- 1Animal and Bioscience Research Centre, Teagasc, Grange, Ireland
- 2School of Medicine, Trinity Biomedical Sciences Institute, Trinity College Dublin, Dublin, Ireland
Diverse commensal populations are now regarded as key to physiological homeostasis and protection against disease. Although bacteria are the most abundant component of microbiomes, and the most intensively studied, the microbiome also consists of viral, fungal, archael, and protozoan communities, about which comparatively little is known. Host-defense peptides (HDPs), originally described as antimicrobial, now have renewed significance as curators of the pervasive microbial loads required to maintain homeostasis and manage microbiome diversity. Harnessing HDP biology to transition away from non-selective, antibiotic-mediated treatments for clearance of microbes is a new paradigm, particularly in veterinary medicine. One family of evolutionarily conserved HDPs, β-defensins which are produced in diverse combinations by epithelial and immune cell populations, are multifunctional cationic peptides which manage the cross-talk between host and microbes and maintain a healthy yet dynamic equilibrium across mucosal systems. They are therefore key gatekeepers to the oral, respiratory, reproductive and enteric tissues, preventing pathogen-associated inflammation and disease and maintaining physiological normality. Expansions in the number of genes encoding these natural antibiotics have been described in the genomes of some species, the functional significance of which has only recently being appreciated. β-defensin expression has been documented pre-birth and disruptions in their regulation may play a role in maladaptive neonatal immune programming, thereby contributing to subsequent disease susceptibility. Here we review recent evidence supporting a critical role for β-defensins as farmers of the pervasive and complex prokaryotic ecosystems that occupy all body surfaces and cavities. We also share some new perspectives on the role of β-defensins as sensors of homeostasis and the immune vanguard particularly at sites of immunological privilege where inflammation is attenuated.
Introduction
Defensins are small cationic peptides, present in a wide range of species across the animal and plant kingdoms. Characterized by a conserved six cysteine signature, three sub-classes of defensins have been identified as α, β, and θ (1). α-defensins arose from β-defensins in some mammals (2) and the bridging pattern between the three pairs of intramolecular disulphide bonds made by the six cysteines structurally separates both classes (3, 4). A high number of functional α-defensins have been described in the equine genome (3, 5) which, surprisingly given the short evolutionary distance were not retained in most artiodactyls (6), including cattle (7). Similarly, only β-defensin genes have been found in birds (8), and β-defensin genes have been identified in all the vertebrate genomes so far sequenced indicating the origin of this gene family very early in mammalian evolution (9). θ-defensins are a recently acquired, primate-specific class of peptide, which are formed by the merging of α and β classes of defensins (10). However, β-defensins have been most extensively studied to date. In the mammalian lineage, multiple gene duplication events and subsequent sequence diversification has resulted in a large family of β-defensin peptides with diverse amino acid sequence and virtually identical tertiary structures based on these characteristic intramolecular disulphide bonds (11). These peptides are the most numerous among vertebrates and are the subject of this review.
β-defensins are usually translated from characteristic two exon gene structures, the first of which encodes a pre-pro-peptide while the mature peptide is encoded by the second exon, containing the six cysteine motif (12). Recent sequencing of many more vertebrate genomes has facilitated a comparative genomics approach to characterizing the β-defensin gene repertoire, and species-specific clades have been identified. Current estimates of the number of β-defensin genes vary from 14 in chicken to 29 in pigs, 38 in dog, 33 in chimp, and 48 in mice and humans (8, 13–15), although final numbers will be subject to change as more genomes are correctly annotated and copy number variation (CNV) is accurately recorded. Comparative immunological analyses have also identified specific amino acid sites under positive selection which is likely to drive additional functional divergence between species. The absence of the classic α-helical region from all bovine β-defensins suggests a divergence in the mechanisms of action which may have contributed to the expansion identified in the ruminant clade (16). The implications of the apparent loss of α-defensins from the genomes of some livestock species and the expansion of the β-defensin family in a species-specific manner are only now becoming apparent, and could potentially be harnessed to improve animal health (17).
From a functional perspective, characterization of β-defensin genes and peptides has been essentially limited to model organisms including mice, rats, and humans. The specific genomic expansions detected in non-model organism species, identified through advances in technology combined with recently improved genome assemblies, have been described but not well characterized to date. Functional characterization in non-model species may shed significant light on the selection pressures that drove the dynamic changes in gene content between species and highlight the multifunctional promise that β-defensins may hold for the development of novel methods to control infection. Here, we build a case here for an intimate relationship between the β-defensin repertoire and microfloral diversity across the mucosal surfaces of the body. Given the constant requirement for appropriate cultivation of the microbiome, and the prevention of inflammation-associated pathology, it is likely that the host defense peptide arsenal in general, and the repertoire of β-defensin peptides in particular represent useful tools with which to maintain health and reduce infectious disease burden in livestock species.
Antimicrobial Meets Immunomodulation in Host Defense
β-defensins were traditionally viewed as exclusively antimicrobial molecules, as their induction in response to diverse bacterial, viral, parasitic and fungal infections was widely reported (18–20). As small cationic peptides, β-defensins are preferentially attracted to the negatively charged outer membranes of bacteria, with reported efficacy against Gram positive and Gram negative bacteria, fungi and enveloped viruses (21). Antimicrobial action is mediated via several mechanisms, including aggregation, pore formation, interference with cell wall synthesis, and prokaryotic membrane depolarization (22). The amphipathic nature of defensins enables them to insert into the phospholipid membrane of pathogens thereby destroying the integrity of the cell wall (10). Whereas, many in vitro studies have confirmed an antimicrobial role of several defensins, only a limited number of studies have verified their role in defense in vivo. Murine defensin 1 (DEFb1) Defb1-knock out mice showed delayed bacterial clearance from the lung (23) and increased Staphylococcal infection in the bladder (24). Expression of porcine β-defensin 1 (pBD1), an ortholog of human β-defensin 2, given at the time of challenge conferred protection against Bordetella pertussis in newborn piglets (25).
An interesting insight, resulting from the study of germ-free mice was the production of β-defensin precursors in the absence of infection (26). The concept of a germ-free animal was recognized more than a century ago by Louis Pasteur, although he also had the foresight to predict that bacteria-free existence is impossible (27). Generation of truly axenic mice requires that the pups remain sterile in the uterus and given what we now know about microbiotic priming in utero (28), these animals are not likely to be sterile.
As additional functions for β-defensins emerged (29), a broader interpretation of these molecules was adopted, leading to the term host defense peptide (HDP) (30, 31). In studies using embryonic kidney cells engineered to express various TLRs, human β-defensin-3 (hBD3) mediated activation of the transcription factor NFκB, depended on the expression of both TLR1 and TLR2 (32) demonstrating that TLR signaling is not restricted to recognition of microbial molecular patterns but also can be initiated by endogenous defensin peptides (33). β-defensins also serve to link the innate and adaptive immune responses—hBD3 can rapidly enter TLR4-stimulated macrophages and dampen the expression of pro-inflammatory genes (34). They also induce expression of the costimulatory molecules on monocytes and myeloid dendritic cells in a TLR-dependent manner by acting as chemoattractants for T-lymphocytes and immature dendritic cells (35). It seems that over the course of evolution, most β-defensins studied have acquired additional roles (a process known as neofunctionalisation) while retaining their original defense role (14). It is now becoming clear that antimicrobial and immunomodulatory functions of β-defensin peptides are not mutually exclusive and it is therefore logical not to compartmentalize their functions in either or, but to view their multifunctionality as an evolutionary work in progress, as critical elements with several roles in the complex function of defense against disease. Importantly, it is their multimodal action (Table 1) which has enabled β-defensins to retain their potency against infectious agents throughout the course of evolution (66). It is also their ubiquity of expression across mucosal surfaces that implicate them as fundamental players and sentinels of homeostasis and health.
