- 1State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, The Innovation Academy of Seed Design, Chinese Academy of Sciences, Graduate University of the Chinese Academy of Sciences, Wuhan, China
- 2Institute of Marine Biology, College of Oceanography, Hohai University, Nanjing, China
Chemokine receptor cxcr4 and its ligand cxcl12 have evolved two paralogs in the teleost lineage. In this study, we have identified four duplicated cxcr4 and cxcl12 genes from hexaploid gibel carp, Carassius gibelio, respectively. Cgcxcr4bs and Cgcxcl12as were dynamically and differentially expressed in immune-related tissues, and significantly up-regulated in head kidney and spleen after crucian carp herpesvirus (CaHV) infection. Blocking Cxcr4/Cxcl12 axis by injecting AMD3100 brought more severe bleeding symptom and lower survival rate in CaHV-infected fish. AMD3100 treatment also suppressed the up-regulation of key antiviral genes in head kidney and spleen, and resulted in more acute replication of CaHV in vivo. Consistently, the similar suppression of up-regulated expression of key antiviral genes were also observed in CAB cells treated by AMD3100 after poly(I:C) stimulation. Finally, MAPK3 and JAK/STAT were identified as the possible pathways that CgCxcr4s and CgCxcl12s participate in to promote the antiviral response in vitro.
Introduction
CXC chemokine receptor 4 (CXCR4) is an exclusive receptor of CXC chemokine ligand 12 (CXCL12), also known as stromal derived factor 1 (SDF-1) (1). CXCR4/CXCL12 axis plays a pivotal role in hematopoiesis, neurogenesis, germ cell development, vascular formation and inflammatory processes in mammals (2–6). During inflammation, CXCL12/CXCR4 axis controls the innate immune cell transporting between bone marrow and blood. CXCL12/CXCR4 interaction is responsible for the neutrophil retention in bone marrow, and both cxcr4b and cxcl12 mutant can increase the neutrophil release into the bloodstream (5, 7, 8). As a weak neutrophil chemoattractant, CXCL12 can increase neutrophil migration to participate in the inflammatory response (9, 10). In mammals, CXCL12/CXCR4 axis diminishes the inflammatory reaction by increasing the production of inflammatory cytokine interferon-γ (IFN-γ) (11, 12), which forms a negative feedback control by decreasing the expression of CXCR4 (13, 14).
In teleost fishes, cxcr4 and cxcl12 also show high expression in immune tissues (15, 16). After virus or bacteria invasion, cxcr4 and/or cxcl12 are significantly up-regulated in Asian swamp eel (Monopterus albus) (17), rock bream (Oplegnathus fasciatus) (18, 19), orange-spotted grouper (Epinephelus coioides) (20, 21), freshwater carp (Catla catla) (22), and yellow croaker (Larimichthys crocea) (16). Therefore, cxcr4 and cxcl12 have been suggested as an important part in fish innate immune system (17–19). In zebrafish, Cxcl12a/Cxcr4b axis controls the neutrophil release into circulation, and antagonizes the wound-induced inflammatory signals (8, 23). In orange-spotted grouper, Cxcl12/Cxcr4 up-regulated their expression after nodavirus (NNV) infection, indicating their contribution to defensing the virus invasion (24). However, the specific role and underneath mechanism of Cxcr4/Cxcl12 axis in this process are still poorly understood in fish.
Gibel carp (Carassius gibelio) (3n = 6x = ~150 chromosomes) has been considered to undergo two extra polyploidy events (an early allopolyploidy followed by a recent autopolyploidy) in addition to the three rounds of whole genome duplication (WGD) shared by all teleost fishes (25–29). As a recurring polyploidy animal, it is an ideal system to investigate divergent expression and function of duplicated genes through WGD. Since 1980s, gibel carp has become an important aquaculture species in China (30). Along with the application of several improved varieties with excellent growth performance, such as allogynogenetic gibel carp “CAS III” (clone A+) and “CAS V” (clone F), the current annual production capacity has increased to about 3 million tons per year in China (31–33). However, due to the highly intensive culture, outbreak of epizootic crucian carp (Carassius auratus) herpesvirus (CaHV) has led to enormous economic losses in main culture areas of Jiangsu province since 2012 (34–36). It is an urgent need to breed novel variety with CaHV resistance. In our previous studies, IFN system genes and immunoglobulin genes were found to play vital roles in defensing CaHV (32, 37, 38). Especially, some genes annotated as cxcl12 and cxcr4 were identified from the comparative transcriptomes between the diseased and control individuals (32), suggesting the antiviral role in response to CaHV infection. In this study, we first analyzed the diversification, evolution and biased expression pattern of cxcr4/cxcl12 genes in hexaploid gibel carp. Then, we explored the role of Cxcr4/Cxcl12 axis in the antiviral immune response by blocking CXCR4 with AMD3100 and revealed the related signaling pathways.
Materials and Methods
CaHV Infection and Sample Collection
Six-month-old individuals of gibel carp clone F with 67.68 (±2.16) g average weight were collected from the GuanQiao Experimental Station, Institute of Hydrobiology, Chinese Academy of Sciences. Ten tissues (spleen, head kidney, kidney, thymus, gill, intestine, brain, heart, muscle and liver) from three individuals were collected for quantitative reverse transcription PCR (qPCR) analysis before CaHV infection. The CaHV challenge experiments were performed as previously described (32, 39). In brief, sixty individuals were randomly divided into two groups, and were intraperitoneally injected with 500 μl CaHV viral suspension (2.915 × 108 virus particles) or PBS per fish in the experimental group or in control group respectively. Head kidney and spleen were collected from three control (0 day) and infected individuals at 1, 3 and 5 days post injection (dpi), respectively. All samples were preserved in RNAlater (Qiagen, Dusseldorf, Germany) and stored at −20°C for nucleic acid extraction. All experiments were performed in triplicate and the results were representative of three independent experiments.
Healthy individuals were gradually acclimatized with aerated water at 24 (±1) °C for 2 weeks before infection, and fed twice a day. After deep and overdosed anesthesia with styrylpyridine (30–50 mg/L; Aladdin, Shanghai, China), the fish were euthanized by immediately cutting off the spinal cord adjacent to the head. All procedures in this study were approved by the Institutional Animal Care and Use Committee of Institute of Hydrobiology, Chinese Academy of Sciences (protocol number 2016-018).
Sequence and Phylogenetic Analyses
The complete cDNA sequences of gibel carp cxcr4s and cxcl12s (GenBank accession numbers MT330400, MT330401, MT330402, MT330403, MT330404, MT330405, MT330406, and MT330407) were amplified by 3′ and 5′ RACE using SMARTer® RACE 5′/3′ Kit (Clontech, San Francisco, USA) from head kidneys of diseased fish after CaHV infection. Amino acid sequences and transmembrane-domain were predicted by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) and SMART (http://smart.embl-heidelberg.de/), respectively. To analyze the evolution of vertebrate Cxcr4 and Cxcl12, the sequence information of 7 species were obtained from the Ensembl Genome browser (http://asia.ensembl.org/index.html) and National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Multiple protein sequences were aligned by Clustal W program and the phylogenetic tree was constructed by bootstrap analysis (1000 replicates) using the neighbor-joining method (NJ) in MEGA 7.0 software. The exon-intron structure prediction was made by mRNA and genomic sequences alignment of cxcr4s and cxcl12s. The conserved synteny was analyzed among the chromosomal regions around cxcr4s and cxcl12s genes in human (Homo sapiens), chicken (Gallus gallus), spotted gar (Lepisosteus oculatus), zebrafish, crucian carp and gibel carp.
AMD3100 Treatment in vivo
One hundred twenty gibel carp individuals were randomly divided into four groups. The healthy and CaHV-infected individuals intraperitoneally injected with 200 μl PBS were considered as negative and positive control group (NC and PC), respectively. In the two experimental groups (E1 and E2), the healthy and CaHV-infected individuals were intraperitoneally injected with CXCR4 antagonist AMD3100 (MedChemExpress, New Jersey, USA) (100 μg in 200 μl PBS). Briefly, PBS or AMD3100 was injected per day until the fish died, and CaHV was injected only once at the 24 h after the first injection of PBS or AMD3100 in PC and E2 group, while NC and E1 group injected PBS instead. The procedure of CaHV infection was described above. After CaHV challenge, the individuals in four groups were monitored every 24 h to score mortality. Head kidney and spleen were collected from individuals at 3dpi to extract RNA for subsequent qPCR assays. The experiments were performed in triplicate.
RNA Extraction and qPCR
RNA extraction and qPCR were performed as previously described (37, 38). Total RNAs from tissues were extracted using SV Total RNA isolation System (Promega, Madison, USA) according to the manufacturer's protocol. The first-strand cDNAs were synthesized in a 20 μL reaction volume following the protocol of GoScript™ Reverse Transcription System (Promega, Madison, USA). qPCR was performed on a CFX96TM Real-Time PCR System (Bio-Rad, California, USA) using an iTaq™ Universal SYBR® Green Supermix (Bio-Rad, California, USA). Primers used for qPCR analysis were designed with Oligo Calc (Oligonucleotide Properties Calculator) (http://biotools.nubic.northwestern.edu/OligoCalc.html) and listed in Supplementary Table 1. The specificity of each pair of primers was confirmed by sequencing. All samples were analyzed in triplicate, and relative expression levels of target genes were calculated using the 2−ΔΔCT method. The optimal reference gene, eukaryotic translation elongation factor 1 alpha 1, like 1 (eef1a1l1) (M value = 0.74 <1.5) was selected as the normalizer for qPCR (37).
AMD3100 Treatment in vitro
Crucian carp (C. auratus) blastula embryonic (CAB) cells were used to perform the Cxcr4 inhibition experiments with a final AMD3100 concentration of 10 μM. Controls were treated with same volumes of ethanol. Cells after 24 h treatment were harvested to extract RNA for subsequent qPCR assays. The experiments were also performed in triplicate and the results were representative of three independent experiments.
Plasmid Constructs
For luciferase assays, expression plasmids were generated by inserting the full length Open-reading frame (ORF) of Cgcxcr4s and Cgcxcl12s into the pCS2+ vector (Invitrogen, Carlsbad, USA). The dominant negative mutant plasmids, gibel carp MAPK1-DN (aa 34–369) and MAPK3-DN (aa 70–396) were generated by cloning the corresponding DNA fragments into pcDNA3.1+ vector. Other plasmids, including CaIFNpro-Luc and CgVIPpro-Luc, and some dominant negative mutant plasmids including STAT1a-ΔC and STAT1b-ΔC were constructed previously (40–42). All constructs were verified by sequencing.
