the folding of the secreted protein (Junker et al., 2009). This hypothesis was further supported by recent structural data obtained with the P. aeruginosa EstA (van den Berg, 2010; Figure 1).
Pore formation across the outer membrane
Secretion systems are also characterized by the existence of a protein that forms an outer membrane pore. The structural characterization of the E. coli TolC protein has been a seminal step forward in the understanding of the T1SS mechanism (Koronakis et al., 2000). This protein forms a trimer with two distinct domains. One domain is a β-barrel which is similar to barrels found in outer membrane proteins such as the porins (Cowan et al., 1992). A main difference is that the barrel forms upon assembly of the trimer whereas with porins the barrel is a monomeric protein. The most striking structural feature of TolC is that it contains a second domain essentially made of helices forming a long channel which spans the periplasm (Koronakis et al., 2000). This periplasmic conduit that connects the outer membrane pore of TolC to the ABC protein (HlyB) within the cytoplasmic membrane is a demonstration that T1SS-dependent substrates may travel through the secretion apparatus and never be exposed to the periplasm. It thus validates the idea of a one-step mechanism. The TolC protein in E. coli is not only involved in T1SS but also in the formation of efflux pumps, such as with ArcAB (Symmons et al., 2009). In P. aeruginosa several efflux pump systems have been identified of which the MexAB-OprM is extensively characterized. The structure of the outer membrane protein OprM is available (Lambert et al., 2005; Phan et al., 2010; Trepout et al., 2010) and suggests a conserved TolC-like structural organization. This family of proteins also includes AprF (Duong et al., 1992) and HasF in the P. aeruginosa T1SS. The TolC protein in P. aeruginosa has not been functionally characterized but is suggested to be PA4974.
The assembly of a β-barrel structure for the formation of a pore in the outer membrane is also the adopted strategy in the case of T5SS. The structure of the EstA autotransporter provides a typical illustration of this organization (Figure 1) although the number of β-strands varies when comparing the T5aSS (12 β-strands; Oomen et al., 2004) and the T5bSS (16 β-strands; Clantin et al., 2007). The structure of the P. aeruginosa T5bSS CdrB could be modeled using the Phyre software (Kelley and Sternberg, 2009) and displays the typical β-barrel fold with POTRA domains at the N-terminus (Figure 5A).
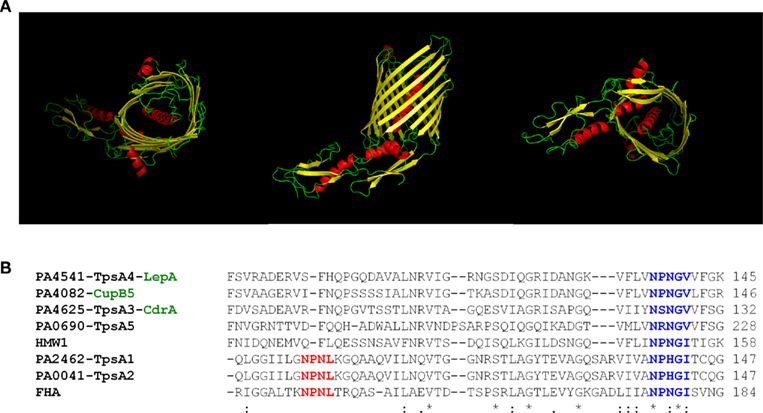
Figure 5. The Two partner secretion systems (T5bSSs). (A) Structural prediction and modeling of P. aeruginosa CdrB, the carrier domain, performed using Phyre (http://www.sbg.bio.ic.ac.uk/phyre). Figures were made using PyMOL (http://www.pymol.org). From left to right, top (facing extracellular milieu), side, and bottom (facing periplasm) view. (B) Amino-acid sequence alignment of the “TPS” domains of several TpsAs passenger including FHA (B. pertussis), HMW1 (H. influenzae), TpsA1–5 and CupB5 (P. aeruginosa). The NPNL motif in FHA and TpsA1–2 (red) or NxxGx motif in all TpsAs (blue) is indicated with bold characters. The alignment was performed using clustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2).
Another fascinating protein, which has been the object of a large number of studies is the secretin found in T2SS and T3SS. The XcpQ protein was one of the first to reveal the donut shape-like structure adopted by these proteins which suggests a homo-multimeric complex with a central channel (Koster et al., 1997; Bitter et al., 1998). Other secretions have since been characterized but none of them could be taken into crystallographic studies (Collins et al., 2003; Chami et al., 2005) except for their periplasmic domain (Korotkov et al., 2009; Spreter et al., 2009). Nevertheless, quality images are obtained from Cryo-EM analysis and 3D reconstruction as with V. cholerae secretin GspD (Reichow et al., 2010; Korotkov et al., 2011). The global view is a cylinder formed by about 12–15 subunits. The cylinder extends in a periplasm via a domain called vestibule. The secretin channel is open on the side facing the outer membrane but gated from the periplasmic end (Korotkov et al., 2011). In P. aeruginosa, the dimension of the central channel formed by XcpQ is 95 Å and is compatible with the transport of folded protein (Bitter et al., 1998). The XcpQ secretin has two domains. The C-terminal domain is inserted in the outer membrane and forms the channel whereas the N-terminus protrudes in the periplasm and forms the vestibule (Brok et al., 1999; Korotkov et al., 2011). Up to now, it has not been possible to understand whether the secretin inserts in the membrane through the formation of a β-barrel or whether it adopts a different mechanism. Recent studies have highlighted that some outer membrane proteins can adopt a unique fold which is a barrel formed by amphipathic α-helices (Dong et al., 2006; Chandran et al., 2009; Kowalska et al., 2010).
None of the T6SS components can be considered as an outer membrane pore. The only putative outer membrane protein is a lipoprotein which is anchored at the inner face of the outer membrane (Aschtgen et al., 2008). It is unlikely to form a pore and the T6SS might involve an unprecedented mechanism for translocation across the outer membrane.
Function of accessory proteins
In addition to ATPases and pore-forming outer membrane proteins, secretion systems involve accessory proteins whose function is far from being elucidated. Some of these proteins could be important to collate the machinery. In the case of T2SS for example, the integral inner membrane protein XcpY is essential for anchoring the ATPase XcpR to the inner face of the cytoplasmic membrane (Ball et al., 1999). In the absence of XcpY, XcpR remains cytosolic. XcpZ, another inner membrane protein, is important for the stability of the XcpY protein and thus for the maintenance of the XcpYR complex (Michel et al., 1998).
The secretin component has also been proposed to require accessory component for proper targeting and insertion in the outer membrane. This component is more currently known as pilotin (Hardie et al., 1996; Shevchik and Condemine, 1998). It is a small lipoprotein able to bind at the C-terminus of the secretin (Daefler et al., 1997). This has been shown for several bacterial species, including Klebsiella and Erwinia. In P. aeruginosa no pilotins have been identified for XcpQ (T2SS) or PscC (T3SS). Recently, it was shown that the secretin HxcQ does not require a pilotin but is self-piloted by a lipid modification at its N-terminus (Viarre et al., 2009). Finally, the inner membrane component XcpP has been shown to interact both with the secretin XcpQ and the XcpYRZ complex (Bleves et al., 1999; Gerard-Vincent et al., 2002; Robert et al., 2005a,b). A proposed function for XcpP is the gate keeping of the secretin channel.
In the T1SS, there is only one additional component that is known as the MFP or periplasmic adaptor. Several structural data are available for MFP from efflux pump systems such as AcrA in E. coli (Mikolosko et al., 2006) or MexA in P. Aeruginosa (Higgins et al., 2004; Trepout et al., 2010). These structures suggest that the periplasmic adaptor is central for the assembly of the whole complex and the interaction between the adaptor and the protruding coiled coil domains of the outer membrane protein TolC may be crucial for controlling the opening of the channel (Lobedanz et al., 2007; Symmons et al., 2009; Pei et al., 2011). The overall structure of a drug efflux pump has been obtained (Symmons et al., 2009) and is likely to apply to T1SS macromolecular complexes.
Protein targeting
Pseudomonas aeruginosa secretes a wide range of proteins and possesses most of the secretion types at the exception of the T4SS. Therefore the bacterial cell needs to select the suitable substrates for each of these machineries using a specific recognition process and avoiding all invalid pairing which may jeopardize the function of the secretion system (Filloux, 2010). Targeting to the Sec or Tat translocon has been largely documented and requires a cleavable N-terminal signal peptide displaying recognizable features (Vonheijne, 1985; Palmer et al., 2010).
In the case of T2SS, the N-terminal signal peptide found on P. aeruginosa exoproteins, e. g. elastase, exotoxin A, or lipase, is sufficient for translocation across the inner membrane in a Sec-dependent manner. In the case of phospholipases, PlcH and PlcN are translocated in a Tat-dependent manner (Voulhoux et al., 2001) whereas PlcB is translocated in a Sec-dependent manner (Barker et al., 2004). In all cases, this first step of translocation is not sufficient to target exoproteins to the T2SS. Additional secretion motifs are needed which provide a strong specificity to the secretion process. For example, there is no cross-exchange of substrates between the two P. aeruginosa T2SS, Xcp, and Hxc. The Hxc-dependent LapA is neither recognized nor secreted by the Xcp system (Ball et al., 2002). Despite many studies on the T2SS in P. aeruginosa and other Gram-negative bacteria, it has no been possible to identify an obvious secretion motif in T2SS substrates. Instead, it is suggested that the motif is conformational and results from the assembly of a patch of residues within the folded protein. Such conclusion has been drawn from studies on elastase or exotoxin A that showed that folding is a prerequisite to secretion and improperly folded proteins are trapped in the periplasm (Braun et al., 1996; Voulhoux et al., 2000).