Farmers of the Microbiome
The numbers of microorganisms across the entire exposed mammalian mucosal surface outnumbers the eukaryotic cell number of the host by orders of magnitude, indicating that constant monitoring, and management of the microfloral diversity and flux is required to prevent systemic colonization and pathology. However, microbial infections are actually the exception in the generally harmonious coexistence of animals with immense numbers of non-pathogenic microorganisms (67). This harmony exists due to many mechanisms including:
1. Secretion of mucus from cells lining the mucosa which reduce direct contact between exogenous antigens, particularly from potential pathogens;
2. Secretion of glycosylated antibodies known as Immunoglobulin A (IgA) found in colostrum and absorbed across the neonatal gut in a brief time after birth and
3. Production of HDPs (68), including β-defensins, predominantly at epithelial surfaces. Commensal bacteria that make it across the epithelium are usually phagocytosed by macrophages within hours but some studies have shown that they can reside within dendritic cells (DCs) for several days (69).
Analysis of the microbiome complexities across tissues and between divergent species is beyond the scope of this review. Instead, we focus on evidence underpinning a role for β-defensins in modulating the host's cross-talk with commensals and how changes in their expression contribute to disease. All mammalian neonates share a common trajectory of immediate exposure to an abundance of microbes in the nutrient rich environment after birth, resulting in colonization across all mucosal surfaces. Although, age-related changes in the development of the microbiome have not been well defined in livestock species, data from humans and mice shows that the early neonatal stages are characterized by high compositional changes which ultimately settles to a core characteristic microbiome (70). Extensive inter-individual variation is also reported, which we contend would likely contribute to significant variation in phenotypic performance across traits in livestock. Whereas, the principal focus to date has been on the intestinal microbiome, commensal populations of microbes are found at all mucosal sites, as well as on the skin. It is now becoming clear that the microbiome precludes pathogens by both inter-species microbial competition and host immune stimulation (71). It is our contention that evolutionarily conserved, multifunctional β-defensins hold the balance of power in farming the microbiome, thereby regulating the host cross-talk with prokaryotes and determining the success with which their eukaryotic subjects can defend themselves against opportunistic disease-causing pathogens.
The Embryome and Immunoeducation
For a neonatal eukaryote to emerge immunologically naive into the world would be a risky evolutionary strategy, particularly for livestock where animal densities, and bacterial loads can be high (72). Whereas, the sterile-at-birth hypothesis was accepted for years without refute, emerging evidence from humans and murine studies now indicate that microbial education of the neonatal immune system begins pre-birth via establishment of commensal microbial populations in utero (28). Their source, the complexities of their transmission to the uterus, and the mechanisms that regulate their proliferation within the nutrient rich environment of the neonate have yet to be established. In humans, bacteria derived from the maternal intestine have been detected in umbilical cord blood, amniotic fluid, meconium, and fetal membranes with no evidence of infection or inflammation (73). Additionally, a recent study in cattle has even postulated that these commensals could be derived from blood (74). It is proposed that this pre-natal introduction to microbial ligands (referred to as immuno-education) is critical for adaptive priming of the immune system and now forms the cornerstone of the Developmental Programming and Fetal Onset of Adult Disease (FOAD) hypothesis (75). This hypothesis holds that a lack of appropriate immuno-education early in childhood may result in dysregulated immune responses and the development of disease in later life. The critical implications of these findings for livestock rearing have yet to be seriously considered.
The environment during pregnancy is considered to be one of immunological privilege, where the introduction of some foreign antigens of paternal origin are tolerated without eliciting an inflammatory immune response (76). Immune privilege is not simply the absence of professional immune cells, but involves immune and non-immune cells acting synergistically to create a unique tolerogenic environment. During normal pregnancy, the fetus grows, and develops in such an environment, while, importantly, the uterus and maternal-fetal interface still retain the intrinsic capacity to respond promptly and efficiently to immunological challenge. Thought to represent an evolutionary strategy to prevent a hyper-inflammatory response immediately after birth, the immune system of the neonate exists in a state of inflammatory anergy. A pre-partum environment of immunological privilege coupled with inflammatory anergy and an immature adaptive immune system immediately post-partum requires a potent innate immune system for protection against inflammation and disease. We advocate that this need is met by the expression of multifunctional suite of HDPs, including β-defensins.
Umbilical vein endothelial cells produce DEFB1, DEFB4, and TAP (77) and additional β-defensin gene expression has been reported in bovine embryos themselves - with expression of DEFB103B at the 8-cell stage and DEFB123 at the 16-cell stage (78). Originally, it was hypothesized that expression of β-defensins in the “sterile” environment of the embryo indicated that these genes may play a role in development. However, in light of the evidence supporting the FOAD hypothesis, it is plausible that embryonic expression of β-defensins may play an important role in the regulation of the maternally-derived microbiome during development. β-defensin expression has also been documented during embryo development in sheep (79), where curiously expression reached a maximum immediately before birth and did not continue to rise into the immediate post-natal period when immune challenge is likely to be highest. Although a restricted number of β-defensin genes have been discovered in the chicken lineage, in ovo developmental changes in expression of these genes has also been reported (80). In poultry, it is already known that some bacteria can traverse the shell (81), so there is no reason to think that some commensals cannot do the same. It is also possible that the repertoire of β-defensins induced by the maternally derived prenatal commensals differ from the suite induced postnatally by environmental microbes, adding another intriguing layer to the protective functions and potential functional complementarity of these molecules.
Whereas, low level constitutive expression is associated with the regulation of commensal bacterial growth, elevated expression may signify the presence of pathogens. The human ortholog of the bovine gene shown earlier to be expressed in the embryo (DEFB103), was increased in inflamed fetal membranes (82), indicating a protective role during embryonic development. Similarly, elevated hBD2 at the time of amniocentesis was positively associated with increased likelihood of preterm birth (83). Human papillomavirus has also been shown to upregulate β-defensin expression by amniotic epithelial cells (84), hinting at an integral ubiquitous protective role for these molecules during pregnancy. In addition to the prenatal microbiome, a prokaryotic “top up” occurs via inoculation from the birth canal during the birth process. Interestingly, where this does not occur, as in cesarean sections, detrimental changes to health can result (85). Although not common in livestock, cesarean section rates are increasing in cattle, some of which are associated with poorer clinical outcomes, especially under field conditions (86).
Immediately after birth, the neonate is coated with vernix caseosa, a creamy biofilm which develops on the skin of the fetus toward the end of pregnancy (87). Although not studied in livestock species, the vernix in humans includes multiple HDPs including defensins (88), which facilitates extra-uterine adaptation of skin. Unlike with humans, most mammals not only eat the fetal membranes but also extensively lick their offspring after birth which possibly spreads the protective effect of the vervix across the newly exposed neonate. Relevantly, artificially reared calves are often removed from their dam early post-partum and thereby forego these potentially protective actions but the consequences for their developing immunity has not been previously considered.
Additional microbes are obtained from the udder or from food, predominantly milk in the early post-natal period (see mammary microbiome later). β-defensin-2 (HBD2) is expressed in human breast milk and was found to be significantly higher in colostrum samples (89). The same study showed that the recombinant BD2 peptide was effective against both Salmonella and E. coli bacterial species. A core microbiome has also been defined in bovine colostrum, and differences in populations documented in milk between primiparous and multiparous cows (90). This finding suggests that calves born to multiparous cows may derive a different starter colonization culture which could differentially affect both the development of their intestinal microbiome, their β-defensin expression profile and potentially their subsequent disease susceptibility (91).