Transfection and Luciferase Activity Assays
CAB cells were seeded overnight in 6-well plates and transfected with CgCxcr4s-pCS2+ and CgCxcl12s-pCS2+ respectively using FuGENE HD Transfection Reagent (Promega, Madison, USA) (40, 43). After 24 h transfection, 1 μg/ml polyinosinic:polycytidylic acid (poly(I:C)), was transfected to induce the expression of immune genes (44, 45). The cells were collected in TRIzol reagent (Invitrogen, Carlsbad, USA) after 24 h stimulation. For luciferase activity assay, CAB cells were seeded overnight in 24-well plates and transfected with various constructs at a ratio of 10:10:1 (250 ng CaIFNpro-Luc or CgVIPpro-Luc plasmid: 250 ng corresponding expression plasmid: 25 ng Renilla luciferase plasmid pRL-TK) (40, 43, 44). For the control, the expression plasmid was replaced by empty vector pCS2+. At 24 h post-transfection, the cells were treated again with poly(I:C) infection and the controls were added serum-free medium instead. After 48 h transfection, the cells were harvested and lysed according to the Dual-Luciferase Reporter Assay System (Promega, Madison, USA). Luciferase activities were measured by a Junior LB9509 luminometer (Berthold, Pforzheim, Germany) and normalized to the amounts of Renilla luciferase activities. All experiments were performed in triplicate and the results were representative of three independent experiments.
Western Blotting Assay
CAB cells were seeded in 6-well plates overnight and treated with ethanol (as a control) and AMD3100 for 24 h. Then, the cells were transfected with 1 μg/ml poly(I:C) or added equal volume serum free medium. After 24 h stimulation, the cell were collected and boiled together with SDS-PAGE protein loading buffer (Beyotime, Wuhan, China) for 8 min, and then analyzed by western blotting using crucian carp IRF3 (CaIRF3) polyclonal Ab (40), zebrafish STAT1 polyclonal Ab (HuaAn, Hangzhou, China) and zebrafish IRF7 monoclonal Ab (HuaAn, Hangzhou, China). The anti-tubulin was purchased from ABclone. To reveal the pathways, CAB cells were seeded in 6-well plates overnight and transfected with different CgCxcl12s and CgCxcr4s combinations. After 24 h transfection, the cells were collected as above described, and analyzed by western blotting using p-JAK2 monoclonal Ab (Cell Signaling Technology, Danvers, USA) and p-MAPK monoclonal Ab (Cell Signaling Technology, Danvers, USA).
Statistical Analysis
qPCR and luciferase activity assay data were shown as means ± SD of three independent experiments, and each performed in triplicate. The statistical analysis was performed by SPSS software (SPSS Inc.) using one-way ANOVA. A probability (p) < 0.05 was considered statistically significant (*), and p < 0.01 was considered extremely significant (**).
Results
Molecular Characterization and Evolution of Duplicated cxcr4/cxcl12 Genes
Four cxcr4 genes were identified in gibel carp (Supplementary Figure 1). Based on the identities, we supposed that gibel carp has two groups of paralogues (cxcr4a and cxcr4b) (caused by the Ts3R), and each of them contains two homoeologues (caused by the allopolyploidy), so they were named as Cgcxcr4a-A, Cgcxcr4a-B, Cgcxcr4b-A and Cgcxcr4b-B respectively. Different from three alleles (≥99.00% identity) of Cgbmp15s or Cgnanos2s (46, 47), only one sequence was identified for each homoeologue, which indicates that the alleles of each Cgcxcr4 gene may be identical. The average identity between homoeologues was 89.65 ± 0.75%, while the average identity between paralogs was about 71.5 ± 0.2% (Supplementary Table 2). The SMART and TMHMM analyses revealed that all CgCxcr4s possessed 7 hydrophobic transmembrane (TM) domains, four extracellular loops (ECL), four intracellular loops (ICL), an extracellular N-terminus and an intracellular C-terminus (Supplementary Figure 3). The major differences existed in the N-terminus and the ECL3 (Supplementary Figure 3). There were also four cxcr4 genes in tetraploid crucian carp (2n = 4x = 100 chromosomes), and their identities to the corresponding genes of gibel carp ranged from 97.5 to 100% (Supplementary Table 2). Same as cxcr4s, four cxcl12 genes were also identified from gibel carp and crucian carp, respectively (Supplementary Figures 2, 3).
Consistent with the accepted species phylogeny, CgCxcr4as and CgCxcr4bs were first clustered with CaCxcr4as and CaCxcr4bs respectively, then clustered with common carp (Cyprinus carpio) and zebrafish Cxcr4a and Cxcr4b, respectively (Figure 1A). Teleost Cxcr4a and Cxcr4b branch were grouped together and then clustered with spot gar Cxcr4, while tetrapod CXCR4s and coelacanth Cxcr4 were clustered into another branch (Figure 1A). Similar phylogenetic tree of vertebrate Cxcl12 was shown in Figure 2A. The phylogenies of Cxcr4/Cxcl12 confirmed the assumption that both gibel carp and crucian carp are originated from a common allotetraploid ancestor (27).
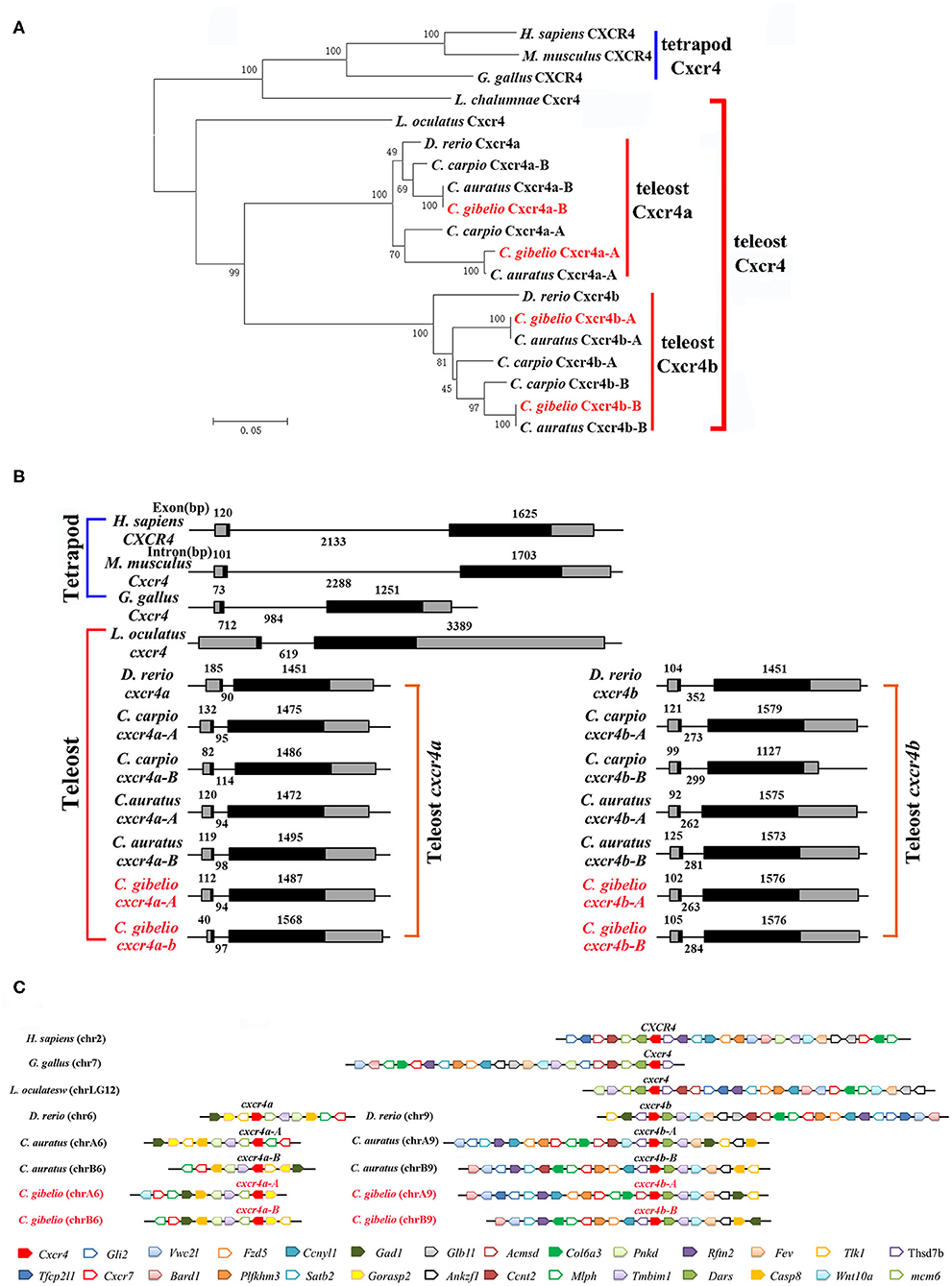
Figure 1. Evolutionary analyses of multiple divergent cxcr4 genes. (A) Phylogenetic tree of Cxcr4 proteins. (B) Genomic structures of cxcr4 genes. Exons and introns are shown by boxes and horizontal lines, respectively. ORFs are highlighted by black boxes. Exon and intron size are indicated above or below as bp. (C) Syntenic alignment of chromosomal regions around cxcr4 genes. Chromosome segments are represented as thick lines. Conserved gene blocks are shown in matching colors and transcription orientations are indicated by arrows. Chr, chromosome.