In the case of T5aSS, no motif other than the signal peptide is needed. The passenger protein is connected to the translocation channel, as described for the P. aeruginosa, EstA (Figure 1), and thus no need for targeting. In the case of T5bSS the passenger protein (TpsA) is distinct from the outer membrane translocation channel (TpsB). The recognition process relies on two motifs, one located in the TpsA protein and called TPS motif, and one in the transporter TpsB called POTRA domain (Mazar and Cotter, 2007). This is well demonstrated with the couple FHA/FhaC in B. pertussis (Clantin et al., 2007) and sequence analysis of the T5bSSs in P. aeruginosa shows that these features are rather well conserved (Figure 5).
The T1SS does not require an initial step of translocation across the cytoplasmic membrane via the Sec or Tat system. Consequently, direct targeting of the exoprotein to the T1SS machine occurs. The secretion motif lies in the C-terminus of the secreted protein and is not cleaved. For example, the secretion signal of hemolysin HlyAlies within the last 53 residues. This region includes a 18-amino-acid amphiphilic α-helix, a cluster of charged residues and a weakly hydrophobic terminal sequence (Koronakis et al., 1989). Furthermore, the NBD of HlyB and a C-terminal 23 kDa fragment of HlyA have been shown to interact with each other in a specific manner (Benabdelhak et al., 2003). In the case of P. aeruginosa AprA, the 50 C-terminal residues constitute an autonomous secretion signal which is specific for AprD recognition but fails to recognize the HlyB ABC-transporter (Duong et al., 1996). Recent studies suggest that additional secretion signals could participate to the earlier steps of recognition, whereas the C-terminal signal might be essential at later stages for the release of the substrate from the T1SS machine (Masi and Wandersman, 2010). In some T1SS-dependent protein, such as P. aeruginosa AprA, a glycine rich repeat domain contributes to yield a roll conformation (Baumann et al., 1993). The secretion signal could be used to promote the secretion of unrelated passenger protein as shown with the C terminal domain of a Pseudomonas fluorescens lipase (TliA; Chung et al., 2009).
In the case of T3SS, many studies converge to the idea that the targeting signal is located at the N-terminus of the exoprotein. The targeting signal could be bipartite with one region located in the very first amino acids, i.e., 1–15 in the case of P. aeruginosa ExoS (Hauser, 2009). Furthermore, for the effectors that require a cognate chaperone, the chaperone-binding site is also located at the N-terminus, i.e., 15–51 in the case of ExoS (Deng and Barbieri, 2008; Hauser, 2009). The structural organization of the signal is likely to be the same with ExoT and ExoU whereas for ExoY no chaperone seems to be required. Strikingly, from other T3SS studies the nature of the signal has been proposed to be located within the secondary structure of the mRNA rather than being directly dependent on the amino-acid sequence (Anderson and Schneewind, 1997). This is a complex issue and both concepts have not been reconciled yet.
Studies on the T6SS do not provide any information on how proteins are targeted to the secretion machine, but very little, if any, investigations have addressed this issue.
Relationship between Secretion Systems and Other Biological Processes
A remarkable feature about secretion systems is that several of them derived from other membrane bound systems that have been adapted for a different purpose. Past studies revealed that T2SS is a copy–paste when compared with the system required for assembly of type IV pili (Hobbs and Mattick, 1993; Filloux, 2004). Whereas type IV pili are polymeric structures attached at the cell surface (Mattick, 2002), the T2SS assembles an abortive pilus, called pseudopilus (Sauvonnet et al., 2000; Durand et al., 2003). Pseudopilus elongation/retraction within the periplasm works as a piston expelling secreted proteins through an outer membrane channel. The T3SS shows exquisite resemblance with the flagellar basal body (Blocker et al., 2003; Journet et al., 2003). The latter is required for secretion and polymerization of the flagellin subunit into a flagellum, an important device for bacterial mobility (Macnab, 2003), whereas the T3SS needle is used for the injection of T3SS effectors within the cytosol of the host cells (Cornelis, 2006). Interestingly, the T4SS, though lacking in P. aeruginosa, was originally identified in A. tumefaciens as a translocating machine targeting bacterial T-DNA into the nucleus of plant host cells (Christie et al., 1989). This machinery is analogous to the system assembling F-pili that are required for DNA exchange in bacterial conjugation (Winans et al., 1996; Cascales and Christie, 2003). The T4SS is now recognized in many bacteria as a genuine protein secretion system, injecting proteins such as Legionella pneumophila RalF, or Helicobacter pylori CagA, into the host cell cytosol (Backert et al., 2000; Nagai et al., 2005). Finally, and this is a major discovery in the field of protein secretion over the last decade, the T6SS shares extensive similarity with the bacteriophage tail which is used to inject the phage DNA by puncturing the bacterial cell envelope (Leiman et al., 2009).
The T2SS Pseudopilus
The P. aeruginosa Xcp secretion is a multiprotein complex comprising an inner membrane platform, XcpSPYZ, a traffic ATPase, XcpR, and a secretin, XcpQ which is the outer membrane pore through which exoproteins are ultimately secreted (Filloux, 2004). Another structure has been characterized which is called pseudopilus after its similarity with type IV pilus (T4P). Type IV pili are long fimbrial structures present at the cell surface of various Gram-negative bacteria (Mattick, 2002). Their formation initiates at the cytoplasmic membrane while polymerization of the subunit PilA results in pilus extrusion through the outer membrane pore PilQ (secretin family; Wolfgang et al., 2000). Type IV pili are involved in adhesion, aggregation, or host cell invasion, whereas pilus retraction promotes type IV pili-dependent mobility on solid surfaces.
In P. aeruginosa, five Xcp proteins (XcpT–X) are classified as type IV pilin-like protein and called pseudopilins (Bally et al., 1992; Bleves et al., 1998). Like PilA, all are substrates for the prepilin peptidase, XcpA/PilD, which has a dual function and contributes both to type IV piliation and type II secretion (Bally et al., 1992; Nunn and Lory, 1992). XcpA cleaves a short consensus leader peptide located at the N-terminus and preceding a hydrophobic region. Because of this similarity, it was proposed that pseudopilins could form a pilus-like structure or pseudopilus (Tommassen et al., 1992; Hobbs and Mattick, 1993). Attempts to visualize an Xcp-dependent pseudopilus were for long unsuccessful, possibly because such structure may be shorter than a type IV pilus and/or only transiently assembled during the secretion process. However, when overproduced, the most abundant pseudopilin (Nunn and Lory, 1993) XcpT is readily assembled into a long pilus (Durand et al., 2003; Figure 6). The 3D structure of XcpT has been resolved and an alphabeta-loop region could be highlighted as a specific motif for identifying major pseudopilins (Alphonse et al., 2010). The long pilus structure may be called hyperpseudopilus or HPP, to emphasize the difference with a physiologically relevant T2SS pseudopilus (Campos et al., 2010, 2011). The extension of the HPP beyond the cell surface is likely to go through the secretin XcpQ. Indeed, formation of the HPP locks the channel and prevents secretion of T2SS substrates which remain inside the bacterial periplasm (Durand et al., 2003).
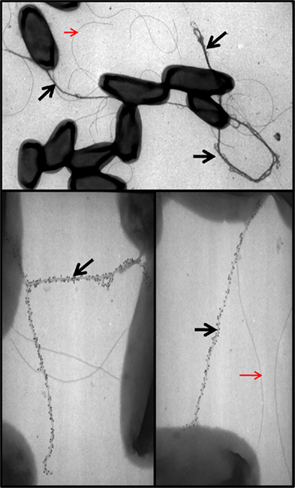
Figure 6. XcpT pseudopilin overexpression results in pseudopilus formation. Shown are the results of TEM analysis of P. aeruginosa PAO1 overexpressing xcpT from pMTWT as described in Durand et al. (2003). The pseudopili are indicated with a black arrow and are labeled with gold particles coupled to antibodies against XcpT. Red arrows indicate unlabeled flagella for comparison.