Oral Microbiome
The neonatal oral cavity is the first point of contact with dietary-derived antigens and the initial colonization cultures are milk derived. In contrast to livestock species, the human oral microbiome has been defined (92). One proteomic analysis has identified over 3,700 human and 2,000 microbial proteins in human saliva samples (93). With now over 1,000 bacterial species identified thus far, the composition and activity of this ecosystem is thought to have enormous relevance to oral health and disease (94). β-defensins have been shown to be extensively expressed in the oral cavity across multiple species. In humans, HDPs are extensively produced by epithelial cells lining the oral cavity, and are referred to as guardians of the oral cavity (95). Expression of β-defensin 1 and 2 has also been documented in biopsies taken from the salivary gland in humans (96). In rats, orthologous genes (RBD-1 and −2) were localized to the acinar and striated duct cells of the major salivary glands, and expression was also shown to be responsive to bacterial endotoxin, LPS (97).
Saliva is key to the maintenance of homeostasis within the oral cavity, and cattle produce over 100 L per day (98). The fact that the majority of livestock do not develop digestive illness routinely in such a high antigen environment, especially in the context of intensive farming, shows how robust oral defense mechanisms must be. Given the studies in humans referred to above, it is likely that the salivary proteins have an enormous role to play in oral and intestinal homeostasis. A recent analysis using three different approaches identified 402 salivary proteins and 45 N-linked glycoproteins in bovine saliva, including multiple HDPs (99). In addition, as a direct result of the licking and suckling processes between mother and calf under natural rearing conditions, this extensive salivary proteome will have important implications for neonatal health as well.
A significant proportion of young calves are artificially reared on milk substitutes or on waste milk, often from mastitic cows whose milk is not suitable for human consumption. Such waste milk is known to contain high levels of pathogenic bacteria, including antibiotic-resistant E. coli, although studies have claimed that its use as a feed for calves do not affect health (diarrhea) or production parameters (100). However, the impact on the microbiome and on immunity in the GI tract was not examined.
Respiratory Tract
Inhalation is a major route of disease transmission in human and livestock populations and the lung microbiome is now well-characterized both in health and disease (101, 102). The respiratory microbiome is of critical importance in livestock species as respiratory disease caused by infection is a major cause of losses, compromised animal welfare and morbidity. Pneumonia is a major respiratory disease caused by bacteria, and as a result, a particular focus has been applied to characterizing the upper respiratory tract in respiratory disease across multiple livestock species (103–105). Stress induced changes (e.g., transport) in the nasopharyngeal microbiota has also been implicated as a contributory factor to disease susceptibility (106, 107).
Tracheal antimicrobial peptide (TAP) was one of the first β-defensin HDPs characterized in bovine lung epithelial cells (108). TAP is now extensively researched for its potential role in resistance to Mycoplasma bovis (109), another major contributor to bovine respiratory disease. Studies using homozygous mBD-1-deficient mice showed that a loss of mBD-1 results in significantly delayed clearance of Haemophilus influenzae from lung, providing a direct link between β-defensin expression and pulmonary immunity (23). Investigations into human airway inflammatory disease have shown that β-defensins participate in antimicrobial defense in the respiratory tract during disease (110), and that the bronchoalveolar lavage fluid concentration of HBD-2 may be a useful marker of airway inflammation (111). One the most recalcitrant respiratory diseases affecting humans and animals is tuberculosis, caused by mycobacterial species of bacteria. Interestingly, artificial induction of β-defensin 2 (mBD2) in bronchial epithelium contributes to improved control Mycobacterium tuberculosis infection in mice (112).
Furthermore, the efficacy of these peptides is not limited to bacterial pathogens. In cattle, expression of multiple β-defensins has been documented in bronchoalveolar lavage from calves infected with bovine respiratory syncytial virus (113). Murine β-defensin 3 has also shown to have anti-viral effects against influenza virus, both in vitro and in vivo (44).
Digestive Tract
The digestive tract has been the site of the most detailed microbiome analyses across all species studied to date, and a diverse repertoire of microeukaryotes have now been identified (114). However, the term “digestive tract” belies the functional complexity that constitutes a number of physiologically distinct regions—esophagus, stomach, duodenum, small, and large intestine and colon, and this is reflected in a very diverse regional-specific microbiome (70). In ruminants, additional complexity exists in the form of a multi-chambered stomach, known as the rumen. Major cellular differentiation exists in the epithelial structures across the intestinal regions between the foregut (rumen-reticulum and omasum) and hindgut (abomasum and small and large intestine). The ruminal epithelium is 4-layer stratified squamous structure, whereas the intestinal epithelium is a single layer of columnar epithelial cells protected by a double layer of mucous. Furthermore, whereas the ruminal epithelial layer lacks an underlying organized lymphoid tissue in the lamina propria, the intestinal lamina propria consists of defined Peyers patches and specialized “M” cells diffused by lymphatic follicles rich in immune cells [for review see Garcia et al. (115)].
In the newborn calf, the rumen is still inactive and rudimental. Instead, the esophageal groove routes the easily digested milk directly to the fourth stomach, the abomasum, which accounts for 70% of the total volume. Development of the rumen occurs during the first 12 weeks of life, during which time the calf transitions from a monogastric to a ruminant, essential for efficient utilization of forage based diets. It entails growth and cellular differentiation of the rumen, and results in a major shift in the pattern of nutrients being delivered to intestines, liver, and peripheral tissues of the animal (116). The physiological transition is stimulated by a defined and progressive sequence of microbial colonization and is essential to enable absorption and utilization of digestion end products from forage. As would be expected, it should be of no surprise given the functional divergence between these tissues that a region-specific microbiome has been identified in the bovine rumen (117). Mucosa-associated bacterial populations are distinct from those inhabiting ingesta, and these divergent microbial populations are associated with signature immune gene expression profiles (118). Epimural bacteria in pre-weaned calves differs significantly from content-associated community (119), and the different bacterial populations have been associated with divergent mRNA and miRNA expression profiles linked with epimural bacterial populations in the neonatal calf (120). In addition to the resident microbiome, environmentally-induced (transport and diet) perturbations in the rumen microbiome have recently been characterized (121).
Within the heterogenous environment of the intestine, β-defensins are known to keep the peace (122) by adjusting the balance among bacterial populations and to control homeostasis (123), although this has predominately been studied in humans. An important role for α-defensins has been documented in the equine intestinal tract (3), and given the intriguing differences in structure and presumably, microbial load between monogastric (horse and pig) and ruminant livestock species, may have contributed to a divergence in function of the defensin sub-classes. The absence of this class may be compensated for by the expansion of β-defensins in the rumen, although expression of β-defensins has been shown in the pig stomach, and in intestinal epithelium (124, 125). How these defensins might perform in the environment of the functional rumen remains an open question.
Short chain fatty acid (SCFAs) are a major source of energy in cattle, initially produced by the beneficial microbiota in the colon of pre-ruminant calves between 2 and 4 weeks of age, when high concentrations of lactate and butyrate are observed. These SCFAs stimulate epithelial cell proliferation leading to longer villi, tight junction, and immune system development. Interestingly, acetate, propionate and butyrate have recently been documented to upregulate BD1 and BD2 in human epithelial cells (126). Amongst the analyses of immune changes induced in response to SCFAs in cattle, a recent study showed decreases in LAP, TAP, and DEFB4A expression in rumen epithelium after infusion with butyrate (127). In sheep, maximal expression of β-defensins (oBD1 and oBD2) was detected in the rumen during the first weeks of life and also in the digestive tract prenatally (128). This evidence would support a role for defensins in managing the microbial interface, especially during initial post-natal colonization of the intestine and rumen.