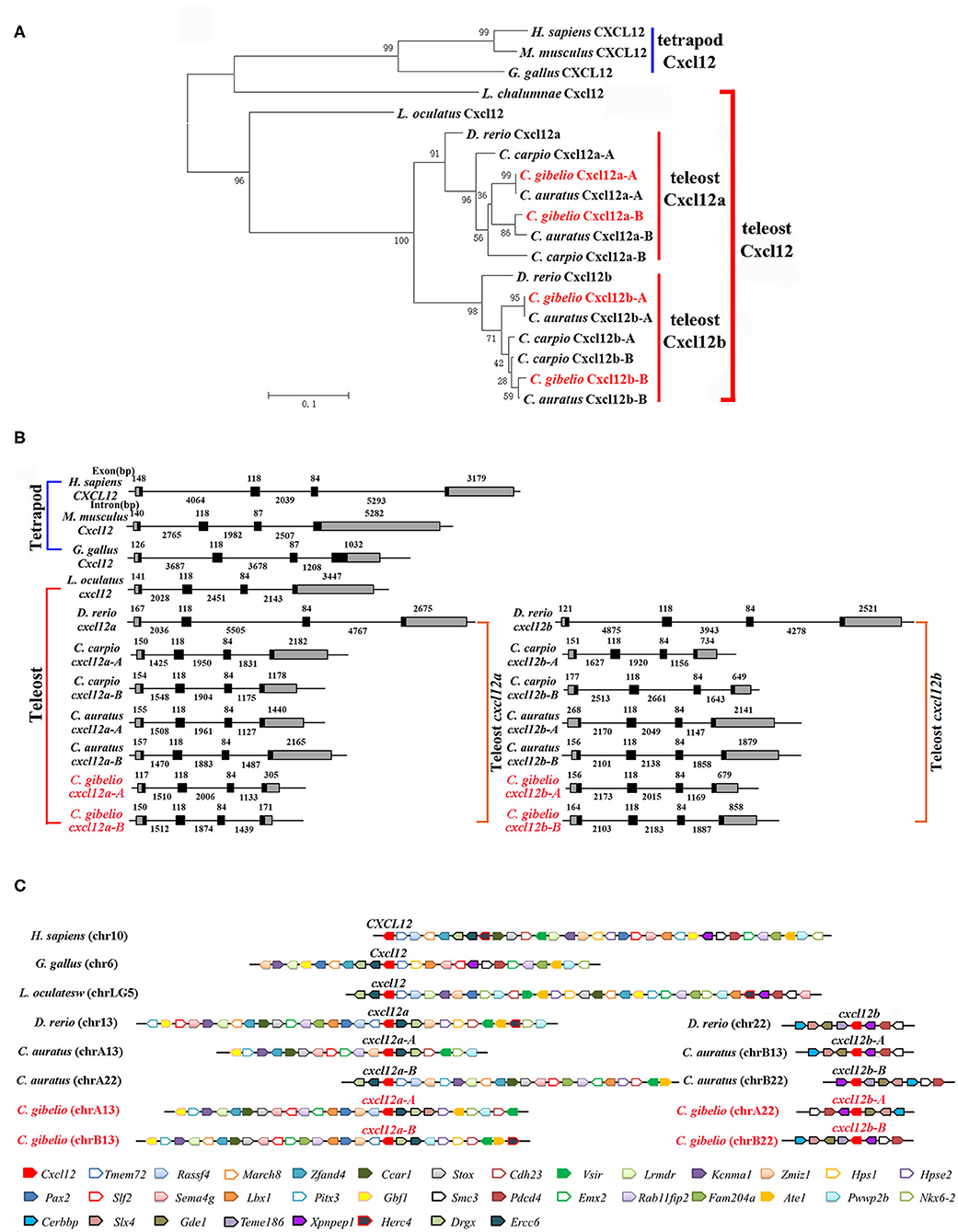
Figure 2. Evolutionary analyses of multiple divergent cxcl12 genes. (A) Phylogenetic tree of Cxcl12 proteins. (B) Genomic structures of cxcl12 genes. Exons and introns are shown by boxes and horizontal lines, respectively. ORFs are highlighted by black boxes. Exon and intron size are indicated above or below as bp. (C) Syntenic alignment of chromosomal regions around cxcl12 genes. Chromosome segments are represented as thick lines. Conserved gene blocks are shown in matching colors and transcription orientations are indicated by arrows. Chr, chromosome.
We also analyzed the genomic structure and gene synteny of gibel carp cxcr4s and cxcl12s with other vertebrates. Same to others, gibel carp cxcr4s all had a bi-exonic structure (Figure 1B) and cxcl12s were all composed of four exons (Figure 2B). Cgcxcr4a-A, Cgcxcr4a-B, Cgcxcr4b-A, and Cgcxcr4b-B were located on chromosome A6 (chrA6), chrB6, chrA9, and chrB9, respectively. Compared to tetrapod, a complementary loss/retention pattern existed in the vicinity genes of cxcr4s between teleost chr6 and chr9 (Figure 1C). Probably owing to the relatively short evolutionary history of allotetraploidy in Carassius, the gene loss did not occur between homoeologous chromosomes (chrA6 and chrB6, chrA9 and chrB9) (Figure 1C). Instead, extensive inversions and rearrangements were observed, such as gene blocks tlk1-gorasp2-gad1, cxcr7-col6a3-mlph, and dars-cxcr4-thsd7b (Figure 1C). The syntenic alignment of cxcl12 came to the similar conclusions (Figure 2C).
Dynamic and Differential Expression Patterns of cxcr4s and cxcl12s
Cgcxcr4s were abundantly expressed in the immune-related tissues, such as spleen, kidney, head kidney and thymus (Figure 3A). Except gill, Cgcxcr4bs showed remarkably higher expression than Cgcxcr4as in the analyzed tissues, suggesting that Cgcxcr4bs may play a vital role in immune regulation. Cxcr4 homoeologues also exhibited divergent expression patterns. Cgcxcr4b-A was expressed significantly higher than Cgcxcr4b-B in most analyzed tissues, while Cgcxcr4a-A and Cgcxcr4a-B showed well-matched expression bias in different tissues. More abundant Cgcxcr4a-A transcripts than Cgcxcr4a-B were detected in head kidney, kidney, gill and heart, while reversely differential expression patterns were observed in spleen, thymus, and intestine (Figure 3A). Consistent with the specific chemotactic interaction of grouper Cxcr4b-Cxcl12a (48), Cgcxcl12a was the mainly expressed paralog in most analyzed immune-related tissues, as well as Cgcxcr4b (Figure 3B). Except in intestine and brain, Cgcxcl12a-B showed significantly higher expression than Cgcxcl12a-A (Figure 3B). These results indicate that the duplicated cxcl12 genes also exhibit a differential expression pattern in gibel carp.
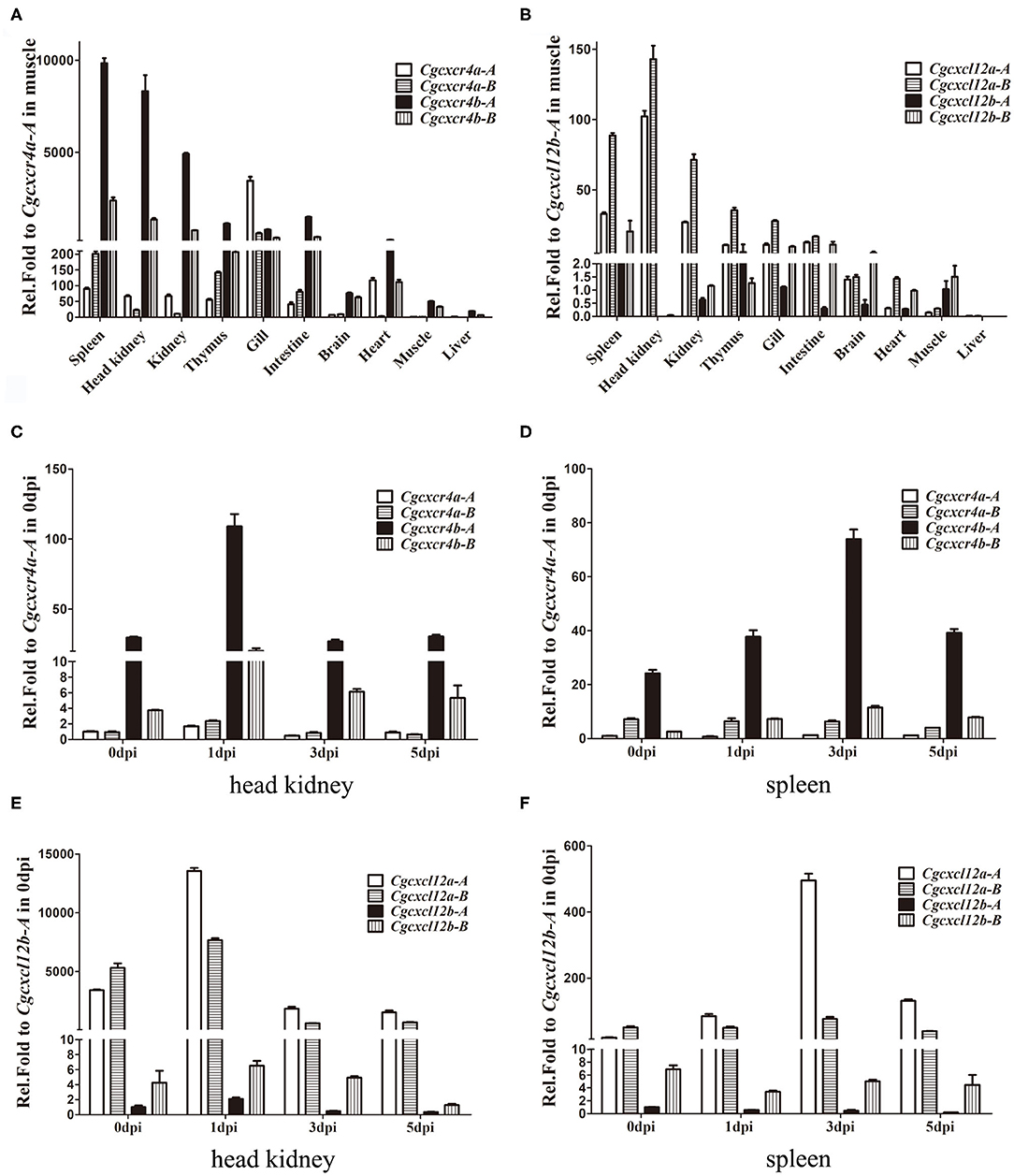
Figure 3. Differential expression patterns of cxcr4 and cxcl12 genes. (A,B) The expression of cxcr4 (A) and cxcl12 (B) in adult tissues. (C–F) The dynamic expression of cxcr4/cxcl12 in head kidney and spleen after CaHV infection, cxcr4 in head kidney (C), in spleen (D), and cxcl12 in head kidney (E), in spleen (F). eef1a1l1 was used as the reference to normalize the templates. Each bar represents the mean ± SEM (n = 3).
Dynamic expression changes of Cgcxcr4s or Cgcxcl12s in gibel carp after CaHV infection were also evaluated by qPCR (Figures 3C–E). At 1 dpi in head kidney, Cgcxcr4b-A and Cgcxcr4b-B increased up to 3.7 and 5.4 folds respectively, and then both returned to the basic expression level (Figure 3C). Although fewer Cgcxcr4a transcripts were detected in head kidney compared to those of Cgcxcr4b, they were also slightly up-regulated (1.7–2.5 folds) at 1 dpi (Figure 3C). In spleen, Cgcxcr4b-A and Cgcxcr4b-B increased their expression and reached a peak at 3 dpi (Figure 3D). No significant expression change of Cgcxcr4as was detected in spleen after CaHV challenge (Figure 3D). For four Cgcxcl12 genes, only Cgcxcl12a-A was significantly up-regulated in head kidney (4.0 folds at 1 dpi) and spleen (25.5 folds at 3 dpi) after CaHV infection (Figures 3E,F).