Among the five pseudopilins, only XcpT is able to form a HPP structure when overproduced, suggesting subtle functions for the other pseudopilins. The unique ability of type IV pili (T4P) to retract and to drive motility relies on the activity of at least two traffic ATPases (Chiang et al., 2008), PilB for elongation (Turner et al., 1993) and PilT for retraction (Misic et al., 2010). In the T2SS, only one ATPase is involved, XcpR (Turner et al., 1993), and thus other mechanism may control the length of the pseudopilus and prevent abnormal elongation such as in the HPP. It has been shown that the pseudopilin XcpX controls the assembly of XcpT into HPP (Durand et al., 2005). Indeed, when the number of XcpX subunits increases within the cell the length of the HPP decreases. Conversely, in the absence of XcpX the pseudopilus length is evenlonger. The impact of XcpX on XcpT polymerization could be part of the molecular mechanism underlying the dynamic control of pseudopilus elongation and retraction. Several recent studies suggest that the other three minor pilins, XcpU–W, together with XcpX, form a quaternary structure present at the tip of the pilus (Korotkov and Hol, 2008; Douzi et al., 2009). It is worth noting that the globular domain found in the XcpX bulk structure may prevent access of the elongating pseudopilus to the surface by blocking the secretin channel (Korotkov and Hol, 2008). A hypothetical model thus suggests that upon contact between the pseudopilus tip and the secretin, the pseudopilus disassembles/retracts and cycles of such event may push T2SS substrates through the secretin channel in a piston-like mechanism. Recent work indicates that in the Hxc system HxcT does not form a HPP. This characteristic could be assigned to the lack of conserved residues in HxcT that are otherwise found in XcpT-like proteins (Durand et al., 2011). This striking difference may distinguish two subtypes of T2SS.
In conclusion, T2S and T4P systems are similar nanomachines involved in distinct functions but likely constructed from a common ancestor. It is interesting to point out that the T2S/T4P dichotomy may break down in some cases where one system can have both functions, such as with the V. cholerae T4P involved in the secretion of a soluble colonization factor, TcpF (Kirn et al., 2003). A major difference is that pili should be long whereas pseudopili should be restricted in length within the space of the periplasm. “To grow or not to grow,” this is the issue between type II secretion and type IV piliation. From an evolution perspective, it is amusing to recall Lamarck’s thoughts about the length of a giraffe neck. Did the giraffe neck grow as the result of a habit to feed on the highest branch trees or long-necked giraffe were selected because they had an obvious advantage to get food and survive during dearth in places where the soil is nearly always arid and barren.
The T3SS Needle
The visualization of the needle-like structure in S. Typhimurium illustrated the idea of an injectisome (Kubori et al., 1998; Figure 2). In P. aeruginosa the component of the needle is called PscF which assembles into small needles 60–80 nm in length and 6–7 nm in width (Pastor et al., 2005). PscF multimerization process is dependent on two other Psc components, known as co-chaperones, PscE and PscG (Quinaud et al., 2005). The formation of the needle/filament is prevented when PscF interacts with PscE and PscG which stabilize the monomeric form of PscF (Ple et al., 2010). The needle is likely to protrude at the cell surface by growing through the secretin channel formed by the PscC secretin. As discussed for the pseudopilus, the length of the needle might be tightly controlled by other T3SS components. Studies in Y. enterocolitica showed that the C-terminal domain of YscP is the molecular ruler that determines the T3SS needle length (Journet et al., 2003; Mota et al., 2005). PscP has a similar function since its C-terminus could functionally replace the YscP C-terminus (Agrain et al., 2005).
The lower part of the injectisome, lying beneath the needle, structurally resembles the basal body of the flagellum (Schraidt et al., 2010). This observation suggests a common evolutionary origin between the two systems especially because flagellar assembly systems secrete other proteins than flagellin in the extracellular medium, as shown in some rare occasions (Young et al., 1999). The basal body of the T3SS or flagellar assembly system is made by the export apparatus, at the basis of which lies the ATPase, and constitutes the MS ring in the inner membrane (Chevance and Hughes, 2008). Another series of rings are found in the periplasm (P ring) and the outer membrane (L ring) that form the channel through which the flagellin or needle component may travel before assembly. A major difference between flagella and T3SS is the presence of a secretin in the T3SS instead of the L ring in the case of flagellum, but the overall structural of the T3SS nanomachine is now well described (Worrall et al., 2011).
The T6SS Tail Tube
The lack of an outer membrane channel in the T6SS suggests an alternative delivery strategy. Perhaps local puncturing of the cell envelope avoids cell lysis whilst allowing transient assembly of the secretion machine? Interestingly, several studies report similarities between T6SS proteins and components of the T4 bacteriophage (Leiman et al., 2009). Bacteriophages are bacterial viruses which inject their DNA into the bacterial cytosol (Rossmann et al., 2004). Once in the cytosol the phage DNA is replicated, the phage gene products (gp) are synthesized and the bacteria can be used as a phage factory. The injection of the DNA requires a so-called tail tube at the tip of which sits a tail spike or puncturing device which perforates the bacterial cell envelope (Leiman et al., 2010). The tube is made by polymerization of the phage protein gp19, whereas the puncturing device results from the trimeric assembly of two phage proteins, gp27 and gp5.
The discovery that components of the T6SS are structurally similar to gp19, gp5, and gp27 has been a major breakthrough and put the scientific community on a trail to understand how the T6SS machine may work. Briefly, the structure of the Hcp1 protein from the P. aeruginosa H1-T6SS was solved in 2006 (Mougous et al., 2006). Hcp1 forms a hexameric ring with an external diameter of 90 Å and a central channel of 40 Å. The Hcp1 hexameric rings can form nanotubes (Figure 4) by stacking on top of each other (Ballister et al., 2008). The dimensions of the Hcp nanotube fit with those of the T4 phage tail tube that is composed of the gp19 protein. Thus, despite a low level of amino-acid identity Hcp1 oligomers resemble bacteriophage tail tubes.
The gp5 and gp27 T4 phage components form a trimeric structure reported as the bacteriophage tail spike (Kanamaru et al., 2002). The gp27 homo-trimer forms a cylinder, and is connected to another homo-trimeric complex formed by gp5. The gp5 N-terminal domain forms an oligonucleotide/oligosaccharide-binding site (OB) involved in binding to the bacterial peptidoglycan. The central region contains a lysozyme domain. Upon oligomerization, the C-terminal domains associate into a triple-stranded β-helix. This helix is considered as the needle part of the phage tail that is responsible for puncturing the bacterial outer membrane. Once the membrane is disrupted the OB and lysozyme domains have access to the peptidoglycan, disrupt it and make way to the tail tube and phage DNA injection (Rossmann et al., 2004). Strikingly, VgrG proteins, essential T6SS components, consist of a fusion between the T4 phage proteins gp27 and gp5 (Figure 4), with the exception of the gp5 lysozyme domain that is lacking in VgrGs (Pukatzki et al., 2007; Leiman et al., 2009).
In summary, VgrGs are similar to the tail-spike puncturing device of the T4 phage and might create a channel across the bacterial cell envelope. The VgrG puncturing device is pushed forward across the bacterial cell envelope by a growing nanotube formed by the Hcp component (Figure 4). In other words, it could be that the T6SS is a translocation machine operating from the inside to the outside of the bacterial cell and mirrors the phage translocation machine which operates from the outside to the inside of the bacterial cell.
Before the T6SS mechanism can be understood, several questions remain to be answered. Firstly, what are the proteins secreted by the T6SS? Initially VgrG and Hcp were considered as the secreted proteins (Mougous et al., 2006; Pukatzki et al., 2006). However, in light of their homology with the T4 phage tail, it is now believed that these components are reaching the cell surface as part of the secretion machine but are not genuine T6SS effectors. In some cases, the VgrG proteins have a C-terminal extension, such as the actin cross-linking domain of the V. cholerae VgrG1(Pukatzki et al., 2007; Ma et al., 2009), or VIP-2 domain of the Aeromonas hydrophila VgrG1(Suarez et al., 2010). In this case the effector domain is transported through the cell envelope together with VgrG1 that acts both as a puncturing device and as a carrier for a secreted passenger protein. In P. aeruginosa, out of the 10 VgrG that could be identified from the PAO1 genome (Hachani et al., 2011), only the VgrG2b protein seem to have a C-terminal extension with similarity to metalloproteases (Pukatzki et al., 2007). In other cases, genuine secreted proteins have been identified, as the P. aeruginosa Tse2 toxin (Hood et al., 2010). One may suggest that Tse2 binds a VgrG protein in a non-covalent manner and is co-transported (Hachani et al., 2011). Alternatively, Tse2 could use the Hcp tube to travel across the cell envelope.
The second important question is: what is the function of the other T6SS components and how much further could the similarity with the phage structure be extended? The answer can be included in the question since it is likely that T6SS components are structurally similar to other proteins from the T4 phage. Indeed, the T4 phage rigid tube (gp19 or Hcp) is surrounded by a contractile sheath and terminated by a multiprotein complex called the baseplate (Kostyuchenko et al., 2003; Yap et al., 2010). The baseplate contains a structure called a wedge (composed of at least seven proteins: gp11, gp10, gp7, gp8, gp6, gp53, and gp25) that is joined around a cylindrical structure that is called the “hub,” which includes the puncturing device, gp5/gp27. Importantly, a gp25-like protein has been identified among the T6SS components and may open the list of a long series of similarity between T6SS and gp proteins (Leiman et al., 2009).
Chaperone–Usher Pathway: The Lost Kid
Secretion in Gram-negative bacteria involves translocation of proteins across the cell envelope and ultimately across the outer membrane that results in proteins being released in the extracellular medium. Somehow it excluded proteins that remain anchored in the outer membrane. Consequently, assembly of pili was not initially considered as a genuine secretion process. However, in light of what is observed with autotransporters that for most of them remains attached to the membrane, the question may deserve to be readdressed. Coining the autotransporter as a secretion system was adequate considering the IgA protease (Pohlner et al., 1987) since autoproteolytic cleavage released the protease in the medium. These questions are now debatable and reviews readdressing this issue are flourishing (Desvaux et al., 2006, 2009).