Comparing the epithelial transcriptome of germ-free vs. conventionally reared mice during intestinal colonization, significantly increased expression of defensins (DEFB37 and DEFB39) in the tip of the colon in the latter group was reported. However, a significant reduction in expression was detected in the ileum, indicating a regional specific β-Defensin response to colonization (129). Interestingly, transgenic mice expressing human defensins protected against intestinal salmonella, again reinforcing a protective role for these molecules (130). Similarly, in chickens, dietary supplementation with butyrate led to a increased defensin gene expression in the caecum and a simultaneous reduction in S. enteritidis carriage (131).
The colon represents the most distal portion of the digestive tract, and the expression of HDPs has been reported in human colon tissue (132). Similarly, bovine enteric β-defensin was named after it was originally found expressed in the small intestine and the colon (133). As a result, it is proposed that their antimicrobial activity can be harnessed as a potential therapy for infectious and non-infectious diseases of the colon.
Mammary Gland
Human breast milk has recently been shown to have a resident microbiome (134) concurrently with β-defensin expression. β-defensin 2 in human breast milk has been shown to have broad antimicrobial activity (89), which is thought to contribute to controlling the proliferation of microbes in this ideal growth medium. It is not surprising that microbial analysis of bovine milk has shown similar commensal bacterial populations and interestingly, changes in diversity have been associated with disease (135). Healthy mammary gland in cows has also been shown to produce β-defensins including TAP (136) and Lingual Antimicrobial Peptide (LAP) (137), and expression is widespread throughout the mammary gland (138). Induction of these genes have also been postulated as markers of the early response to inflammation (20) and mastitis (135).
It is likely that protection of the mammary gland is a primary function of β-defensin expression in milk. While the analysis of HDP action usually focuses on known pathogens, these peptides have not been tested for efficacy against newly characterized commensal species, so their role in regulating mammary gland microbial homeostasis remains unclear. Is has also been suggested that β-defensins in milk may help regulate the intestinal development in the neonate but this remains speculative until further functional characterization has been performed (91).
Reproductive Tract
The female reproductive tract (FRT) is well endowed with HDPs including β-defensins (139, 140) and during each window of physiological transitions (pregnancy to non-gravid), the cross-talk between the immune and reproductive systems provided by endogenous HDPs may play important roles in dampening the immune response to foreign antigen such as sperm but also in regulating immune tolerance of an allogenic fetus during pregnancy (141). In mice, β-defensins are more highly expressed in the vagina than the uterus, with uterine levels peaking during the estrogen-dominant phase of the cycle (142, 143). The role for hormonal regulation has been supported with findings that estrogen increases DEFB4A expression by primary human uterine epithelial cells in vitro (144). In the ovine oviductal epithelium, SBD1 expression is also increased by estrogen (145). However, estrogen appears to have an opposing effect on DEFB4A regulation in the human vagina, with expression in epithelial cultures decreasing when estrogen is used alone (146). A separate study indicates estrogen increases LPS-driven DEFB4A expression in the same model, with progesterone decreasing it (147).
Comparisons of virgin and pregnant bovine uteri showed divergent dominant bacterial phyla between groups (148), suggesting that pregnancies are established and maintained in the presence of a uterine microbiome in cattle too. For the majority of the year, livestock species are pregnant, and it is around reproduction when follicles are growing and estrogen is secreted but examination of their influence on β-defensin expression has not been examined to date. It is well established that after pregnancy the female (and specifically the cow) must shift from an immunosuppressive (or immunotolerant) state during pregnancy to a state of heightened immune activation with concurrent inflammation to expel fetal membranes, clear infection and restore homeostasis (149). Pro-inflammatory signals also regulate β-defensin expression in the FRT. Sperm and seminal plasma can activate inflammatory cytokines, and physiological inflammation in the female tract can promote beneficial pregnancy outcomes. IL-1β and TNFα drive expression of DEFB4A by primary human trophoblast cells (150) and DEFB4A and DEFB103 in endometrial epithelial cells (151, 152). These cytokines may mediate the observed innate immune response to pathogens detected in the FRT. Bacterial vaginosis drives an increase in secretion of hBD-2, the peptide encoded by DEFB4A (153).
In humans, unprotected sex is also known to change the microbiome of the vagina and sexual transmission of commensal and potentially pathogenic bacteria (154). Expression of human and mouse BD1 has been documented in the lower urinary tract (155) and increased β-defensin (HBD2 and HBD3) expression has also been documented with inflammation of the cervix (156), supporting an important role for peptides in mucus for defense of the FRT. It is probable that β-defensins evolved to regulate this microfloral influx during natural reproduction. Interestingly, a range of HDPs, including hBD-1 are found in the cervical mucus plug and this pregnancy-generated structure exerts antimicrobial activity against a number of relevant pathogens (157).
Bovine endometrial cells produce β-defensins (158) and these are significantly elevated during uterine disease (159). Viral infections are also thought to play a role in endometritis (160) and therefore it is of interest that β-defensins may also mediate the anti-viral response in the FRT, as hBD-2 levels in the cervico-vaginal lavage fluid of HIV-positive women correlate with anti-HIV activity (161). In contrast, gammaherpesvirus infection of the murine cervix attenuates β-defensin expression, which may explain the predisposition toward bacterial infection of the cervix observed in virally-infected animals (162).
Some additional intriguing insights have emerged regarding the role of commensal microbe populations in the human reproductive tract which have important implications for fertility in livestock species (163). For example, a commensal bacteria, Staphylococcus epidermidis was found in the oviductal fluid of parous mice and abolition of these populations by intraperitoneal antibiotic injection led to disturbed uterus- embryo interaction and derangement of embryo spacing (164). This new paradigm in our understanding also extends to the role of β-defensins within the male reproductive tract. Extensive expression of these molecules has been documented across the male tract of multiple mammalian species, particularly in the epididymis of male rats, mice, rams, horses and cattle (51, 165, 166) [for review see: (167)]. The epididymis is a single, convoluted duct, through which sperm progressively acquire functional competency for fertilization. Highly regionalised expression expression profiles of proteins including β-defensins give rise to a dynamic intraluminal environment (168). One particular β-defensin, (BD126) has been documented to play a number of roles critical to sperm survival, motility, and interaction with the female reproductive tract (49, 50, 169). In agreement with their broad role in reproductive physiology, a recent study has also shown that β-defensin gene knock-out male mice are infertile (170). Interference with DEFB1 function also decreases both sperm function but also with bactericidal activity (171), and interestingly adding back DEFB1 restored both functions. Investigations in livestock species are again preliminary but recently a β-defensin haplotype has been associated with sperm function and fertility in bulls (51, 172).
A remarkable recent discovery has been the identification of a seminal fluid microbiome in mice (166) and humans. Seminal fluid bacterial diversity has been linked to semen quality and HIV viral load in humans (173, 174). It is logical, given the abundance of amino acids and other nutritional substrates in basic seminal fluid, that it would promote bacterial growth and this microbiome is likely to influence the colonization of the FRT (175). In fact, researchers now propose that this microbiome impacts directly on the etiology of infertility (174). In this context, it now seems plausible to propose that β-defensins evolved to regulate the microbiome in seminal fluid and prevent the growth of bacterial populations that may be detrimental to either sperm quality or uterine health. However, detailed follow-on studies are required to further investigate this hypothesis.