CgCxcr4/CgCxcl12 Axis Controls Antiviral Response Against CaHV
To explore the role of CgCxcr4/CgCxcl12 axis in antiviral response to CaHV, we investigated the symptoms and the CaHV abundance among four gibel carp groups treated with/without AMD3100 that specifically blocks the binding between CXCR4 and CXCL12 (49, 50). AMD3100 injection did not affect the survival of the healthy individuals (not challenged by CaHV) (Figures 4A,B). After CaHV infection, the individuals exhibited hemorrhage at the base of fins and on abdomen at 3 dip (arrows in Figure 4A). Cxcr4-blocked individuals showed more severe bleeding and lower survival rate (Figures 4A,B), indicating more susceptibility to CaHV infection than control individuals (injecting PBS). The death started at 3 dpi in Cxcr4-blocked group and all died at 5 dpi, while the first death of control group occurred at 4 dpi and about 7% survivors appeared healthy at 10 dpi (Figure 4B). To further confirm this susceptibility induced by Cxcr4 inhibition, we detected the abundances of five CaHV genes to evaluate the CaHV replication (38). After 35 amplification cycles, no DNA fragment of CaHV was detected from the healthy individuals, while the specific bands were amplified in the CaHV-infected individuals (Figure 4C). Accordant with more severe symptoms, the Cxcr4-blocked fish had markedly stronger bands than the control individuals at 3 dpi (Figure 4C).
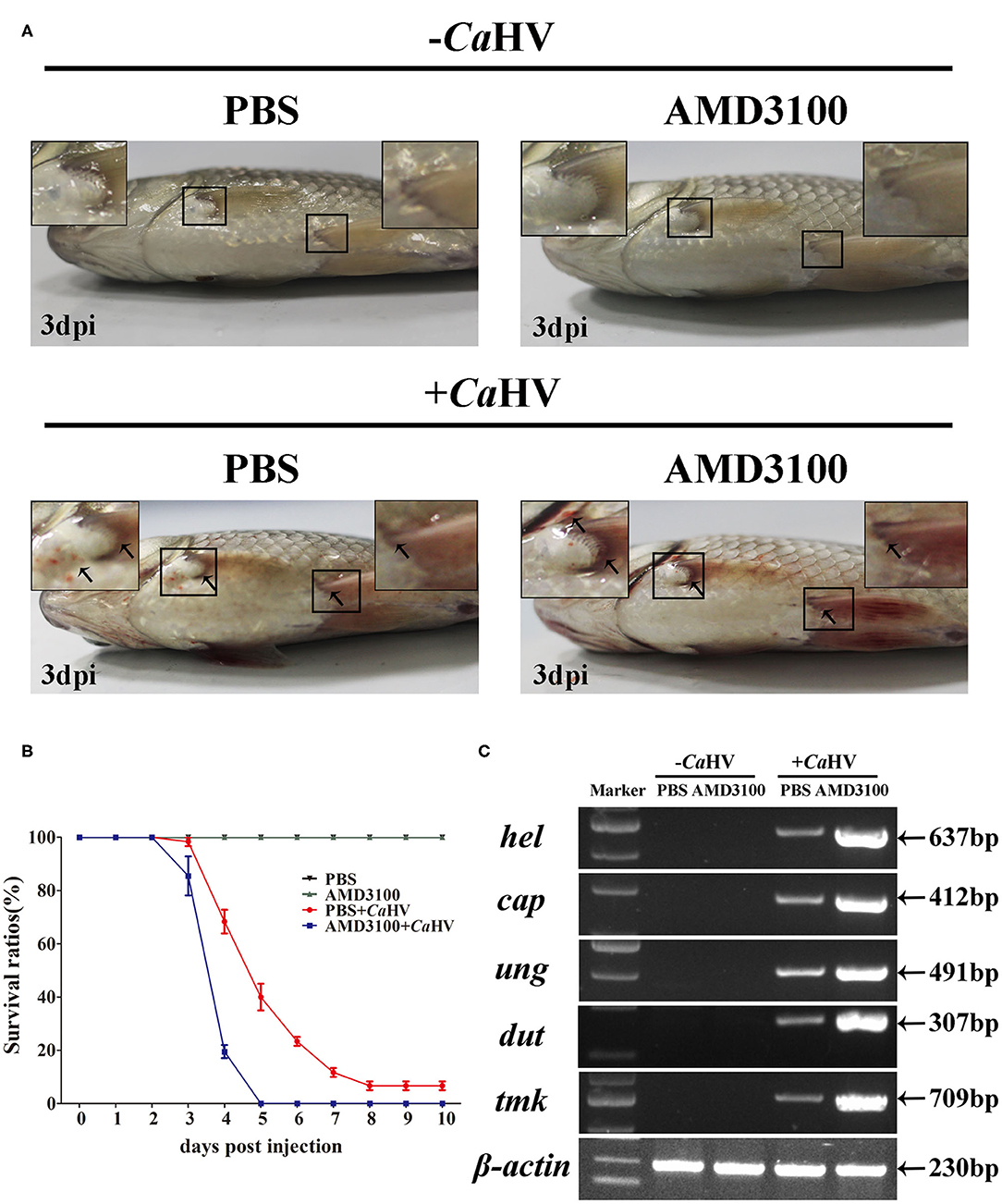
Figure 4. Gibel carp treated with AMD3100 were more sensitive to CaHV infection. (A) Representative images of gibel carp, both un-infected group (–CaHV) and infected group (+CaHV) were photographed at third day after intraperitoneal injection PBS (–CaHV) or CaHV (+CaHV), along with the treatment of vehicle (PBS; the control) or AMD3100 (100 μg in 200 μl PBS) every 24 h. Un-infected gibel individuals appeared normal while infected individuals exhibited hemorrhage at the base of fins and on abdomen (black arrows), and AMD3100 treated gibel carp had more severe bleeding symptom. (B) Survival ratios after CaHV infection. We counted the numbers of dead fish every 24 h post-infection. The values are the mean ± SEM from three replicate tanks. (C) Electrophoretogram of the PCR amplified products of five CaHV genes (hel, cap, ung, dut, and tmk) in four groups (NC, E1, PC, and E2). Gene symbols and the sizes of amplified products are indicated by the left and right side of the figure, respectively.
We also analyzed the expression of several key antiviral genes in head kidney and spleen (37), such as interferon (ifnφ1 and ifnφ3), interferon regulatory factor (irf3 and irf7), host pattern recognition receptors PRRs [toll-like receptor 9 (tlr9) and retinoic acid induce gene I (rig1)] and PRR-mediated IFN signal pathway genes [mediator of irf3 activation (mita) and myeloid differentiation primary-response gene 88 (myd88)]. Similar to cxcr4 and cxcl12, they all possessed A and B homoeologues with divergent sequences and with biased expression in gibel carp (Figure 5). After CaHV infection, they were all up-regulated, and the up-regulation folds were significantly suppressed when Cxcr4-Cxcl12 binding was blocked (Figure 5 and Supplementary Figure 4). Together with above results, we suggest that blocking Cxcr4/Cxcl12 axis should suppress gibel carp antiviral response to CaHV.
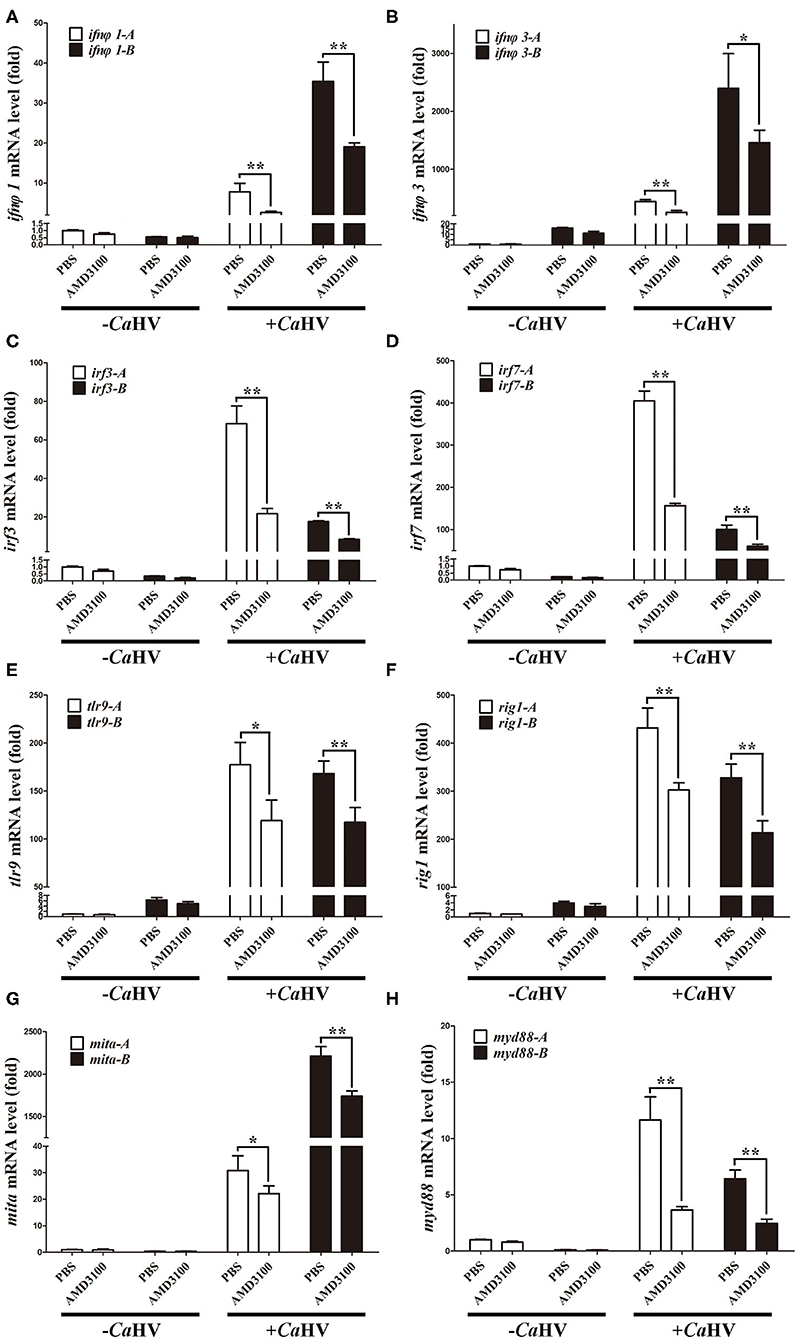
Figure 5. AMD3100 suppressed CaHV-induced activation of key antiviral genes in gibel carp head kidney. (A–H) qPCR analysis of ifnφ1 (A), ifnφ3 (B), irf3 (C), irf7 (D), tlr9 (E), rig1 (F), mita (G), and myd88 (H) in AMD3100 treated gibel carp after CaHV infextion. eef1a1l1 was used as control. Each bar represents mean ± SEM (n = 3). The asterisks indicate the significant differences (*p < 0.05, **p < 0.01).