In this respect, the chaperone–usher pathway has been for years forgotten in secretion stories, though it is likely the pathway which is best characterized in terms of molecular mechanisms and structural details (Waksman and Hultgren, 2009). The system involves an outer membrane protein, called usher, which oligomerizes and forms a channel. The fimbrial subunits are targeted to the usher by periplasmic chaperones. These chaperones bind the fimbrial subunits emerging from the Sec system and prevent their misfolding in the periplasm. Finally, the fimbrial subunits reach the extracellular medium but they remain bound together to form the filament while the filament remains cell-anchored. Nevertheless, considering the usher as a secretion channel for fimbrial subunits makes sense in light of the description we made of the CupB3 P-usher (Ruer et al., 2008).
In P. aeruginosa the pili best described are the type IV pili. However, since their discovery in 2001(Vallet et al., 2001) the importance of chaperone–usher pathways (Cup) in surface attachment and biofilm formation has continuously increased. Several P. aeruginosa cup gene clusters have been functionally studied. On the PAO1 genome, four clusters, cupA, cupB, cupC, and cupE are identified (Vallet et al., 2001; Giraud et al., 2011). In all cases the cluster includes a gene encoding an usher and one or two genes encoding a chaperone. The cupA-C gene clusters encode typical fimbrial subunits whereas the cupE gene cluster encodes fimbrial subunits with different characteristics and domains which were first identified in the fimbrial subunits of the chaperone–usher system in Acinetobacter baumanii (Tomaras et al., 2003). Both CupB6 and CupE6, display the characteristic two-domain organization of fimbrial adhesins, and CupA4 might also be an adhesin. No gene encoding a putative fimbrial adhesin has been associated with the cupC gene cluster. From one P. aeruginosa isolate to another the composition in cup genes may vary. Remarkably, in the PA14 strain there is an additional cup gene cluster, cupD (Mikkelsen et al., 2009; Nicastro et al., 2009), which is located on the pathogenicity island PAPI-1(He et al., 2004) and which is very similar in composition and structure to the cupA gene cluster. Of note the exoU gene is also located on PAPI-1(Kulasekara et al., 2006).
Concluding Remarks
The diversity of secretion systems is fascinating and likely results from an evolutionary process which has optimized various biological functions into nanomachines whose role is to give access to the extracellular medium for a large variety of proteins.
The diversity may exist within one single organism such as P. aeruginosa that possesses all the secretion systems known to date but the T4SS (Figure 7). The lack of T4SS in P. aeruginosa may be interpreted by the fact that a similar function is exerted by the T3SS, i.e., injecting proteins in eukaryotic host cells.
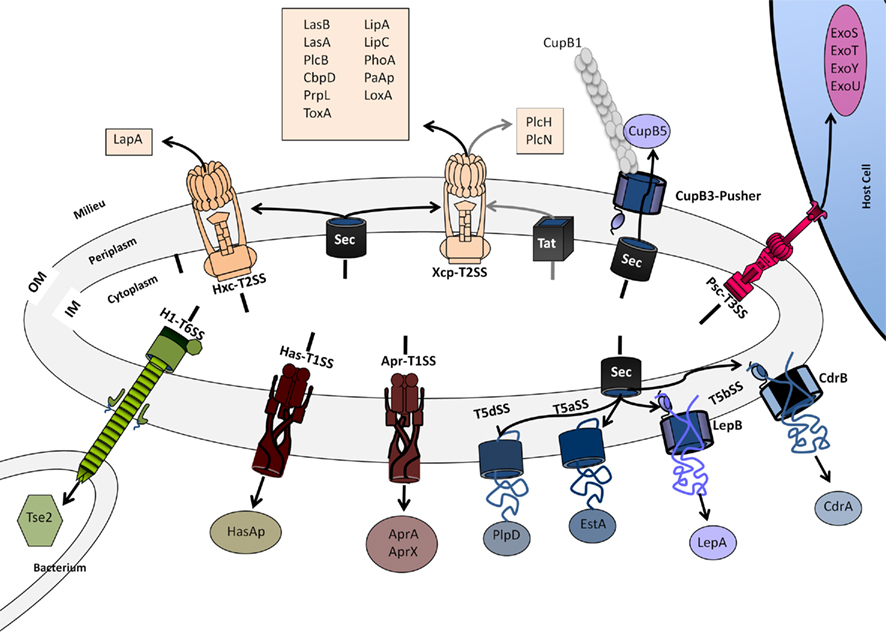
Figure 7. Secretion systems in P. aeruginosa. The systems shown are discussed in the main text. For characteristics of exoproteins see also Table 1. The design of the secretion systems is as previously presented in Bleves et al. (2010). Color code is: brown (T1SS), light Orange (T2SS), red (T3SS), blue/purple (T5SS), green (T6SS).
The specificity of a single system may not be a luxury but a fine-tuned secretion mechanism adapted to the characteristics and features of a subset of exoproteins. Some systems may have evolved after duplication while others have been acquired via horizontal gene transfer. In some cases the determination of the architecture reached exquisite details such as with the T3SS. However, in other cases the assembly and overall architecture is now emerging, like with the T2SS. The visualization of these impressive macromolecular complexes always raises the question on how the cell envelope, and particularly the peptidoglycan, are reorganized to make them fit. One obvious answer is the use of specific transglycosylases, or the assembly of the nanomachine at the pole (Senf et al., 2008), region where the peptidoglycan mesh is loose.
In conclusion, though much need to be discovered to understand secretion systems, basic rules are well established and should allow a better understanding of every individual system at the molecular level.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Alain Filloux is supported by the Royal Society, Wellcome Trust grant WT091939, and MRC grant G0800171/ID86344.
References
Agrain, C., Callebaut, I., Journet, L., Sorg, I., Paroz, C., Mota, L. J., and Cornelis, G. R. (2005). Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56, 54–67.
Akoh, C. C., Lee, G. C., Liaw, Y. C., Huang, T. H., and Shaw, J. F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552.
Akrim, M., Bally, M., Ball, G., Tommassen, J., Teerink, H., Filloux, A., and Lazdunski, A. (1993). Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol. Microbiol. 10, 431–443.
Alphonse, S., Durand, E., Douzi, B., Waegele, B., Darbon, H., Filloux, A., Voulhoux, R., and Bernard, C. (2010). Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J. Struct. Biol. 169, 75–80.
Anderson, D. M., and Schneewind, O. (1997). A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278, 1140–1143.
Arnoux, P., Haser, R., Izadi, N., Lecroisey, A., Delepierre, M., Wandersman, C., and Czjzek, M. (1999). The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 6, 516–520.
Arts, J., van Boxtel, R., Filloux, A., Tommassen, J., and Koster, M. (2007). Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076.
Aschtgen, M. S., Bernard, C. S., De Bentzmann, S., Lloubes, R., and Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531.
Backert, S., Ziska, E., Brinkmann, V., Zimny-Arndt, U., Fauconnier, A., Jungblut, P. R., Naumann, M., and Meyer, T. F. (2000). Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2, 155–164.
Bageshwar, U. K., and Musser, S. M. (2007). Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 179, 87–99.
Ball, G., Chapon-Herve, V., Bleves, S., Michel, G., and Bally, M. (1999). Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181, 382–388.
Ball, G., Durand, E., Lazdunski, A., and Filloux, A. (2002). A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43, 475–485.
Ballister, E. R., Lai, A. H., Zuckermann, R. N., Cheng, Y., and Mougous, J. D. (2008). In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738.
Bally, M., Ball, G., Badere, A., and Lazdunski, A. (1991). Protein secretion in Pseudomonas aeruginosa: the xcpA gene encodes an integral inner membrane protein homologous to Klebsiella pneumoniae secretion function protein PulO. J. Bacteriol. 173, 479–486.
Bally, M., Filloux, A., Akrim, M., Ball, G., Lazdunski, A., and Tommassen, J. (1992). Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6, 1121–1131.
Banerji, S., and Flieger, A. (2004). Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150, 522–525.
Barker, A. P., Vasil, A. I., Filloux, A., Ball, G., Wilderman, P. J., and Vasil, M. L. (2004). A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol. Microbiol. 53, 1089–1098.
Baumann, U., Wu, S., Flaherty, K. M., and McKay, D. B. (1993). Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 12, 3357–3364.
Benabdelhak, H., Kiontke, S., Horn, C., Ernst, R., Blight, M. A., Holland, I. B., and Schmitt, L. (2003). A specific interaction between the NBD of the ABC-transporter HlyB and a C-terminal fragment of its transport substrate haemolysin A. J. Mol. Biol. 327, 1169–1179.
Berthelot, P., Attree, I., Plesiat, P., Chabert, J., de Bentzmann, S., Pozzetto, B., and Grattard, F. (2003). Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exo genes. J. Infect. Dis. 188, 512–518.
Bever, R. A., and Iglewski, B. H. (1988). Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170, 4309–4314.