Future Perspectives—β-Defensins and Reproduction—Sperm Wastage or a Hidden Immune Agenda?
The male reproductive tract is home to the germ cells and reproductive fluids are ideal environments for microbial growth and particularly viral transmission (176). Of the millions of sperm ejaculated, very few make it successfully to the upper female reproductive tract, where fertilization occurs. Logically therefore, the majority of sperm are non-fertilizing sperm and are presumed wasted. We propose that on the contrary, it may be the case that these β-defensin-coated sperm may play a secondary role in the prevention of ascending infection through neutrophil recruitment and neutrophil mediated killing of microbes in the female tract. Recently a direct link between the microbiome and neutrophil recruitment has been established (177) and a role for β-defensins in neutrophil chemoattraction has been previously documented, albeit in a disease context (178). Further studies are urgently required.
β-Defensins—diversity, Dysbiosis, and Disease
The healthy microbiome can be characterized in terms of diversity, stability, resistance and resilience (179). Compositional and functional alterations that compromise any of these parameters is referred to as dysbiosis reflected by a bloom of pathology-associated microbiota (referred to as pathobionts) and/or a loss of commensals and consequential reduction in microfloral diversity (180). Whereas, diversity is beneficial against disease (105), dysbiosis is associated with a range of pathological conditions in mice (181), humans (182), and cattle (183). The resulting dysbiosis would provide opportunities for opportunistic pathogens to invade mucosal sites, cause excessive inflammation accompanied by an associated loss of metabolites leading to dysregulated immune cell responses, pathology and disease (see Figure 1).
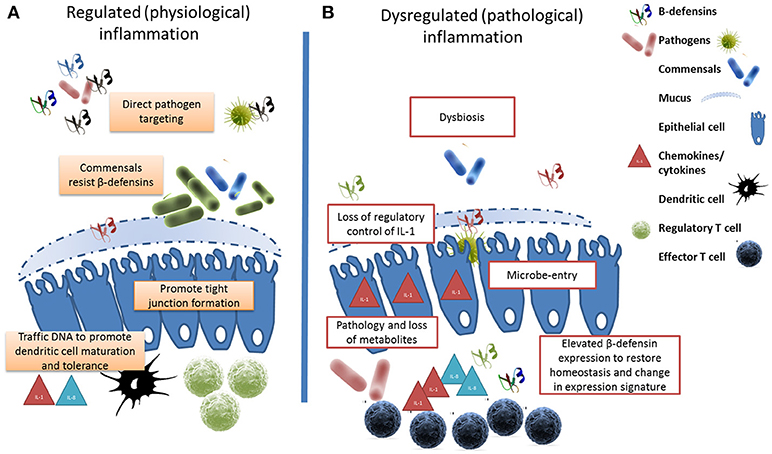
Figure 1. β-defensins manage the microbial interface. (A) Under homeostatic conditions, β-defensin-mediated preferential trafficing of microbes to dendritic cells promotes the induction of tolerance and the control of inflammation. Similarly, prenatal expression of β-defensins induces inflammatory anergy while education of the immune system occurs. (B) Stessor-induced dysbiosis results in dysregulation of inflammation and the loss of metabolites due to cell damage leads to a bloom of pathobionts. In an effort to restore homeostasis, elevated β-defensin expression occurs resulting in a changed expression signature. The genes encoding these host defense peptides vary in number between individuals resulting in a personal β-defensin signature which may be more or less effective at preventing a switch to pathological inflammation.
The current dogma suggests that colonization of mucosal surfaces begins with a period of cell-mediated immunity which is followed by the development of regulatory cell populations (e.g., regulatory T cells), which prevent excessive inflammation-associated pathology. A direct relationship has now been established between the commensal microbiome and the regulation of inflammation as the proinflammatory cytokine Interleukin 1-alpha (IL-1α) is a key regulatory target of commensal bacteria (184). Although, the mechanisms involved in immune tolerance to first colonizers of mucosal tissues are currently unknown (116), commensal (and probiotics) bacteria are now thought to contribute directly to the regulation of inflammation by the secretion of metabolites, via the prevention of LPS binding to host cells, and through the attenuation of NFKB mediated IL-1β production (185, 186). In fact, whereas a recent studies have shown commensal-induced IL-1β via the NLRP3 inflammasome (187), activation via another inflammasome complex (NLRC4) has been proposed to discriminate between pathogenic and commensal bacteria (188). Macrophage-derived IL-1α has been shown to significantly increase the expression levels of the DEFB4 in intestinal epithelium (64). This and related work in the human intestine also suggests that defensins are key regulators of bacterial diversity and thereby tissue homeostasis (122). Intestinal phagocytes are anergic to TLR ligands or commensals but constitutively express pro-IL1β, and it is now thought that HDPs initiate IL-1β posttranslational processing (189). The close association between a dysregulated microbiome and altered β-defensin expression has been most intensively studied in the context of the chronic inflammatory diseases of the human intestine known as Inflammatory Bowel Disease (IBD) (190). Clinical studies have linked the defective expression of β-defensins to the reduced killing of certain microorganisms by the intestinal mucosa of patients and directly couple dysbiosis to primary β-defensin immunodeficiency (182, 191).
The epithelium in inflamed intestinal segments of patients with Crohn's disease is characterized by a change in tight junction protein content and composition (192), resulting in barrier defects leading to luminal antigen uptake which causes mucosal inflammation. The tight junction is dynamic, multi-protein complex that forms a selective permeable seal between adjacent epithelial cells and demarcates the boundary between apical and basolateral membrane domains. Apical tight junction proteins are critical in the maintenance of epithelial barrier function and control of paracellular permeability to prevent disease (193) and an established virulence mechanism for pathogens is to limit pathogens by stabilizing tight junctions (194). HDPs, and specifically defensins are now thought to promote the resolution of inflammation and endotoxin resolution via the formation and maintenance of tight junctions (195). Defensins have also been proposed as potential markers of mucosal permeability (196). HBD-3 increased the expression of several claudins, elevated the transepithelial electrical resistance, and reduced the paracellular permeability of keratinocyte layers (36).
Although it seems the consensus from most studies is that these peptides are immunosuppressive (197) and attentuate inflammation (198), there are reports that some β-defensins amplify the immune response [as reviewed by Semple and Dorin (199)]. Some studies report a positive correlation between HBD2 expression levels and that of the potent chemokine IL-8, leading to additional inflammatory cell recruitment and acceleration of the pathogenesis of IBD (200). HBD2 has also been shown to activate pro-inflammatory cytokine expression (201) in peripheral blood cells, with similar reports for HBD3 (32) in macrophages. However, it is difficult to effectively compare between studies with various disease models, as the induction of β-defensins may be a protective attempt by the host to reduce exacerbation of the inflammatory cascade and will likely be affected by the stage of disease, the epigenetic landscape and microbial load. Furthermore, studies have also demonstrated that the structure of the β-defensin peptide will determine the biological effects detected—for example, the canonical structure of hBD3 is required for its to enter macrophages and exert its immunosuppressive effects (34).
β-Defensin Immunogenetics
Host genetic variation has a significant effect on the microbiome across multiple body sites (202, 203), and we contend that the extensive variation uncovered in β-defensin genes contributes to phenotypic diversity in several livestock-relevant traits. Recent studies have associated individual SNPs in β-defensin genes with health and production phenotypes including somatic cell count (204) and milk constituents (205). More recently, in cattle, haplotypes have been uncovered which regulate important traits like bull fertility (172). The widespread expression profile for these genes in cattle is shown in Figure 2.