CgCxcl12/CgCxcr4 Axis Enhances Cellular Antiviral Response Through MAPK3 and JAK/STAT Pathway
The function and signal pathways of about 20 IFN system genes have been well documented in CAB cells (40, 41, 51–53), so we used CAB cells to explore the antiviral response pathways related to CgCxcr4/CgCxcl12 axis. After treatment with poly(I:C) that mimics the double-strand RNA (dsRNA) virus infection, all of the analyzed IFN system genes, including ifn, ifi58 (interferon inducible protein 58), pkr (dsRNA-activated protein kinase R), irf1 (interferon regulatory factor 1), irf3, irf7, viperin, mx1 (MX dynamin-like GTPase 1), tbk1 (TANK binding kinase 1), stat1(signal transducer and activator of transcription 1), tlr3 (toll-like receptor 3) and rig1, were significantly up-regulated (Figure 6A). Consistent with the results in vivo (Figure 5), the similar suppression effects were also observed in the CAB cells treated with AMD3100 (Figure 6A). Furthermore, ADM3100 pre-treatment also inhibited the activities of CaIFN and CgVip promoters, as well as elevating the amounts of STAT1, IRF7 and phosphorylated IRF3 (40), induced by poly(I:C) stimulation (Figures 6B,C). Finally, both CgCxcr4 or CgCxcl12 overexpression (Supplementary Figure 5) promoted the activation of IFN promoter induced by poly(I:C) (Figure 6D). In addition, The up-regulation folds affected by CgCxcr4bs or CgCxcl12as were bigger than their paralogues respectively, and no obviously differences observed between each homoeologous pair (CgCxcr4b-A: 33.7, CgCxcr4b-B: 34.2, CgCxcr4a-A: 29.2, CgCxcr4a-B: 28.7; CgCxcl12a-A: 36.2, CgCxcl12a-B: 34.1, CgCxcl12b-A: 31.3 and CgCxcl12b-B:30.2) (Figure 6D). Considering the bias expression in immune organs, we suppose that Cxcr4b and Cxcl12a might be the major antiviral couple in gibel carp. Altogether, Cxcr4/Cxcl12 axis facilitates the cellular antiviral response stimulated by poly(I:C).
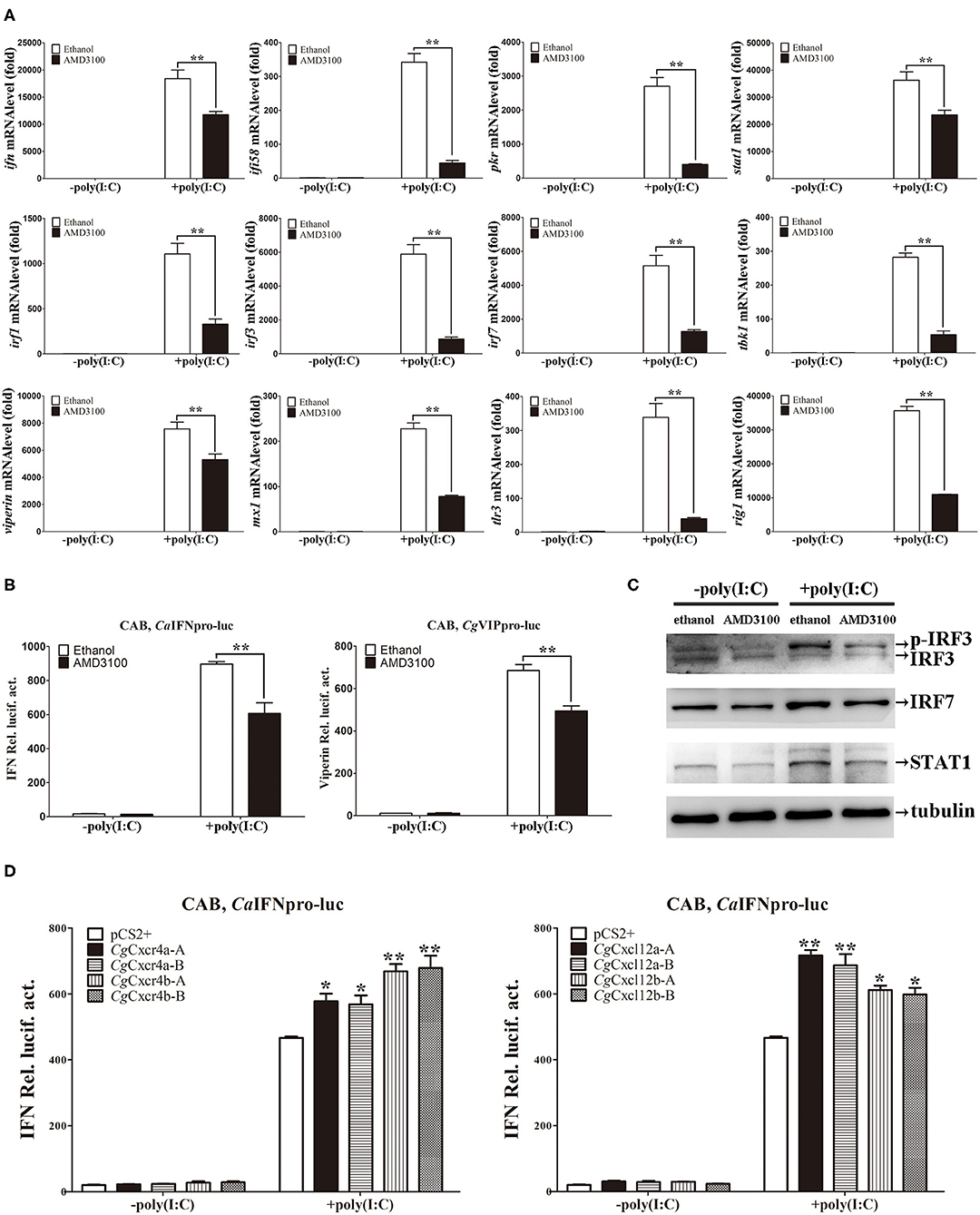
Figure 6. CgCxcl12s and CgCxcr4s facilitated cellular antiviral response. (A) Treatment of CAB cells with 1μM AMD3100 after poly(I:C) stimulation suppressed the up-regulation expression of ifn, ifi58, pkr, stat1, irf1, irf3, irf7, tbk1, viperin, mx1, tlr3, and rig1. (B) AMD3100 inhibited the activation of fish IFN and viperin promoters in CAB cells after poly(I:C) stimulation. (C) The up-regulation protein expression of p-IRF3, IRF7, and STAT1 were suppressed in AMD3100-treated CAB cells after poly(I:C) stimulation. (D) Poly(I:C)-triggered activation of IFN promoter was enhanced by overexpression of Cxcr4s or Cxcl12s. Each bar represents mean ± SEM (n = 3). The asterisks indicate the significant differences (*p < 0.05, **p < 0.01).
To avoid the possible bias effects of poly(I:C), we co-transfected different CgCxcr4s-CgCxcl12s combinations in CAB cells to directly detect the CaIFN promoter luciferase activities. Compared to the control, co-transfection of CgCxcl12a-A-CgCxcr4b-A combination resulted in stronger activation of CaIFN promoter (Figure 7A), and other different ligand-receptor combinations showed the similar results (Supplementary Figure 6). CXCR4/CXCL12 axis can activate PI3K/Akt, PLC/IP3, ERK1/2 pathways (14, 54), and β-arrestin/JAK/STAT pathway in mammals (55, 56). To reveal the molecular mechanisms by which Cxcr4/Cxcl12 axis regulates IFN expression, we co-transfected CgCxcr4b-A and CgCxcl12a-A with dominant negative mutants of MAPK1, MAPK3 or STAT1 (MAPK1-DN, MAPK3-DN or STAT1a-ΔC+STAT1b-ΔC) in above vitro system. After 24 h of transfection, the activity of CaIFN promoter was diminished by the co-overexpression of MAPK3-DN or STAT1a-ΔC+STAT1b-ΔC, while overexpression of MAPK1-DN did not obviously affect the induction of IFN expression (Figure 7B). To further verify the results, we used western blotting to detect the expression of phosphorylation JAK2 (p-JAK2) and p-MAPK3 proteins. As shown in Figure 7C, overexpression of CgCxcr4b-A and CgCxcl12a-A promoted the expression of p-JAK2 and p-MAPK3. The increased expression was significantly reduced by co-transfecting MAPK3-DN or STAT1a-ΔC+STAT1b-ΔC (Figure 7D). These results indicate that CgCxcl12/CgCxcr4 axis can regulate IFN expression through MAPK3 and JAK/STAT pathways.
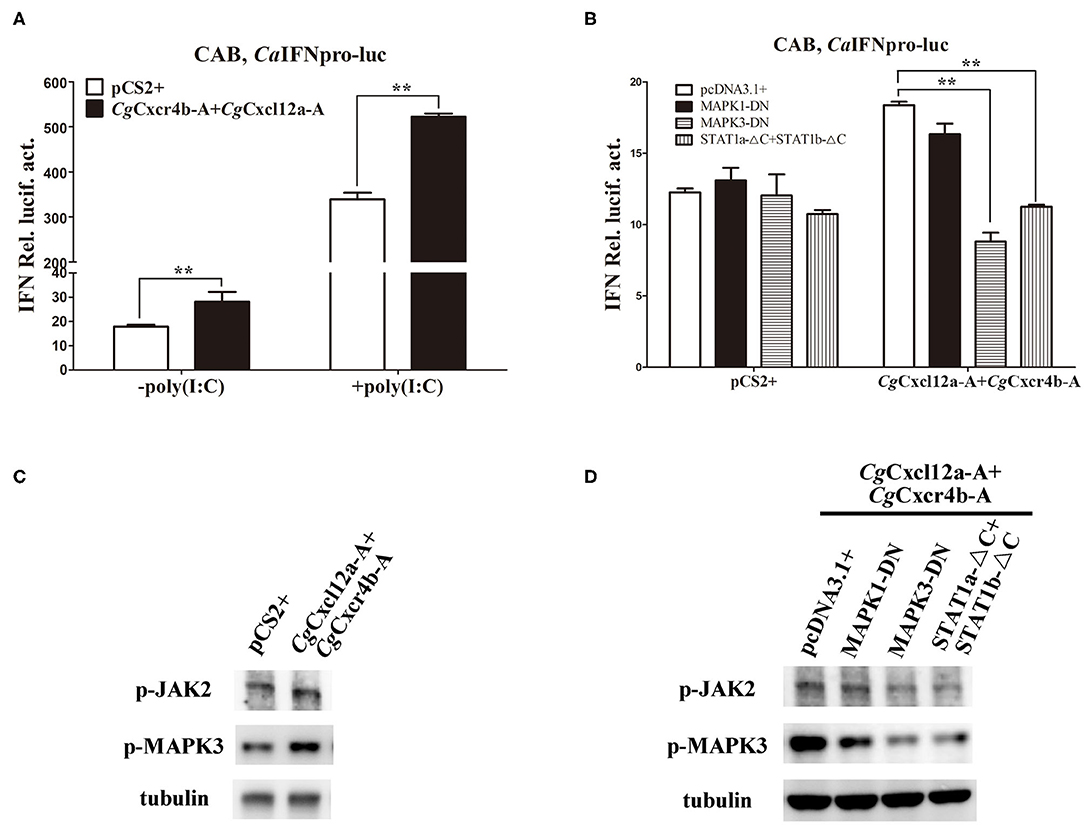
Figure 7. Cxcr4/Cxcl12 axis induced IFN activity via MAPK3 and JAK/STAT pathways. (A) CgCxcl12a-A and CgCxcr4b-A combination activated the activity of CaIFN promoter. (B) The activation of CaIFN promoter induced by CgCxcl12a-A and CgCxcr4b-A overexpressing was inhibited by overexpression of dominant negative mutants of MAPK and JAK/STAT signaling molecules. (C) Overexpression of CgCxcl12a-A and CgCxcr4b-A increased the protein expression of p-JAK2, STAT1, p-MAPK3 and MAPK3. (D) The up-regulation protein expression of p-JAK2, STAT1, p-MAPK3, and MAPK3 were suppressed by transfecting dominant negative mutants MAPK3-DN and STAT1a-ΔC+STAT1b-ΔC. Each bar represents mean ± SEM (n = 3). The asterisks indicate the significant differences (**p < 0.01).