Bitter, W., Koster, M., Latijnhouwers, M., de Cock, H., and Tommassen, J. (1998). Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27, 209–219.
Bleves, S., Gerard-Vincent, M., Lazdunski, A., and Filloux, A. (1999). Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181, 4012–4019.
Bleves, S., Lazdunski, A., and Filloux, A. (1996). Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J. Bacteriol. 178, 4297–4300.
Bleves, S., Viarre, V., Salacha, R., Michel, G. P., Filloux, A., and Voulhoux, R. (2010). Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543.
Bleves, S., Voulhoux, R., Michel, G., Lazdunski, A., Tommassen, J., and Filloux, A. (1998). The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol. 27, 31–40.
Blocker, A., Komoriya, K., and Aizawa, S. (2003). Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U.S.A. 100, 3027–3030.
Blocker, A. J., Deane, J. E., Veenendaal, A. K., Roversi, P., Hodgkinson, J. L., Johnson, S., and Lea, S. M. (2008). What’s the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 105, 6507–6513.
Bonemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H., and Mogk, A. (2009). Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325.
Bonemann, G., Pietrosiuk, A., and Mogk, A. (2010). Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76, 815–821.
Borlee, B. R., Goldman, A. D., Murakami, K., Samudrala, R., Wozniak, D. J., and Parsek, M. R. (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842.
Braun, P., de Groot, A., Bitter, W., and Tommassen, J. (1998). Secretion of elastinolytic enzymes and their propeptides by Pseudomonas aeruginosa. J. Bacteriol. 180, 3467–3469.
Braun, P., Tommassen, J., and Filloux, A. (1996). Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol. Microbiol. 19, 297–306.
Brok, R., Van Gelder, P., Winterhalter, M., Ziese, U., Koster, A. J., de Cock, H., Koster, M., Tommassen, J., and Bitter, W. (1999). The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294, 1169–1179.
Broms, J. E., Forslund, A. L., Forsberg, A., and Francis, M. S. (2003). PcrH of Pseudomonas aeruginosa is essential for secretion and assembly of the type III translocon. J. Infect. Dis. 188, 1909–1921.
Broz, P., Mueller, C. A., Muller, S. A., Philippsen, A., Sorg, I., Engel, A., and Cornelis, G. R. (2007). Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65, 1311–1320.
Brutinel, E. D., Vakulskas, C. A., Brady, K. M., and Yahr, T. L. (2008). Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 68, 657–671.
Campos, M., Francetic, O., and Nilges, M. (2011). Modeling pilus structures from sparse data. J. Struct. Biol. 173, 436–444.
Campos, M., Nilges, M., Cisneros, D. A., and Francetic, O. (2010). Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc. Natl. Acad. Sci. U.S.A. 107, 13081–13086.
Cascales, E., and Christie, P. J. (2003). The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149.
Chami, M., Guilvout, I., Gregorini, M., Remigy, H. W., Muller, S. A., Valerio, M., Engel, A., Pugsley, A. P., and Bayan, N. (2005). Structural insights into the secretin PulD and its trypsin-resistant core. J. Biol. Chem. 280, 37732–37741.
Chandran, V., Fronzes, R., Duquerroy, S., Cronin, N., Navaza, J., and Waksman, G. (2009). Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015.
Chevance, F. F., and Hughes, K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465.
Chiang, P., Sampaleanu, L. M., Ayers, M., Pahuta, M., Howell, P. L., and Burrows, L. L. (2008). Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126.
Choo, K. H., Tan, T. W., and Ranganathan, S. (2009). A comprehensive assessment of N-terminal signal peptides prediction methods. BMC Bioinformatics 10(Suppl. 15), S2. doi: 10.1186/1471-2105-10-S15-S2
Christie, P. J., Ward, J. E. Jr., Gordon, M. P., and Nester, E. W. (1989). A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc. Natl. Acad. Sci. U.S.A. 86, 9677–9681.
Chung, C. W., You, J., Kim, K., Moon, Y., Kim, H., and Ahn, J. H. (2009). Export of recombinant proteins in Escherichia coli using ABC transporter with an attached lipase ABC transporter recognition domain (LARD). Microb. Cell Fact. 8, 11.
Clantin, B., Delattre, A. S., Rucktooa, P., Saint, N., Meli, A. C., Locht, C., Jacob-Dubuisson, F., and Villeret, V. (2007). Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961.
Collins, R. F., Ford, R. C., Kitmitto, A., Olsen, R. O., Tonjum, T., and Derrick, J. P. (2003). Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185, 2611–2617.
Cornelis, G. R. (2010). The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol. Chem. 391, 745–751.
Cossart, P., and Sansonetti, P. J. (2004). Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248.
Cowan, S. W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R. A., Jansonius, J. N., and Rosenbusch, J. P. (1992). Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733.
Dacheux, D., Goure, J., Chabert, J., Usson, Y., and Attree, I. (2001). Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40, 76–85.
Daefler, S., Guilvout, I., Hardie, K. R., Pugsley, A. P., and Russel, M. (1997). The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol. Microbiol. 24, 465–475.
Das, S., and Chaudhuri, K. (2003). Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In silico Biol. 3, 287–300.
Dautin, N., and Bernstein, H. D. (2007). Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 61, 89–112.
de Groot, A., Filloux, A., and Tommassen, J. (1991). Conservation of xcp genes, involved in the two-step protein secretion process, in different Pseudomonas species and other gram-negative bacteria. Mol. Gen. Genet. 229, 278–284.
de Groot, A., Koster, M., Gerard-Vincent, M., Gerritse, G., Lazdunski, A., Tommassen, J., and Filloux, A. (2001). Exchange of Xcp (Gsp) secretion machineries between Pseudomonas aeruginosa and Pseudomonas alcaligenes: species specificity unrelated to substrate recognition. J. Bacteriol. 183, 959–967.
de Groot, A., Krijger, J. J., Filloux, A., and Tommassen, J. (1996). Characterization of type II protein secretion (xcp) genes in the plant growth-stimulating Pseudomonas putida, strain WCS358. Mol. Gen. Genet. 250, 491–504.
d’Enfert, C., Ryter, A., and Pugsley, A. P. (1987). Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6, 3531–3538.
Deng, Q., and Barbieri, J. T. (2008). Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu. Rev. Microbiol. 62, 271–288.
Desvaux, M., Hebraud, M., Henderson, I. R., and Pallen, M. J. (2006). Type III secretion: what’s in a name? Trends Microbiol. 14, 157–160.
Desvaux, M., Hebraud, M., Talon, R., and Henderson, I. R. (2009). Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17, 139–145.
Dong, C., Beis, K., Nesper, J., Brunkan-Lamontagne, A. L., Clarke, B. R., Whitfield, C., and Naismith, J. H. (2006). Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229.
Douzi, B., Durand, E., Bernard, C., Alphonse, S., Cambillau, C., Filloux, A., Tegoni, M., and Voulhoux, R. (2009). The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J. Biol. Chem. 284, 34580–34589.
Duong, F., Bonnet, E., Geli, V., Lazdunski, A., Murgier, M., and Filloux, A. (2001). The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene 262, 147–153.
Duong, F., Lazdunski, A., Cami, B., and Murgier, M. (1992). Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene 121, 47–54.
Duong, F., Lazdunski, A., and Murgier, M. (1996). Protein secretion by heterologous bacterial ABC-transporters: the C-terminus secretion signal of the secreted protein confers high recognition specificity. Mol. Microbiol. 21, 459–470.
Durand, E., Alphonse, S., Brochier-Armanet, C., Ball, G., Douzi, B., Filloux, A., Bernard, C., and Voulhoux, R. (2011). The assembly mode of the pseudopilus: a hallmark to distinguish a novel secretion system subtype. J. Biol. Chem. 286, 24407–24416.
Durand, E., Bernadac, A., Ball, G., Lazdunski, A., Sturgis, J. N., and Filloux, A. (2003). Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758.
Durand, E., Michel, G., Voulhoux, R., Kurner, J., Bernadac, A., and Filloux, A. (2005). XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J. Biol. Chem. 280, 31378–31389.
Durand, E., Verger, D., Rego, A. T., Chandran, V., Meng, G., Fronzes, R., and Waksman, G. (2009). Structural biology of bacterial secretion systems in gram-negative pathogens – potential for new drug targets. Infect. Disord. Drug Targets 9, 518–547.
Feldman, M. F., Muller, S., Wuest, E., and Cornelis, G. R. (2002). SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol. Microbiol. 46, 1183–1197.
Filloux, A. (2004). The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694, 163–179.
Filloux, A. (2010). Secretion signal and protein targeting in bacteria: a biological puzzle. J. Bacteriol. 192, 3847–3849.
Filloux, A., Bally, M., Ball, G., Akrim, M., Tommassen, J., and Lazdunski, A. (1990). Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 9, 4323–4329.
Filloux, A., Bally, M., Murgier, M., Wretlind, B., and Lazdunski, A. (1989). Cloning of Xcp-genes located at the 55-min region of the chromosome and involved in protein secretion in Pseudomonas aeruginosa. Mol. Microbiol. 3, 261–265.
Filloux, A., Bally, M., Soscia, C., Murgier, M., and Lazdunski, A. (1988). Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol. Gen. Genet. 212, 510–513.
Filloux, A., Hachani, A., and Bleves, S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583.