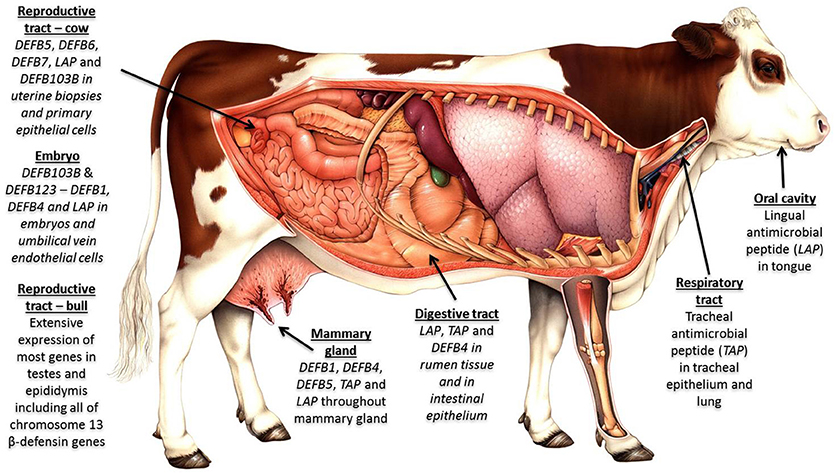
Figure 2. β-defensins as regulators of the microbiome and biosensors of immune homeostasis: Expression of β-defensin genes in bovine tissues—four genomic clusters of these genes exist in cattle on chromosome 8 (4 genes); chromosome 13 (19 genes); chromosome 23 (5 genes); and chromosome 27 (30 genes). The β-defensin genes on chromosome 27 are the least well conserved between species, are known to be CNV and show an expansion in number in cattle. Tissues with immunological privilege (reproductive tract) show particularly extensive β-defensin expression patterns.
β-defensins evolved through a complex mechanism of duplication which has led to highly polymorphic gene CNV (206). β-defensin gene CNV can affect disease resistance (207, 208)–it is hypothesized that increased gene-copy number contributes to susceptibility to inflammatory or autoimmune diseases but is protective against infectious disease (209). Although data in cattle are limited, β-defensin loci account for 7 of the top 25 most CNV regions across the bovine genome (210, 211). Upwards of 20 gene copies are seen for some immune genes in various cattle breeds sequenced, including for LAP, TAP, and DEFB5, genes which have been previously shown to be upregulated in response to infection, particularly in mammary, lung and uterine tissues [for review see Meade et al. (17)].
Several multigene families are subject to birth-and-death evolution and rates of gene gain and gene loss vary considerably between closely related species or even between individuals of the same species (212–215). These genes represent a major source of new (or additional) biological functions through a process known as neofunctionalisation (216). Selection for noncoding regulatory regions in hBD103 in human populations from Asia has also recently been described (217), which is thought to be a response to selection pressure from influenza viruses in the region. In cattle, the selection pressure thought to contribute to the β-defensin expansion was originally the evolution of the rumen (218). Rumen specific defense mechanisms are important to ensure the balance between immune surveillance of the diverse gut microbiota (219) and the maintenance of the integrity of the gastrointestinal epithelial barrier (123). However, the presence of these genes in monogastrics including horses (220) and pigs (221) supports an alternative rationale. Furthermore, biological systems most affected by changes in the number and organization of genes in the cattle lineage during evolution include reproduction and immunity as well as lactation and digestion (218). The intensive artificial selection that cattle have been selected to over the last 60 years, is likely to have further shaped the β-defensin genetic variation remaining in extant breeds.
The extensive expression of the expanded repertoire of β-defensins across the reproductive tract of mammalian species highlights the importance of protecting the gametes over the course of evolution. The herd breeding structure of most mammalian species has been postulated as a potential selection pressure where an expanded β-defensin peptide family would provide enhanced protection against ascending infections of the reproductive tract. Intriguingly, the genomes of avian species have not undergone expansion of these genes, but the loss of the penis in males of most avian species (222) may preclude the requirement for an expanded β-defensins family. Male ducks and Ostrich members of this clade have retained their penis and although annotation is far from complete, the duck genome is currently estimated to contain 16 defensins distributed over 3 scaffolds, a number that was slightly higher than that of the 14 defensins found in chicken (223). A preliminary search of available Ostrich genome sequence identified a number of conserved 6 cysteine sequences in addition to the currently identified β-defensin family members (unpublished data) potentially representing a β-defensin expansion. However, conclusive evidence is not yet available and it is likely that other selective pressures account for the expansion detected in the mammalian lineage.
β-Defensins—Decoupling Inflammation From Defense
A more convincing selection pressure contributing to the expansion of the β-defensin repertoire in mammals is their requirement to maintain homeostasis in environments with a high microbe diversity as well as immunoprotection at sites of immune privilege. Inflammation in specific body sites (e.g., brain, eyes, testes) would be detrimental to the survival of the species causing pathology and possibly death. In immunopriviledged sites, immune molecules (including IL-1) can be present at high concentrations without causing inflammation indicating important immunoregulatory mechanisms. Immune cell phenotype, including regulatory T cells and macrophages, as well as the expression of anti-inflammatory cytokines is known to be critical to maintaining a functional state of hypoactivity (224). Therefore, defense against pathogens is orchestrated by alternative means, and β-defensins are likely to have a principal role.
The testes are one such site (225), where haploid gametes require protection and isolation from inflammatory immune cells and in this environment, regular inflammatory mechanisms are attenuated. The blood-testis barrier is maintained by several types of cell junctions, which limit the access of systemic immune contents to the inner reproductive compartments (225). Cytokines and chemokines are expressed in healthy tissue and function during normal testicular development (226), and increased concentrations of inflammatory cytokines have been associated with increased reactive oxygen species and histone abnormalities in sperm (227). Whereas, levels of the anti-inflammatory cytokine IL-10 in bull seminal fluid is positively associated with sperm motility, inverse correlations was detected with proinflammatory cytokines (228). High levels of the chemokine IL8 has also been positively associated with human sperm defects (229). It is our contention that the requirement for protection of the gametes and reproductive tract over the course of evolution led to the expansion of β-defensins in this regulated environment, and this thesis is supported by the extensive expression of these molecules in the epididymis across all species in which they have been studied to date (230). The requirement for β-defensins in immunopriviledged sites is not limited to the male reproductive tract and may also explain their documented expression in female reproductive tissues during pregnancy and also in the brain and eye (231).
Interestingly, studies on LPS stimulated macrophages shows that TLR-induced genes fall into two distinct categories on the basis of their functions and regulatory requirements. Referred to as “proinflammatory” and “antimicrobial genes,” a tailored innate immune response is controlled via epigenetic modifications to individual promoters (232). Methylation and chromatin modifications thereby permit the decoupling of proinflammatory from antimicrobial effector mechanisms of the innate immune system which maintains protection of the host whilst simultaneously minimizing inflammation-associated pathological damage. Such mechanisms are now known to form the cornerstone of the developing paradigm of innate memory, and have enormous relevance for the understanding of LPS tolerance, immune cell anergy and immunosuppression.
Evidence for epigenetic regulation of β-defensin genes supports this concept. The epigenetic enzyme, HDAC1 has been shown to controls BD in lung epithelial cells (233). Further work identified that DEFB1 expression was associated with specific histone marks (234). Interestingly, ablation of the microbiota has genome-wide epigenetic effects (235), and can affect transcription factor binding (236) thereby providing a direct mechanism by which commensal bacteria can regulate the immune response in a gene-specific manner. However, epigenetic changes at HDP gene promoters have not been hitherto examined.