Discussion
Polyploidy is considered as a major driver of species diversification and frequently occurred during teleost evolution (25, 57, 58). Although the consequences are complex and variable, it might increase short-term adaptive potential and long-term evolvability for polyploid organisms along with subsequent rediploidization (59–61). Ts3R was supposed to result in two paralogs of each gene compared to tetrapods (28, 62–64). Gibel carp retains two cxcr4 and cxcl12 paralogs (cxcr4a and cxcr4b, cxcl12a, and cxcl12b) (Figures 1, 2; Supplementary Figures 1–3). In addition, polyploidy occurred repeatedly in Cypriniformes which further increase genetic variation (28, 65, 66). We identified two homoeologues (A and B) of each paralog from common carp, crucian carp and gibel carp (Figures 1, 2; Supplementary Figures 1–3). Similar to salmons that experienced an extra WGD 95 million years ago, diversification is a major evolution force of duplication genes (67).
During rediploidization, the sub-functionalization and neo-functionalization are the major maintenance ways of gene duplicates (68, 69). The dynamic expression pattern was observed in some fishes after pathogen infection, elucidating that cxcr4/cxcl12 is not only important in immune defense against bacterial and viral infection but also may restrain the immune system in some other physiological processes (17–20). In gibel carp, both paralogous and homoeologous pairs of cxcr4b and cxcl12a show dynamic and differential expression patterns and antiviral responses. Our results indicate that cxcr4bs and cxcl12as should play a dominant role in immune response and confirm the specific ligand-receptor recognitions of Cxcl12b and Cxcr4a (48, 70). Although derived from the common ancestor (71), the expression patterns of duplicated genes evolved divergently after separation. For example, a common carp cxcl12b (Cccxcl12b) exhibits highest identity with Cgcxcl12b-B, and they are both highly expressed in the brain (72). But besides, Cgcxcl12b-B also shows high expression in spleen (Figure 3B). In gibel carp, all cxcr4 and cxcl12 paralogues/homoeologues can promote IFN gene expression in CAB cells, although Cxcl12as and Cxcr4bs show a bit higher ability (Figure 6D). It seems that the sub-functionalization and neo-functionalization of cxcr4s or cxcl12s mainly occur at the transcriptional level, and cxcr4 or cxcl12 copies may still overlap for some functions (68, 73).
Although CXCR4 blockade benefits to autoimmune diseases, it is deleterious in acute inflammation and ischemia, and has a negative impact on negative physiological defense (74–76). In mammals, CXCR4 triggered by CXCL12 binding activates anti-inflammatory signaling pathways and inhibits inflammation (77, 78). It increases the production of pro-inflammatory cytokines, including IFN-γ, TNF-α, IL-6 and so on (79, 80). AMD3100 administration also results in the significant reduction of IFN-γ, TNF-α and IL-6 in the acute stage of ischemic stroke or after spinal cord injury (11, 12). In addition, interruption of the CXCR4/CXCL12 axis not only down-regulates the expression of IFN-γ and TNF-α, but also negatively influences TLR4 signaling and leads to the decrease of inflammatory responses in the LPS treatment mice (76, 81). In zebrafish, Cxcl12/Cxcr4 axis plays an important role to antagonize wound-induced inflammatory signals, but Cxcl12/Cxcr4 signaling also leads to the inappropriate retention of neutrophils at the inflammatory sites (8, 23, 82). Interestingly, impairment in the Cxcl12a/Cxcr4b signaling axis does not affect the recruitment of neutrophils when cxcr4b mutants are infected by Salmonella typhimurium (8). Therefore, the role of Cxcl12/Cxcr4 signaling axis against infection may be not affected by the retention and/or migration of leukocyte population. In this study, Cxcr4 blockade not only promotes the duplication of CaHV, but also suppresses the up-regulation expression of the virus recognition receptors tlr9, tlr3 and the following downstream antiviral genes, both in gibel carp after CaHV infection and CAB cells with poly(I:C) stimulation (Figures 4–6). Moreover, overexpression of CgCxcr4s or CgCxcl12s both instigate the induction of IFN by poly(I:C) (Figure 6D). The innate immune response triggered by virus infection increases the expression of IFN and downstream IFN-stimulated genes (ISGs) to protect the host both in mammals and teleost (53, 83, 84). Taken together, we suggest that Cxcr4/Cxcl12 axis may be important for the innate immune responses to resist virus invasion in teleost.
Cxcl12 was reported to enhance dimerization of Cxcr4, and then to activate the downstream pathways (85). Activating some signaling pathways such as ERK, JAK/STAT and PI3K can regulate the transcription factors, including AP1, NF-κB and NFAT, to regulate the expression of cytokines (86–89). They also play some roles in IFN induction by virus infection, as their domain negative mutants markedly inhibited the induction of IFN (Figure 7B). Following CXCl12 binding, CXCR4 activates the Janus Kinases by changing their conformation to promote the JAK/STAT signaling pathway and the downstream transcription factor NF-κB that stimulates IFN secretion (85, 90, 91). In addition, CXCR4 internalization also activates the β-arrestin pathway, which severs as a scaffold for ERK and enhance the ERK activation (56, 92). In our study, blocking MAPK3 and STAT1 has a significant suppression effect on IFN induction by CgCxcr4b-A and CgCxcl12a-A over-expression (Figure 7), indicating MAPK and JAK/STAT maybe the major pathways that CgCxcr4s/CgCxcl12s axis participates in antiviral response. However, blocking MAPK1 does not affect the induction of IFN, and the MAPK1 protein is much less than MAPK3 in CAB cells (data not shown), which suggest that MAPK1 might play a different role from MAPK3 in teleost.
In conclusion, the current study represents the diversification and dominant expression in immune tissues of Cxcl12/Cxcr4 axis in hexaploid gibel carp. It is the first time to report its function in teleost antiviral response, which regulates IFN expression mainly through MAPK3 and JAK/STAT pathways. These findings would expand our knowledge about the complex function of Cxcl12/Cxcr4 axis in teleost, which is a link between the innate and the adaptive immune system and play a key role to improve the adaptive immunity (22, 93). Understanding the regulation mechanisms of Cxcl12/Cxcr4 signaling can provide useful information for disease control with effective immune protection in gibel carp. Further study of Cxcl12/Cxcr4 signaling and its downstream factors will help us to better understand the mechanism of host against CaHV infection, and provide resistant-related candidate genes for disease-resistance breeding of gibel carp.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Institute of Hydrobiology, Chinese Academy of Sciences (protocol number 2016-018).
Author Contributions
W-JL and F-XG performed the experiments, analyzed the data, and drafted the manuscript. YW participated in the data analysis and revised manuscript. ZL participated in the sample collection. Y-LZ and X-JZ participated in the data analysis. LZ designed the studies, analyzed the data, and drafted the manuscript. J-FG conceived the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24030203 and XDA24030104), the National Natural Science Foundation (31772838), and China Agriculture Research System (CARS-45-07). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Yi-Bing Zhang (Institute of Hydrobiology, Chinese Academy of Sciences) for providing some plasmids used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.02176/full#supplementary-material
Supplementary Figure 1. Nucleotide sequences and deduced amino acid sequences of four Cgcxcr4 genes.
Supplementary Figure 2. Nucleotide sequences and deduced amino acid sequences of four Cgcxcl12 genes.
Supplementary Figure 3. Multiple amino acid sequence alignment of Cxcr4/Cxcl12 proteins.
Supplementary Figure 4. AMD3100 suppresses CaHV-induced activation of key antiviral genes in gibel carp spleen.
Supplementary Figure 5. The transfection efficiency and subcellular localization of CgCxcr4s.
Supplementary Figure 6. Different CgCxcl12 and CgCxcr4 combinations activated the activity of CaIFN promoter.
Supplementary Table 1. Primers used in this study.
Supplementary Table 2. The amino acid sequence identities of gibel carp and other vertebrate Cxcr4s/Cxcl12s.