Filloux, A., and Hardie, K. R. (1998). A systematic approach to the study of protein secretion in Gram-negative bacteria. Methods Microbiol. 27, 301–318.
Filloux, A., Murgier, M., Wretlind, B., and Lazdunski, A. (1987). Characterization of 2 Pseudomonas aeruginosa mutants with defective secretion of extracellular proteins and comparison with other mutants. FEMS Microbiol. Lett. 40, 159–163.
Finck-Barbancon, V., Goranson, J., Zhu, L., Sawa, T., Wiener-Kronish, J. P., Fleiszig, S. M., Wu, C., Mende-Mueller, L., and Frank, D. W. (1997). ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25, 547–557.
Finck-Barbancon, V., Yahr, T. L., and Frank, D. W. (1998). Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J. Bacteriol. 180, 6224–6231.
Fleiszig, S. M., Wiener-Kronish, J. P., Miyazaki, H., Vallas, V., Mostov, K. E., Kanada, D., Sawa, T., Yen, T. S., and Frank, D. W. (1997). Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 65, 579–586.
Folders, J., Algra, J., Roelofs, M. S., van Loon, L. C., Tommassen, J., and Bitter, W. (2001). Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183, 7044–7052.
Folders, J., Tommassen, J., van Loon, L. C., and Bitter, W. (2000). Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182, 1257–1263.
Fox, A., Haas, D., Reimmann, C., Heeb, S., Filloux, A., and Voulhoux, R. (2008). Emergence of secretion-defective sublines of Pseudomonas aeruginosa PAO1 resulting from spontaneous mutations in the vfr global regulatory gene. Appl. Environ. Microbiol. 74, 1902–1908.
Francetic, O., and Pugsley, A. P. (2005). Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J. Bacteriol. 187, 7045–7055.
Frank, D. W. (1997). The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26, 621–629.
Gerard-Vincent, M., Robert, V., Ball, G., Bleves, S., Michel, G. P., Lazdunski, A., and Filloux, A. (2002). Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 44, 1651–1665.
Ghigo, J. M., Letoffe, S., and Wandersman, C. (1997). A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179, 3572–3579.
Giraud, C., Bernard, C. S., Calderon, V., Yang, L., Filloux, A., Molin, S., Fichant, G., Bordi, C., and de Bentzmann, S. (2011). The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ. Microbiol. 13, 666–683.
Goodman, A. L., Merighi, M., Hyodo, M., Ventre, I., Filloux, A., and Lory, S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259.
Goure, J., Broz, P., Attree, O., Cornelis, G. R., and Attree, I. (2005). Protective anti-V antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J. Infect. Dis. 192, 218–225.
Goure, J., Pastor, A., Faudry, E., Chabert, J., Dessen, A., and Attree, I. (2004). The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72, 4741–4750.
Guzzo, J., Murgier, M., Filloux, A., and Lazdunski, A. (1990). Cloning of the Pseudomonas aeruginosa alkaline protease gene and secretion of the protease into the medium by Escherichia coli. J. Bacteriol. 172, 942–948.
Guzzo, J., Pages, J. M., Duong, F., Lazdunski, A., and Murgier, M. (1991a). Pseudomonas aeruginosa alkaline protease: evidence for secretion genes and study of secretion mechanism. J. Bacteriol. 173, 5290–5297.
Guzzo, J., Duong, F., Wandersman, C., Murgier, M., and Lazdunski, A. (1991b). The secretion genes of Pseudomonas aeruginosa alkaline protease are functionally related to those of Erwinia chrysanthemi proteases and Escherichia coli alpha-haemolysin. Mol. Microbiol. 5, 447–453.
Hachani, A., Lossi, N. S., Hamilton, A., Jones, C., Bleves, S., Albesa-Jove, D., and Filloux, A. (2011). Type VI secretion system in Pseudomonas Aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327.
Hardie, K. R., Lory, S., and Pugsley, A. P. (1996). Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15, 978–988.
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665.
Hauser, A. R., Cobb, E., Bodi, M., Mariscal, D., Valles, J., Engel, J. N., and Rello, J. (2002). Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30, 521–528.
He, J., Baldini, R. L., Deziel, E., Saucier, M., Zhang, Q., Liberati, N. T., Lee, D., Urbach, J., Goodman, H. M., and Rahme, L. G. (2004). The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U.S.A. 101, 2530–2535.
Heikkila, M. P., Honisch, U., Wunsch, P., and Zumft, W. G. (2001). Role of the Tat ransport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183, 1663–1671.
Higgins, C. F. (2001). ABC transporters: physiology, structure and mechanism – an overview. Res. Microbiol. 152, 205–210.
Higgins, M. K., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2004). Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. U.S.A. 101, 9994–9999.
Hinsa, S. M., Espinosa-Urgel, M., Ramos, J. L., and O’Toole, G. A. (2003). Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49, 905–918.
Hobbs, M., and Mattick, J. S. (1993). Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243.
Hodak, H., Clantin, B., Willery, E., Villeret, V., Locht, C., and Jacob-Dubuisson, F. (2006). Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol. Microbiol. 61, 368–382.
Hodgkinson, J. L., Horsley, A., Stabat, D., Simon, M., Johnson, S., da Fonseca, P. C., Morris, E. P., Wall, J. S., Lea, S. M., and Blocker, A. J. (2009). Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16, 477–485.
Holland, I. B., Schmitt, L., and Young, J. (2005). Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review). Mol. Membr. Biol. 22, 29–39.
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., Silverman, J. M., Ohlson, B. B., Hicks, K. G., Plemel, R. L., Li, M., Schwarz, S., Wang, W. Y., Merz, A. J., Goodlett, D. R., and Mougous, J. D. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37.
Jacob-Dubuisson, F., Locht, C., and Antoine, R. (2001). Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40, 306–313.
Jaeger, K. E., Ransac, S., Dijkstra, B. W., Colson, C., van Heuvel, M., and Misset, O. (1994). Bacterial lipases. FEMS Microbiol. Rev. 15, 29–63.
Jepkorir, G., Rodriguez, J. C., Rui, H., Im, W., Lovell, S., Battaile, K. P., Alontaga, A. Y., Yukl, E. T., Moënne-Loccoz, P., and Rivera, M. (2010). Structural, NMR spectroscopic, and computational investigation of hemin loading in the hemophore HasAp from Pseudomonas aeruginosa. J. Am. Chem. Soc. 132, 9857–9872.
Journet, L., Agrain, C., Broz, P., and Cornelis, G. R. (2003). The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760.
Junker, M., Besingi, R. N., and Clark, P. L. (2009). Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol. Microbiol. 71, 1323–1332.
Kajava, A. V., and Steven, A. C. (2006). The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of Type V secretory proteins. J. Struct. Biol. 155, 306–315.
Kanamaru, S., Leiman, P. G., Kostyuchenko, V. A., Chipman, P. R., Mesyanzhinov, V. V., Arisaka, F., and Rossmann, M. G. (2002). Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557.
Kelley, L. A., and Sternberg, M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371.
Kessler, E., and Safrin, M. (1994). The propeptide of Pseudomonas aeruginosa elastase acts an elastase inhibitor. J. Biol. Chem. 269, 22726–22731.
Kida, Y., Higashimoto, Y., Inoue, H., Shimizu, T., and Kuwano, K. (2008). A novel secreted protease from Pseudomonas aeruginosa activates NF-kappaB through protease-activated receptors. Cell. Microbiol. 10, 1491–1504.
Kirn, T. J., Bose, N., and Taylor, R. K. (2003). Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49, 81–92.
Koronakis, V., Koronakis, E., and Hughes, C. (1989). Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 8, 595–605.
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919.
Korotkov, K. V., Gonen, T., and Hol, W. G. (2011). Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. PMID: 21565514. [Epub ahead of print].
Korotkov, K. V., and Hol, W. G. (2008). Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 15, 462–468.
Korotkov, K. V., Pardon, E., Steyaert, J., and Hol, W. G. (2009). Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265.
Koster, M., Bitter, W., de Cock, H., Allaoui, A., Cornelis, G. R., and Tommassen, J. (1997). The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26, 789–797.
Kostyuchenko, V. A., Leiman, P. G., Chipman, P. R., Kanamaru, S., van Raaij, M. J., Arisaka, F., Mesyanzhinov, V. V., and Rossmann, M. G. (2003). Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 10, 688–693.
Kowalska, K., Soscia, C., Combe, H., Vasseur, P., Voulhoux, R., and Filloux, A. (2010). The C-terminal amphipathic alpha-helix of Pseudomonas aeruginosa PelC outer membrane protein is required for its function. Biochimie 92, 33–40.
Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., Galán, J. E., and Aizawa, S. I. (1998). Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605.
Kulasekara, B. R., Kulasekara, H. D., Wolfgang, M. C., Stevens, L., Frank, D. W., and Lory, S. (2006). Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188, 4037–4050.
Lambert, O., Benabdelhak, H., Chami, M., Jouan, L., Nouaille, E., Ducruix, A., and Brisson, A. (2005). Trimeric structure of OprN and OprM efflux proteins from Pseudomonas aeruginosa, by 2D electron crystallography. J. Struct. Biol. 150, 50–57.