β-Defensins and the Response to Vaccination
β-defensins may also prove useful directly as broad-spectrum adjuvants which are required for improved vaccine design in cattle (237). As an endogenous ligand for Toll-like receptor 4 (TLR-4), inducing up-regulation of costimulatory molecules (33), β-defensin expression is thought to contribute to the establishment of a beneficial Th1 response via dendritic cell activation and increased expression of cytokines such as IFNy, IL-12 and IL-6 (238). It has been suggested that β-defensins form antigen complexes (in a manner similar to how they bind to sperm) which may be an important mechanisms of microbe trafficking to antigen presenting cells [as reviewed by (239)] which would enable appropriate education of the neonatal immune system as well as their priming during the formation of innate and adaptive immune memory. In vivo murine models have shown that β-defensin 2 promotes anti-tumor NK and beneficial T cell responses (240).
β-defensins are known to share significant structural similarity with chemokines and a major known mechanism by which defensins work is as chemoattractants for immune cells. Mediated via both CCR2 and CCR6 receptors (35, 241), β-defensins thereby attract myeloid and lymphoid cells to mucosal sites where they are expressed, and thereby linking innate, and adaptive immunity. They also function to increase the uptake of DNA, and promote Interferon α expression (242) which further suggests their utility in enhancing vaccine responses. DCs are usually found in mucosal tissues where they monitor the local environment for signs of pathogenic (or commensal) invasion and are integral to ensuring that pathological immune responses to harmless antigens do not develop (243). It was commonly considered that these commensal containing DCs were confined to the respective mucosal immune systems, where they prime tolerant responses of B and T cells (244). However, more recent studies have shown that homing of these DCs to the lymph nodes can lead to the development of regulatory cell populations which promote systemic tolerance (245).
Defensins are also found in B cells (246, 247), confirming that these peptides are capable of contributing to a prolonged cellular and humoral response to a pathogen (248), and this has potentially important consequences for vaccine development across all species. Therefore β-defensins are the recruitment portal through which systemic immunoregulatory DCs are recruited and thereby directs the priming of the adaptive immune response (249). This implies that neonatal β-defensin expression profiles, in regulating microbiome development, can regulate vaccine adjuvancy and thereby the efficacy and duration of protection provided (245). These studies have critical relevance for the design of next generation vaccines and adjuvants in livestock species.
Commensal Mechanisms to Resist HDP Antimicrobial Action
A key focus of future research will identify the multiple mechanisms by which commensals can tolerate or resist the antimicrobial effects β-defensin HDPs. Large structural variations in LPS antigens have been documented across bacterial phyla, leading to different host efficacy of LPS sensing and resulting immunogenicity (250). Predominant commensal members of the human gut microbiota show temporal stability despite exposure to the host inflammatory response, but the mechanisms involved are poorly understood. A recent study identified that the dominant gut phyla in the human intestine are resistant to HDPs, and that the resistance mechanism of the common gut commensal Bacteroides thetaiotaomicron, is via an LPS modification (251). The authors found that the bacterial phosphatase LpxF catalyzes the removal of a phosphate group from LPS, thereby reducing AMP-dependent membrane disruption. As LpxF orthologs are widespread, this strategy may be common across gut commensals to resist host inflammation.
Furthermore, the absence of virulence factors in commensal bacteria within the bovine rumen has been suggested as key to adaptive processes that result in their exploitation of epithelial tissue for nutritional benefit as well as subversion of a detrimental immune response (252). However, full characterization of the diverse bacterial strains which exist within the microbiome will take some time.
Concluding Remarks
Re-evaluation of the interrelationships between the microbial world and their eukaryotic subjects is well underway. It is now clear that physiological sterility is essentially impossible and microbiomes are being documented across sites from the developing fetus to healthy breast milk, tears and semen. Although these findings could have major implications for the design of new therapeutics, care is warranted and careful validation is required (253) before translation of these findings from a limited number of studies to other species can be advocated. However, untargeted, black box approaches to disease prevention or treatment are no longer seen as likely to provide long term protection. In contrast, a new appreciation for our resident prokaryotic kingdom is emerging and it is increasingly seen as key to homeostasis and health. Although bacteria are the most abundant component of the microbiome, and the most intensively studied, the microbiome actually also consists of viral, fungal, archael and protozoan communities (184, 254–256), about which comparatively little is known (184, 257–259), even in model species.
β-defensins may even have a neurological role (260). The ability of the microbiome to regulate serotonin production identifies mechanisms by which they might also regulate neural chemistry (254). Through their inhibition of glucocorticoid elevation in response to stress, defensins are even thought to prevent stress-induced immunosuppression (255). Although at first glance, the psychological effects of the microbiome in livestock species may not seem relevant, stress is of increasing concern in ethical animal production systems. Stress is a critical causal factor that not only contributes to lost production and risks to human welfare, it can also affect disease susceptibility. One study in cattle found that dexamethasone-treated calves had significantly lower expression of TAP and LAP β-defensin expression in the lungs, which was claimed as a critical contributing factor to disease susceptibility (256). β-defensin expression has been documented in human cerumen of the ear (261) and the recent discovery of β-defensin expression in the pituitary gland of fish (262) and in the murine brain (263, 264), suggests that defining the full range of β-defensin activities have yet to be defined.
Consequentially, in the absence of sterility, basal (or constitutive) expression in vivo may not exist. Perhaps, basal expression, of β-defensins for example, tells us a lot more than we realized previously and may provide insights into the status of the microbiome which will inevitably affect every phenotype of interest and ultimately disease outcomes. High basal expression may actually represent an induced response indicative of colonization (immunoeducation), infection or indeed identify eukaryotes with a more robust prokaryotic management system, and therefore an immune advantage. Furthermore, the occurrence of mysterious “natural antibodies” at birth may reflect priming that occurs during in utero immune programming.
The use of antibiotics has been shown to enhance intestinal colonization of enteric pathogens (71), and have enormous significance for animal production systems. New green antimicrobial drugs are urgently required in medicine and in veterinary medicine in particular (265). Although significant limitations, both technological and economic hamper the current therapeutic use of β-defensins, these are not insurmountable and still offer hope for the rationale design of new effective drugs (266). HDPs have been proposed as potentially useful alternatives to antibiotics in feed (267), and they exhibit significant potential to improve intestinal barrier function, animal health and productivity (268). A recent study showed that feeding them to juvenile ruminants led to increased body weight, average daily weight gain, enzymatic activity and positively influenced on ruminal fermentation (269). In addition, goats treated with AMPs had higher rumen microorganism diversity indices than the control groups. For new treatments to be successful, selectivity is required—and all available evidence to date suggests that HDPs are ideally suited to this role.
β-defensins have important roles not just for the prevention of infectious disease, metabolic diseases are a major limiting factor to livestock production systems (270), and are set to grow in importance as agricultural production intensifies, and are regarded as perturbations associated with production. An exquisite symbiotic relationship therefore exists between the microbiome and the metabolome where metabolites secreted by both commensal bacteria and their host are key to maintaining homeostasis (271). As regulators of the microbiome and thereby the cellular metabolomic milieu, β-defensins have a key role in this regard (272). Nutrients (like SCFAs) directly regulate systemic energy homeostasis is key to productive and healthy livestock for human food production (273). They also directly influence immune cell development, as butyrate was previously shown to induce a murine Th1 response (274).
The relevance to agriculture is not limited to improving livestock health, but are also critically important to reproduction. Interference with DEFB1 function has been shown to decrease human sperm function and bactericidal activity (171), and interestingly adding back DEFB1 restored both functions. Deletion of a number of murine β-defensin genes resulted in abnormal sperm structure, function and sterility in male mice (275). Similar work in cattle identified a β-defensin haplotype has been associated with sperm function and fertility in bulls (51, 172) making them molecules of major interest to agriculture.