References
1. Nazari A, Khorramdelazad H, Hassanshahi G. Biological/pathological functions of the CXCL12/CXCR4/CXCR7 axes in the pathogenesis of bladder cancer. Int J Clin Oncol. (2017) 22:991–1000. doi: 10.1007/s10147-017-1187-x
2. Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. (1998) 393:595–9. doi: 10.1038/31269
3. Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. (2010) 11:37–49. doi: 10.1038/nrm2815
4. Cui L, Qu H, Xiao T, Zhao M, Jolkkonen J, Zhao C. Stromal cell-derived factor-1 and its receptor CXCR4 in adult neurogenesis after cerebral ischemia. Restor Neurol Neurosci. (2013) 31:239–51. doi: 10.3233/RNN-120271
5. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. (2014) 32:659–702. doi: 10.1146/annurev-immunol-032713-120145
6. Janssens R, Struyf S, Proost P. The unique structural and functional features of CXCL12. Cell Mol Immunol. (2018) 15:299–311. doi: 10.1038/cmi.2017.107
7. Hendrix CW, Collier AC, Lederman MM, Schols D, Pollard RB, Brown S, et al. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. Jaids J Acq Imm Def. (2004) 37:1253–62. doi: 10.1097/01.qai.0000137371.80695.ef
8. Paredes-Zúñiga S, Morales RA, Muñoz-Sánchez S, Muñoz-Montecinos C, Parada M, Tapia K, et al. CXCL12a/CXCR4b acts to retain neutrophils in caudal hematopoietic tissue and to antagonize recruitment to an injury site in the zebrafish larva. Immunogenetics. (2017) 69:341–9. doi: 10.1007/s00251-017-0975-9
9. Gouwy M, Schiraldi M, Struyf S, Van Damme J, Uguccioni M. Possible mechanisms involved in chemokine synergy fine tuning the inflammatory response. Immunol Lett. (2012) 145:10–4. doi: 10.1016/j.imlet.2012.04.005
10. Gouwy M, Struyf S, Leutenez L, Portner N, Sozzani S, Van Damme J. Chemokines and other GPCR ligands synergize in receptor-mediated migration of monocyte-derived immature and mature dendritic cells. Immunobiology. (2014) 219:218–29. doi: 10.1016/j.imbio.2013.10.004
11. Wang GD, Liu YX, Wang X, Zhang YL, Zhang YD, Xue F. The SDF-1/CXCR4 axis promotes recovery after spinal cord injury by mediating bone marrow-derived from mesenchymal stem cells. Oncotarget. (2017) 8:11629–40. doi: 10.18632/oncotarget.14619
12. Li D, Sun HH, Li WZ, Zhang X. Protective effects of AMD3100 on ischemic stroke by inhibiting inflammation and promoting angiogenesis and neuranagenesis. Int J Clin Exp Med. (2018) 11:7971–8.
13. Dewan MZ, Ahmed S, Iwasaki Y, Ohba K, Toi M, Yamamoto N. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother. (2006) 60:273–6. doi: 10.1016/j.biopha.2006.06.004
14. Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. (2007) 1768:952–63. doi: 10.1016/j.bbamem.2006.11.002
15. Fu Q, Yang Y, Li C, Zeng Q, Zhou T, Li N, et al. The CC and CXC chemokine receptors in channel catfish (Ictalurus punctatus) and their involvement in disease and hypoxia responses. Dev Comp Immunol. (2017) 77:241–51. doi: 10.1016/j.dci.2017.08.012
16. Liu X, Kang L, Liu W, Lou B, Wu C, Jiang L. Molecular characterization and expression analysis of the large yellow croaker (Larimichthys crocea) chemokine receptors CXCR2, CXCR3, and CXCR4 after bacterial and poly I: C challenge. Fish Shellfish Immunol. (2017) 70:228–39. doi: 10.1016/j.fsi.2017.08.029
17. Gao W, Fang L, Yang D, Ai K, Luo K, Tian G, et al. Cloning and expression of Asian swamp eel (Monopterus albus) cxcr4 paralogues, and their modulation by pathogen infection. Aquaculture. (2016) 457:50–60. doi: 10.1016/j.aquaculture.2016.02.021
18. Thulasitha WS, Umasuthan N, Revathy KS, Whang I, Lee J. Molecular characterization, genomic structure and expressional profiles of a CXC chemokine receptor 4 (CXCR4) from rock bream Oplegnathus fasciatus. Fish Shellfish Immunol. (2015) 44:471–7. doi: 10.1016/j.fsi.2015.03.012
19. Thulasitha WS, Umasuthan N, Whang I, Lim BS, Jung HB, Noh JK, et al. A CXC chemokine gene, CXCL12, from rock bream, Oplegnathus fasciatus: molecular characterization and transcriptional profile. Fish Shellfish Immunol. (2015) 45:560–6. doi: 10.1016/j.fsi.2015.05.002
20. Lin CY, Chen YM, Hsu HH, Shiu CT, Kuo HC, Chen TY. Grouper (Epinephelus coioides) CXCR4 is expressed in response to pathogens infection and early stage of development. Dev Comp Immunol. (2012) 36:112–20. doi: 10.1016/j.dci.2011.06.009
21. Wang Q, Wang S, Zhang Y, Yu Y, Zhao H, Yang H, et al. The CXC chemokines and CXC chemokine receptors in orange-spotted grouper (Epinephelus coioides) and their expression after Singapore grouper iridovirus infection. Dev Comp Immunol. (2019) 90:10–20. doi: 10.1016/j.dci.2018.08.015
22. Banerjee R, Roy S, Samanta M, Das S. Molecular cloning, characterization and expression analysis of MHCI and chemokines CXCR3 and CXCR4 gene from freshwater carp, Catla catla. Microbiol Immunol. (2019) 63:379–91. doi: 10.1111/1348-0421.12728
23. Isles HM, Herman KD, Robertson AL, Loynes CA, Prince LR, Elks PM, et al. The CXCL12/CXCR4 Signaling axis retains neutrophils at inflammatory sites in zebrafish. Front Immunol. (2019) 10:1784. doi: 10.3389/fimmu.2019.01784
24. Wu CS, Wang TY, Chen TY. CXCL12-CXCR4 signaling pathways of orange-spotted, grouper (Epinephelus coioides) contribute to nodavirus infection. Fish Shellf Immunol. (2016) 53:103. doi: 10.1016/j.fsi.2016.04.058
25. Gui J, Zhou L. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci China Life Sci. (2010) 53:409–15. doi: 10.1007/s11427-010-0092-6
26. Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. (2013) 45:415–21. doi: 10.1038/ng.2568
27. Li XY, Zhang XJ, Li Z, Hong W, Liu W, Zhang J, et al. Evolutionary history of two divergent Dmrt1 genes reveals two rounds of polyploidy origins in gibel carp. Mol Phylogenet Evol. (2014) 78:96–104. doi: 10.1016/j.ympev.2014.05.005
28. Zhou L, Gui J. Natural and artificial polyploids in aquaculture. Aquacult Fish. (2017) 2:103–11. doi: 10.1016/j.aaf.2017.04.003
29. Li XY, Gui JF. Diverse and variable sex determination mechanisms in vertebrates. Sci China Life Sci. (2018) 61:1503–14. doi: 10.1007/s11427-018-9415-7
30. Zhou L, Wang ZW, Wang Y, Gui JF. Crucian carp and gibel carp culture. In: Gui JF, Tang QS, Li ZJ, Liu JS, De Silva SS, editors. Aquaculture in China: Success Stories and Modern Trends. Oxford: John Wiley & Sons Ltd Press (2018). p. 149–57.
31. Wang ZW, Zhu HP, Wang D, Jiang F-F, Guo W, Zhou L, et al. A novel nucleo-cytoplasmic hybrid clone formed via androgenesis in polyploid gibel carp. BMC Res Notes. (2011) 4:82. doi: 10.1186/1756-0500-4-82
32. Gao FX, Wang Y, Zhang QY, Mou CY, Li Z, Deng YS, et al. Distinct herpesvirus resistances and immune responses of three gynogenetic clones of gibel carp revealed by comprehensive transcriptomes. BMC Genomics. (2017) 18:561. doi: 10.1186/s12864-017-3945-6
33. Chen F, Li XY, Zhou L, Yu P, Wang Z-W, Li Z, et al. Stable genome incorporation of sperm-derived DNA fragments in gynogenetic clone of gibel carp. Mar Biotechnol. (2020) 22:54–66. doi: 10.1007/s10126-019-09930-w
34. Zhang QY, Gui JF. Virus genomes and virus-host interactions in aquaculture animals. Sci China Life Sci. (2015) 58:156–69. doi: 10.1007/s11427-015-4802-y
35. Zeng XT, Chen ZY, Deng YS, Gui JF, Zhang QY. Complete genome sequence and architecture of crucian carp Carassius auratus herpesvirus (CaHV). Arch Virol. (2016) 161:3577–81. doi: 10.1007/s00705-016-3037-y
36. Zhang QY, Gui JF. Diversity, evolutionary contribution and ecological roles of aquatic viruses. Sci China Life Sci. (2018) 61:1486–502. doi: 10.1007/s11427-018-9414-7
37. Mou CY, Wang Y, Zhang QY, Gao FX, Li Z, Tong JF, et al. Differential interferon system gene expression profiles in susceptible and resistant gynogenetic clones of gibel carp challenged with herpesvirus CaHV. Dev Comp Immunol. (2018) 86:52–64. doi: 10.1016/j.dci.2018.04.024
38. Lu WJ, Gao FX, Wang Y, Zhang QY, Li Z, Zhang XJ, et al. Differential expression of innate and adaptive immune genes in the survivors of three gibel carp gynogenetic clones after herpesvirus challenge. BMC Genomics. (2019) 20:432. doi: 10.1186/s12864-019-5777-z
39. Gao FX, Lu WJ, Wang Y, Zhang QY, Zhang YB, Mou CY, et al. Differential expression and functional diversification of diverse immunoglobulin domain-containing protein (DICP) family in three gynogenetic clones of gibel carp. Dev Comp Immunol. (2018) 84:396–407. doi: 10.1016/j.dci.2018.03.013
40. Sun F, Zhang YB, Liu TK, Gan L, Yu FF, Liu Y, et al. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J Immunol. (2010) 185:7573–82. doi: 10.4049/jimmunol.1002401
41. Wang B, Zhang Y-B, Liu T-K, Shi J, Sun F, Gui J-F. Fish viperin exerts a conserved antiviral function through RLR-triggered IFN signaling pathway. Deve Comp Immunol. (2014) 47:140–9. doi: 10.1016/j.dci.2014.07.006
42. Feng H, Zhang YB, Zhang QM, Li Z, Zhang QY, Gui JF. Zebrafish IRF1 regulates IFN antiviral response through binding to IFNφ1 and IFNφ3 promoters downstream of MyD88 signaling. J Immunol. (2015) 194:1225–38. doi: 10.4049/jimmunol.1402415
43. Sun F, Zhang YB, Liu TK, Shi J, Wang B, Gui JF. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J Immunol. (2011) 187:2531–9. doi: 10.4049/jimmunol.1100642
44. Zhang QM, Zhao X, Li Z, Wu M, Gui JF, Zhang YB. Alternative splicing transcripts of zebrafish LGP2 gene differentially contribute to IFN antiviral response. J Immunol. (2018) 200:688–703. doi: 10.4049/jimmunol.1701388
45. Wu M, Zhao X, Gong XY, Wang Y, Gui JF, Zhang YB. FTRCA1, a species-specific member of finTRIM family, negatively regulates fish IFN response through autophage-lysosomal degradation of TBK1. J Immunol. (2019) 202:2407–20. doi: 10.4049/jimmunol.1801645
46. Jiang SY, Wang Y, Zhou L, Chen F, Li Z, Gui JF. Molecular characteristics, genomic structure and expression patterns of diverse bmp15 alleles in polyploid gibel carp clone F. Acta Hydrobiol Sin. (2020) 44:518–27. doi: 10.7541/2020.063
47. Zhang QQ, Zhou L, Li Z, Gan RH, Yu ZX, Gui JF, et al. Allelic diversification, syntenic alignment and expression patterns of nanos2 in polyploid gibel carp. Acta Hydrobiol Sin. (2020) 161:252–65. doi: 10.1104/pp.112.205161
48. Lu WJ, Zhou L, Gao FX, Sun ZH, Li Z, Liu XC, et al. Divergent expression patterns and function of two cxcr4 paralogs in hermaphroditic Epinephelus coioides. Int J Mol Sci. (2018) 19:2943. doi: 10.3390/ijms19102943
49. Zabel BA, Wang Y, Lewen S, Berahovich RD, Penfold ME, Zhang P, et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J Immunol. (2009) 183:3204–11. doi: 10.4049/jimmunol.0900269
50. Mercurio L, Ajmone-Cat MA, Cecchetti S, Ricci A, Bozzuto G, Molinari A, et al. Targeting CXCR4 by a selective peptide antagonist modulates tumor microenvironment and microglia reactivity in a human glioblastoma model. J Exp Clin Cancer Res. (2016) 35:55. doi: 10.1186/s13046-016-0326-y
51. Zhang YB, Hu CY, Zhang J, Huang GP, Wei LH, Zhang QY, et al. Molecular cloning and characterization of crucian carp (Carassius auratus L.) interferon regulatory factor 7. Fish Shellf Immunol. (2003) 15:453–66. doi: 10.1016/S1050-4648(03)00025-1
52. Yu FF, Zhang YB, Liu TK, Liu Y, Sun F, Jiang J, et al. Fish virus-induced interferon exerts antiviral function through Stat1 pathway. Mol Immunol. (2010) 47:2330–41. doi: 10.1016/j.molimm.2010.05.282
53. Zhang YB, Gui JF. Molecular regulation of interferon antiviral response in fish. Dev Comp Immunol. (2012) 38:193–202. doi: 10.1016/j.dci.2012.06.003
54. Ward SG. T lymphocytes on the move: chemokines, PI3-kinase and beyond. Trends Immunol. (2006) 27:80–7. doi: 10.1016/j.it.2005.12.004
55. Mellado M, Rodríguez-Frade JM, Vila-Coro AJ, Fernández S, Martin de Ana A, Jones DR, et al. Chemokine receptor homo - or heterodimerization activates distinct signaling pathways. EMBO J. (2001) 20:2497–507. doi: 10.1093/emboj/20.10.2497
56. Sun Y, Cheng Z, Ma L, Pei G. β-Arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J Biol Chem. (2002) 277:49212–9. doi: 10.1074/jbc.M207294200
57. Volff JN, Schartl M. Evolution of signal transduction by gene and genome duplication in fish. In: Van de Peer Y, Meyer A, editors. Genome Evolution. Berlin: Springer (2003). p. 139–50.