Lee, V. T., Pukatzki, S., Sato, H., Kikawada, E., Kazimirova, A. A., Huang, J., Li, X., Arm, J. P., Frank, D. W., and Lory, S. (2007). Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 75, 1089–1098.
Leiman, P. G., Arisaka, F., van Raaij, M. J., Kostyuchenko, V. A., Aksyuk, A. A., Kanamaru, S., and Rossmann, M. G. (2010). Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355.
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., Burley, S. K., Almo, S. C., and Mekalanos, J. J. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159.
Letoffe, S., Delepelaire, P., and Wandersman, C. (1996). Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15, 5804–5811.
Letoffe, S., Nato, F., Goldberg, M. E., and Wandersman, C. (1999). Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol. Microbiol. 33, 546–555.
Letoffe, S., Redeker, V., and Wandersman, C. (1998). Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol. 28, 1223–1234.
Lin, H. T., Bavro, V. N., Barrera, N. P., Frankish, H. M., Velamakanni, S., van Veen, H. W., Robinson, C. V., Borges-Walmsley, M. I., and Walmsley, A. R. (2009). MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J. Biol. Chem. 284, 1145–1154.
Lobedanz, S., Bokma, E., Symmons, M. F., Koronakis, E., Hughes, C., and Koronakis, V. (2007). A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. U.S.A. 104, 4612–4617.
Loo, T. W., Bartlett, M. C., and Clarke, D. M. (2002). The “LSGGQ” motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J. Biol. Chem. 277, 41303–41306.
Lu, H. M., Mizushima, S., and Lory, S. (1993). A periplasmic intermediate in the extracellular secretion pathway of Pseudomonas aeruginosa exotoxin A. J. Bacteriol. 175, 7463–7467.
Ma, A. T., McAuley, S., Pukatzki, S., and Mekalanos, J. J. (2009). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243.
Mackman, N., Nicaud, J. M., Gray, L., and Holland, I. B. (1985). Identification of polypeptides required for the export of haemolysin 2001 from E. coli. Mol. Gen. Genet. 201, 529–536.
Martinez, A., Ostrovsky, P., and Nunn, D. N. (1999). LipC, a second lipase of Pseudomonas aeruginosa, is LipB and Xcp dependent and is transcriptionally regulated by pilus biogenesis components. Mol. Microbiol. 34, 317–326.
Masi, M., and Wandersman, C. (2010). Multiple signals direct the assembly and function of a type 1 secretion system. J. Bacteriol. 192, 3861–3869.
Mazar, J., and Cotter, P. A. (2007). New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 15, 508–515.
McIver, K. S., Kessler, E., and Ohman, D. E. (2004). Identification of residues in the Pseudomonas aeruginosa elastase propeptide required for chaperone and secretion activities. Microbiology 150, 3969–3977.
McIver, K. S., Kessler, E., Olson, J. C., and Ohman, D. E. (1995). The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol. 18, 877–889.
Meng, G., Surana, N. K., St Geme, J. W. III, and Waksman, G. (2006). Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 25, 2297–2304.
Merrell, D. S., and Falkow, S. (2004). Frontal and stealth attack strategies in microbial pathogenesis. Nature 430, 250–256.
Michel, G., Bleves, S., Ball, G., Lazdunski, A., and Filloux, A. (1998). Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology 144(Pt 12), 3379–3386.
Michel, G. P., Durand, E., and Filloux, A. (2007). XphA/XqhA, a novel GspCD subunit for type II secretion in Pseudomonas aeruginosa. J. Bacteriol. 189, 3776–3783.
Michel, L. O., Sandkvist, M., and Bagdasarian, M. (1995). Specificity of the protein secretory apparatus: secretion of the heat-labile enterotoxin B subunit pentamers by different species of gram- bacteria. Gene 152, 41–45.
Michiels, T., Wattiau, P., Brasseur, R., Ruysschaert, J. M., and Cornelis, G. (1990). Secretion of Yop proteins by Yersiniae. Infect. Immun. 58, 2840–2849.
Mikkelsen, H., Ball, G., Giraud, C., and Filloux, A. (2009). Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS ONE 4, e6018. doi: 10.1371/journal.pone.0006018
Mikolosko, J., Bobyk, K., Zgurskaya, H. I., and Ghosh, P. (2006). Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14, 577–587.
Misic, A. M., Satyshur, K. A., and Forest, K. T. (2010). P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 400, 1011–1021.
Mota, L. J., Journet, L., Sorg, I., Agrain, C., and Cornelis, G. R. (2005). Bacterial injectisomes: needle length does matter. Science 307, 1278.
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., Goodman, A. L., Joachimiak, G., Ordoñez, C. L., Lory, S., Walz, T., Joachimiak, A., and Mekalanos, J. J. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530.
Nagai, H., Cambronne, E. D., Kagan, J. C., Amor, J. C., Kahn, R. A., and Roy, C. R. (2005). A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U.S.A. 102, 826–831.
Nicastro, G. G., Boechat, A. L., Abe, C. M., Kaihami, G. H., and Baldini, R. L. (2009). Pseudomonas aeruginosa PA14 cupD transcription is activated by the RcsB response regulator, but repressed by its putative cognate sensor RcsC. FEMS Microbiol. Lett. 301, 115–123.
Nunn, D. N., and Lory, S. (1991). Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285.
Nunn, D. N., and Lory, S. (1992). Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc. Natl. Acad. Sci. U.S.A. 89, 47–51.
Nunn, D. N., and Lory, S. (1993). Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175, 4375–4382.
Oomen, C. J., van Ulsen, P., van Gelder, P., Feijen, M., Tommassen, J., and Gros, P. (2004). Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266.
Ostroff, R. M., and Vasil, M. L. (1987). Identification of a new phospholipase C activity by analysis of an insertional mutation in the hemolytic phospholipase C structural gene of Pseudomonas aeruginosa. J. Bacteriol. 169, 4597–4601.
Palmer, T., and Berks, B. C. (2003). Moving folded proteins across the bacterial cell membrane. Microbiology 149, 547–556.
Palmer, T., Berks, B. C., and Sargent, F. (2010). Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli. Methods Mol. Biol. 619, 191–216.
Papanikou, E., Karamanou, S., and Economou, A. (2007). Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5, 839–851.
Parsot, C., Hamiaux, C., and Page, A. L. (2003). The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14.
Pastor, A., Chabert, J., Louwagie, M., Garin, J., and Attree, I. (2005). PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 253, 95–101.
Patel, S., and Latterich, M. (1998). The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8, 65–71.
Pei, X. Y., Hinchliffe, P., Symmons, M. F., Koronakis, E., Benz, R., Hughes, C., and Koronakis, V. (2011). Structures of sequential open states in a symmetrical opening transition of the TolC exit duct. Proc. Natl. Acad. Sci. U.S.A. 108, 2112–2117.
Phan, G., Benabdelhak, H., Lascombe, M. B., Benas, P., Rety, S., Picard, M., Ducruix, A., Etchebest, C., and Broutin, I. (2010). Structural and dynamical insights into the opening mechanism of P. aeruginosa OprM channel. Structure 18, 507–517.
Planet, P. J., Kachlany, S. C., DeSalle, R., and Figurski, D. H. (2001). Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508.
Ple, S., Job, V., Dessen, A., and Attree, I. (2010). Cochaperone interactions in export of the type III needle component PscF of Pseudomonas aeruginosa. J. Bacteriol. 192, 3801–3808.
Pohlner, J., Halter, R., Beyreuther, K., and Meyer, T. F. (1987). Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325, 458–462.
Possot, O., and Pugsley, A. P. (1994). Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol. 12, 287–299.
Pozidis, C., Chalkiadaki, A., Gomez-Serrano, A., Stahlberg, H., Brown, I., Tampakaki, A. P., Lustig, A., Sianidis, G., Politou, A. S., Engel, A., Panopoulos, N. J., Mansfield, J., Pugsley, A. P., Karamanou, S., and Economou, A. (2003). Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J. Biol. Chem. 278, 25816–25824.
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513.
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., Heidelberg, J. F., and Mekalanos, J. J. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533.
Qiu, X., Gurkar, A. U., and Lory, S. (2006). Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19830–19835.
Quinaud, M., Chabert, J., Faudry, E., Neumann, E., Lemaire, D., Pastor, A., Elsen, S., Dessen, A., and Attree, I. (2005). The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280, 36293–36300.
Reichow, S. L., Korotkov, K. V., Hol, W. G., and Gonen, T. (2010). Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232.
Robert, V., Filloux, A., and Michel, G. P. (2005a). Subcomplexes from the Xcp secretion system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 252, 43–50.
Robert, V., Filloux, A., and Michel, G. P. (2005b). Role of XcpP in the functionality of the Pseudomonas aeruginosa secreton. Res. Microbiol. 156, 880–886.
Robien, M. A., Krumm, B. E., Sandkvist, M., and Hol, W. G. (2003). Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333, 657–674.
Rossmann, M. G., Mesyanzhinov, V. V., Arisaka, F., and Leiman, P. G. (2004). The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 14, 171–180.