58. Lu M, Li XY, Li Z, Du WX, Zhou L, Wang Y, et al. Regain of sex determination system and sexual reproduction ability in a synthetic octoploid male fish. Sci China Life Sci. (2020). doi: 10.1007/s11427-020-1694-7. [Epub ahead of print].
59. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. (2016) 533:200–5. doi: 10.1038/nature17164
60. Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. (2017) 18:411–24. doi: 10.1038/nrg.2017.26
61. Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. (2018) 4:258–68. doi: 10.1038/s41477-018-0136-7
62. Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. (2004) 431:946–57. doi: 10.1038/nature03025
63. Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. (2007) 447:714–9. doi: 10.1038/nature05846
64. Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. (2016) 48:427–37. doi: 10.1038/ng.3526
65. Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fisher. (2003) 13:237–46. doi: 10.1023/B:RFBF.0000033049.00668.fe
66. Van de Peer Y, Taylor JS, Meyer A. Are all fishes ancient polyploids? In: Van de Peer Y, Meyer A, editors. Genome Evolution. Berlin: Springer (2003). p. 65–73.
67. Grimholt U, Hauge H, Hauge AG, Leong J, Koop BF. Chemokine receptors in Atlantic salmon. Dev Comp Immunol. (2015) 49:79–95. doi: 10.1016/j.dci.2014.11.009
68. Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. (2009) 100:605–17. doi: 10.1093/jhered/esp047
69. Liu S, Li Z, Gui JF. Fish-specific duplicated dmrt2b contributes to a divergent function through Hedgehog pathway and maintains left-right asymmetry establishment function. PLoS ONE. (2009) 4:7261. doi: 10.1371/journal.pone.0007261
70. Bussmann J, Raz E. Chemokine-guided cell migration and motility in zebrafish development. EMBO J. (2015) 34:1309–18. doi: 10.15252/embj.201490105
71. Lu XJ, Zhu K, Shen HX, Nie L, Chen J. CXCR4s in teleosts: two paralogous chemokine receptors and their roles in hematopoietic stem/progenitor cell homeostasis. J Immunol. (2020) 204:1225–41. doi: 10.4049/jimmunol.1901100
72. Huising MO, van der Meulen T, Flik G, Verburg-van Kemenade BML. Three novel carp CXC chemokines are expressed early in ontogeny and at nonimmune sites. Eur J Biochem. (2004) 271:4094–106. doi: 10.1111/j.1432-1033.2004.04347.x
73. Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. (1999) 151:1531–45.
74. Dai S, Yuan F, Mu J, Li C, Chen N, Guo S, et al. Chronic AMD3100 antagonism of SDF-1α-CXCR4 exacerbates cardiac dysfunction and remodeling after myocardial infarction. J Mol Cell Cardiol. (2010) 49:587–97. doi: 10.1016/j.yjmcc.2010.07.010
75. Devi S, Wang Y, Chew WK, Lima R, A-González N, Mattar CN, et al. Neutrophil mobilization via plerixafor-mediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J Exp Med. (2013) 210:2321–36. doi: 10.1084/jem.20130056
76. Seemann S, Lupp A. Administration of AMD3100 in endotoxemia is associated with pro-inflammatory, pro-oxidative, and pro-apoptotic effects in vivo. J Biomed Sci. (2016) 23:68. doi: 10.1186/s12929-016-0286-8
77. Poznansky MC, Olszak IT, Foxall R, Evans RH, Luster AD, Scadden DT. Active movement of T cells away from a chemokine. Nat Med. (2000) 6:543–8. doi: 10.1038/75022
78. Fan H, Wong D, Ashton SH, Borg KT, Halushka PV, Cook JA. Beneficial effect of a CXCR4 agonist in murine models of systemic inflammation. Inflammation. (2012) 35:130–7. doi: 10.1007/s10753-011-9297-5
79. Lu DY, Tang CH, Yeh WL, Wong KL, Lin CP, Chen YH, et al. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. Eur J Pharmacol. (2009) 613:146–54. doi: 10.1016/j.ejphar.2009.03.001
80. Chen HT, Tsou HK, Hsu CJ, Tsai CH, Kao CH, Fong YC, et al. Stromal cell-derived factor-1/CXCR4 promotes IL-6 production in human synovial fibroblasts. J Cell Biochem. (2011) 112:1219–27. doi: 10.1002/jcb.23043
81. Triantafilou M, Lepper PM, Briault CD, Ahmed MAE, Dmochowski JM, Schumann C, et al. Chemokine receptor 4 (CXCR4) is part of the lipopolysaccharide “sensing apparatus”. Eur J Immunol. (2008) 38:192–203. doi: 10.1002/eji.200636821
82. Rankin SM. The bone marrow: a site of neutrophil clearance. J Leukocyte Biol. (2010) 88:241–51. doi: 10.1189/jlb.0210112
83. Gürtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. (2013) 21:413–20. doi: 10.1016/j.tim.2013.04.004
84. Tan X, Sun L, Chen J, Chen ZJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol. (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605
85. Vila-Coro AJ, Rodríguez-frade JM, Martin De Ana A, Moreno-Ortìz MC, Martìnez-A C, Mellado M. The chemokine SDF-1α triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. (1999) 13:1699–710. doi: 10.1096/fasebj.13.13.1699
86. Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. (2013) 121:1951–60. doi: 10.1182/blood-2012-09-435057
87. Pelham CJ, Agrawal DK. Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert Rev Clin Immunol. (2014) 10:243–56. doi: 10.1586/1744666X.2014.866519
88. Tian L, Choi SC, Murakami Y, Allen J, Morse HC III, Qi CF, et al. (2014). p85alpha recruitment by the CD300f phosphatidylserine receptor mediates apoptotic cell clearance required for autoimmunity suppression. Nat Commun. 5:3146. doi: 10.1038/ncomms4146
89. Gao S, Yi Y, Xia G, Yu C, Ye C, Tu F, et al. The characteristics and pivotal roles of triggering receptor expressed on myeloid cells-1 in autoimmune diseases. Autoimmun Rev. (2019) 18:25–35. doi: 10.1016/j.autrev.2018.07.008
90. Wong D, Korz W. Translating an antagonist of chemokine receptor CXCR4: from bench to bedside. Clin Cancer Res. (2008) 14:7975–80. doi: 10.1158/1078-0432.CCR-07-4846
91. Pozzobon T, Goldoni G, Viola A, Molon B. CXCR4 signaling in health and disease. Immunol Lett. (2016) 177:6–15. doi: 10.1016/j.imlet.2016.06.006
92. Cheng ZJ, Zhao J, Sun Y, Hu W, Wu YL, Cen B, et al. β-arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between β-arrestin and CXCR4. J Biol Chem. (2000) 275:2479–85. doi: 10.1074/jbc.275.4.2479
Keywords: CXCR4, CXCL12, AMD3100, CaHV, antiviral response, innate immunity
Citation: Lu W-J, Zhou L, Gao F-X, Zhou Y-L, Li Z, Zhang X-J, Wang Y and Gui J-F (2020) Dynamic and Differential Expression of Duplicated Cxcr4/Cxcl12 Genes Facilitates Antiviral Response in Hexaploid Gibel Carp. Front. Immunol. 11:2176. doi: 10.3389/fimmu.2020.02176
Received: 23 April 2020; Accepted: 10 August 2020;
Published: 11 September 2020.
Edited by:
Tiehui Wang, University of Aberdeen, United KingdomReviewed by:
Shih-Chu Chen, National Pingtung University of Science and Technology, TaiwanBrian Dixon, University of Waterloo, Canada
Magdalena Chadzińska, Jagiellonian University, Poland
Copyright © 2020 Lu, Zhou, Gao, Zhou, Li, Zhang, Wang and Gui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Wang, wangyang@ihb.ac.cn; Jian-Fang Gui, jfgui@ihb.ac.cn
 Wei-Jia Lu1
Wei-Jia Lu1 Jian-Fang Gui
Jian-Fang Gui