Roy, P. H., Tetu, S. G., Larouche, A., Elbourne, L., Tremblay, S., Ren, Q., Dodson, R., Harkins, D., Shay, R., Watkins, K., Mahamoud, Y., and Paulsen, I. T. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 5, e8842. doi: 10.1371/journal.pone.0008842
Ruer, S., Ball, G., Filloux, A., and de Bentzmann, S. (2008). The “P-usher”, a novel protein transporter involved in fimbrial assembly and TpsA secretion. EMBO J. 27, 2669–2680.
Sagulenko, E., Sagulenko, V., Chen, J., and Christie, P. J. (2001). Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183, 5813–5825.
Saier, M. H. Jr. (2006). Protein secretion and membrane insertion systems in gram-negative bacteria. J. Membr. Biol. 214, 75–90.
Salacha, R., Kovacic, F., Brochier-Armanet, C., Wilhelm, S., Tommassen, J., Filloux, A., Voulhoux, R., and Bleves, S. (2010). The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel Type V secretion system. Environ. Microbiol. 12, 1498–1512.
Sardis, M. F., and Economou, A. (2010). SecA: a tale of two protomers. Mol. Microbiol. 76, 1070–1081.
Sargent, F. (2007). The twin-arginine transport system: moving folded proteins across membranes. Biochem. Soc. Trans. 35, 835–847.
Sato, H., and Frank, D. W. (2004). ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53, 1279–1290.
Sauri, A., Soprova, Z., Wickström, D., de Gier, J. W., Van der Schors, R. C., Smit, A. B., Jong, W. S., and Luirink, J. (2009). The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155, 3982–3991.
Sauvonnet, N., Vignon, G., Pugsley, A. P., and Gounon, P. (2000). Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19, 2221–2228.
Sawa, T., Yahr, T. L., Ohara, M., Kurahashi, K., Gropper, M. A., Wiener-Kronish, J. P., and Frank, D. W. (1999). Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5, 392–398.
Schmitt, L., Benabdelhak, H., Blight, M. A., Holland, I. B., and Stubbs, M. T. (2003). Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J. Mol. Biol. 330, 333–342.
Schoehn, G., Di Guilmi, A. M., Lemaire, D., Attree, I., Weissenhorn, W., and Dessen, A. (2003). Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22, 4957–4967.
Schraidt, O., Lefebre, M. D., Brunner, M. J., Schmied, W. H., Schmidt, A., Radics, J., Mechtler, K., Galán, J. E., and Marlovits, T. C. (2010). Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824. doi: 10.1371/journal.ppat.1000824
Schraidt, O., and Marlovits, T. C. (2011). Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195.
Senf, F., Tommassen, J., and Koster, M. (2008). Polar secretion of proteins via the Xcp type II secretion system in Pseudomonas aeruginosa. Microbiology 154, 3025–3032.
Sexton, J. A., and Vogel, J. P. (2002). Type IVB secretion by intracellular pathogens. Traffic 3, 178–185.
Shen, D. K., Quenee, L., Bonnet, M., Kuhn, L., Derouazi, M., Lamotte, D., Toussaint, B., and Polack, B. (2008). Orf1/SpcS chaperones ExoS for type three secretion by Pseudomonas aeruginosa. Biomed. Environ. Sci. 21, 103–109. PMID: 18548848
Shevchik, V. E., and Condemine, G. (1998). Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 144 (Pt 11): 3219–3228.
Shevchik, V. E., Robert-Baudouy, J., and Condemine, G. (1997). Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16, 3007–3016.
Spreter, T., Yip, C. K., Sanowar, S., Andre, I., Kimbrough, T. G., Vuckovic, M., Pfuetzner, R. A., Deng, W., Yu, A. C., Finlay, B. B., Baker, D., Miller, S. I., and Strynadka, N. C. (2009). A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476.
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., Garber, R. L., Goltry, L., Tolentino, E., Westbrock-Wadman, S., Yuan, Y., Brody, L. L., Coulter, S. N., Folger, K. R., Kas, A., Larbig, K., Lim, R., Smith, K., Spencer, D., Wong, G. K., Wu, Z., Paulsen, I. T., Reizer, J., Saier, M. H., Hancock, R. E., Lory, S., and Olson, M. V. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964.
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2010). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168.
Symmons, M. F., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2009). The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178.
Thanabalu, T., Koronakis, E., Hughes, C., and Koronakis, V. (1998). Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17, 6487–6496.
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., and Actis, L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484.
Tommassen, J., Filloux, A., Bally, M., Murgier, M., and Lazdunski, A. (1992). Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 9, 73–90.
Trepout, S., Taveau, J. C., Benabdelhak, H., Granier, T., Ducruix, A., Frangakis, A. S., and Lambert, O. (2010). Structure of reconstituted bacterial membrane efflux pump by cryo-electron tomography. Biochim. Biophys. Acta 1798, 1953–1960.
Turner, L. R., Lara, J. C., Nunn, D. N., and Lory, S. (1993). Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175, 4962–4969.
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A., and Filloux, A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 98, 6911–6916.
van den Berg, B. (2010). Crystal structure of a full-length autotransporter. J. Mol. Biol. 396, 627–633.
van Ulsen, P., Rutten, L., Feller, M., Tommassen, J., and van der Ende, A. (2008). Two-partner secretion systems of Neisseria meningitidis associated with invasive clonal complexes. Infect. Immun. 76, 4649–4658.
Vance, R. E., Hong, S., Gronert, K., Serhan, C. N., and Mekalanos, J. J. (2004). The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proc. Natl. Acad. Sci. U.S.A. 101, 2135–2139.
Viarre, V., Cascales, E., Ball, G., Michel, G. P., Filloux, A., and Voulhoux, R. (2009). HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J. Biol. Chem. 284, 33815–33823.
Voulhoux, R., Ball, G., Ize, B., Vasil, M. L., Lazdunski, A., Wu, L. F., and Filloux, A. (2001). Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20, 6735–6741.
Voulhoux, R., Taupiac, M. P., Czjzek, M., Beaumelle, B., and Filloux, A. (2000). Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa. J. Bacteriol. 182, 4051–4058.
Waksman, G., and Hultgren, S. J. (2009). Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7, 765–774.
Wandersman, C., and Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647.
Wilhelm, S., Tommassen, J., and Jaeger, K. E. (1999). A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181, 6977–6986.
Winans, S. C., Burns, D. L., and Christie, P. J. (1996). Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4, 64–68.
Wohlfarth, S., Hoesche, C., Strunk, C., and Winkler, U. K. (1992). Molecular genetics of the extracellular lipase of Pseudomonas aeruginosa PAO1. J. Gen. Microbiol. 138, 1325–1335.
Wolfgang, M., van Putten, J. P., Hayes, S. F., Dorward, D., and Koomey, M. (2000). Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19, 6408–6418.
Wolfgang, M. C., Kulasekara, B. R., Liang, X., Boyd, D., Wu, K., Yang, Q., Miyada, C. G., and Lory, S. (2003). Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 8484–8489.
Worrall, L. J., Lameignere, E., and Strynadka, N. C. (2011). Structural overview of the bacterial injectisome. Curr. Opin. Microbiol. 14, 3–8.
Wretlind, B., and Pavlovskis, O. R. (1984). Genetic mapping and characterization of Pseudomonas aeruginosa mutants defective in the formation of extracellular proteins. J. Bacteriol. 158, 801–808.
Yahr, T. L., Goranson, J., and Frank, D. W. (1996). Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22, 991–1003.
Yahr, T. L., Mende-Mueller, L. M., Friese, M. B., and Frank, D. W. (1997). Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179, 7165–7168.
Yahr, T. L., Vallis, A. J., Hancock, M. K., Barbieri, J. T., and Frank, D. W. (1998). ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U.S.A. 95, 13899–13904.
Yap, M. L., Mio, K., Leiman, P. G., Kanamaru, S., and Arisaka, F. (2010). The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J. Mol. Biol. 395, 349–360.
Yen, Y. T., Kostakioti, M., Henderson, I. R., and Stathopoulos, C. (2008). Common themes and variations in serine protease autotransporters. Trends Microbiol. 16, 370–379.
Young, G. M., Schmiel, D. H., and Miller, V. L. (1999). A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U.S.A. 96, 6456–6461.
Yukl, E. T., Jepkorir, G., Alontaga, A. Y., Pautsch, L., Rodriguez, J. C., Rivera, M., and Moenne-Loccoz, P. (2010). Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry 49, 6646–6654.
Zhang, L. H., Fath, M. J., Mahanty, H. K., Tai, P. C., and Kolter, R. (1995). Genetic analysis of the colicin V secretion pathway. Genetics 141, 25–32.
Keywords: cell envelope, nanomachine, macromolecular complex, channel, targeting
Citation: Filloux A (2011) Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front. Microbio. 2:155. doi: 10.3389/fmicb.2011.00155
Received: 26 April 2011;
Paper pending published: 25 May 2011;
Accepted: 01 July 2011;
Published online: 18 July 2011.
Edited by:
Dara Frank, Medical College of Wisconsin, USAReviewed by:
Maria Sandkvist, University of Michigan, USAKatrina Forest, University of Wisconsin-Madison, USA
Copyright: © 2011 Filloux. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Alain Filloux, Division of Cell and Molecular Biology, Centre for Molecular Microbiology and Infection, Imperial College London, South Kensington Campus, Flowers Building, SW72AZ London, UK. e-mail: a.filloux@imperial.ac.uk