In conclusion, any definition of a commensal or pathogen is problematic given what we now know about microbiomes (276, 277). To view the ecosystem of a microbiome as a ‘battle of good vs. bad bugs' is simplistic in the extreme. We contend that regulating the composition and diversity of commensal populations is just as important a feature of HDP function as preventing invasion by pathogens. We therefore view β-defensins as sensors of the microbial equilibrium and guardians of homeostasis. It is possible that the neonatal β-defensin gene or protein signatures may be diagnostic for homeostasis across mucosal sites and prognostic for dysregulated immunity and disease later in life. What is clear is that colonization by a diverse microbiome occurs across eukaryotic surfaces, both internal and external, begins pre-birth, and incessant quantitative and qualitative pruning of the microbiome is critical to health and homeostasis (278). The cumulative evidence presented here provides strong support that without active cultivation of the microbiome by β-defensin HDPs, pathology and disease would be inevitable. We predict that stimulating β-defensin expression will emerge as a key therapeutic mechanism to enhance natural immunity, restore homeostasis and reduce the burden of infectious disease in livestock.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
Research conducted on β-defensins performed in the laboratories of KM and COF was supported by funding from the Department of Agriculture, Food and the Marine under Stimulus grant 11/S/104.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors appreciate the critical reading of the manuscript and constructive comments from Dr. Paul Cormican, Teagasc, Grange. The authors would also like to thank both reviewers of the manuscript for their contributions.
References
1. Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. (2004) 327:539–49. doi: 10.1016/j.crvi.2003.12.007
2. Xiao Y, Hughes AL, Ando J, Matsuda Y, Cheng JF, Skinner-Noble D, et al. A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics (2004) 5:56. doi: 10.1186/1471-2164-5-56
3. Bruhn O, Paul S, Tetens J, Thaller G. The repertoire of equine intestinal alpha-defensins. BMC Genomics (2009) 10:631. doi: 10.1186/1471-2164-10-631
4. Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. (2002) 14:96–102. doi: 10.1016/S0952-7915(01)00303-X
5. Bruhn O, Regenhard P, Michalek M, Paul S, Gelhaus C, Jung S, et al. A novel horse alpha-defensin: gene transcription, recombinant expression and characterization of the structure and function. Biochem J. (2007) 407:267–76. doi: 10.1042/BJ20070747
6. Lynn DJ, Bradley DG. Discovery of alpha-defensins in basal mammals. Dev Compar Immunol. (2007) 31:963–7. doi: 10.1016/j.dci.2007.01.007
7. Fjell CD, Jenssen H, Fries P, Aich P, Griebel P, Hilpert K, et al. Identification of novel host defense peptides and the absence of alpha-defensins in the bovine genome. Proteins (2008) 73:420–30. doi: 10.1002/prot.22059
8. Lynn DJ, Higgs R, Gaines S, Tierney J, James T, Lloyd AT, et al. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics (2004) 56:170–7. doi: 10.1007/s00251-004-0675-0
9. Zou J, Mercier C, Koussounadis A, Secombes C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol. (2007) 44:638–47. doi: 10.1016/j.molimm.2006.01.012
11. Bauer F, Schweimer K, Kluver E, Conejo-Garcia JR, Forssmann WG, Rosch P, et al. Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity. Prot Sci. (2001) 10:2470–9. doi: 10.1110/ps.24401
12. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. (2003) 3:710–20. doi: 10.1038/nri1180
13. Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Cross-species analysis of the mammalian beta-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol Genom. (2005) 23:5–17. doi: 10.1152/physiolgenomics.00104.2005
14. Radhakrishnan Y, Fares MA, French FS, Hall SH. Comparative genomic analysis of a mammalian beta-defensin gene cluster. Physiol Genom. (2007) 30:213–22. doi: 10.1152/physiolgenomics.00263.2006
15. Choi MK, Le MT, Nguyen DT, Choi H, Kim W, Kim JH, et al. Genome-level identification, gene expression, and comparative analysis of porcine ss-defensin genes. BMC Genet. (2012) 13:98. doi: 10.1186/1471-2156-13-98
16. Tu J, Li D, Li Q, Zhang L, Zhu Q, Gaur U, et al. Molecular evolutionary analysis of beta-defensin peptides in vertebrates. Evol Bioinform Online (2015) 11:105–14. doi: 10.4137/EBO.S25580
17. Meade KG, Cormican P, Narciandi F, Lloyd A, O'Farrelly C. Bovine beta-defensin gene family: opportunities to improve animal health? Physiol Genomics. (2014) 46:17–28. doi: 10.1152/physiolgenomics.00085.2013
18. Meade KG, O'Gorman GM, Hill EW, Narciandi F, Agaba M, Kemp SJ, et al. Divergent antimicrobial peptide (AMP) and acute phase protein (APP) responses to Trypanosoma congolense infection in trypanotolerant and trypanosusceptible cattle. Mol Immunol. (2009) 47:196–204. doi: 10.1016/j.molimm.2009.09.042
19. Swanson K, Gorodetsky S, Good L, Davis S, Musgrave D, Stelwagen K, et al. Expression of a beta-defensin mRNA, lingual antimicrobial peptide, in bovine mammary epithelial tissue is induced by mastitis. Infect Immun. (2004) 72:7311–4. doi: 10.1128/IAI.72.12.7311-7314.2004
20. Caverly JM, Diamond G, Gallup JM, Brogden KA, Dixon RA, Ackermann MR. Coordinated expression of tracheal antimicrobial peptide and inflammatory-response elements in the lungs of neonatal calves with acute bacterial pneumonia. Infect Immun. (2003) 71:2950–5. doi: 10.1128/IAI.71.5.2950-2955.2003
21. Sorensen OE, Borregaard N, Cole AM. Antimicrobial peptides in innate immune responses. Contrib Microbiol. (2008) 15:61–77. doi: 10.1159/000136315
22. Sahl HG, Pag U, Bonness S, Wagner S, Antcheva N, Tossi A. Mammalian defensins: structures and mechanism of antibiotic activity. J Leukocyte Biol. (2005) 77:466–75. doi: 10.1189/jlb.0804452
23. Moser C, Weiner DJ, Lysenko E, Bals R, Weiser JN, Wilson JM. beta-Defensin 1 contributes to pulmonary innate immunity in mice. Infect Immun. (2002) 70:3068–72. doi: 10.1128/IAI.70.6.3068-3072.2002
24. Morrison G, Kilanowski F, Davidson D, Dorin J. Characterization of the mouse beta defensin 1, Defb1, mutant mouse model. Infect Immun. (2002) 70:3053–60. doi: 10.1128/IAI.70.6.3053-3060.2002
25. Elahi S, Buchanan RM, Attah-Poku S, Townsend HG, Babiuk LA, Gerdts V. The host defense peptide beta-defensin 1 confers protection against Bordetella pertussis in newborn piglets. Infect Immun. (2006) 74:2338–52. doi: 10.1128/IAI.74.4.2338-2352.2006
26. Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. (2000) 275:40478–82. doi: 10.1074/jbc.M007816200
27. Al-Asmakh M, Zadjali F. Use of germ-free animal models in microbiota-related research. J Microbiol Biotechnol. (2015) 25:1583–8. doi: 10.4014/jmb.1501.01039
28. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
29. Brogden KA, Ackermann M, McCray PB Jr, Tack BF. Antimicrobial peptides in animals and their role in host defences. Int J Antimicrob Agents. (2003) 22:465–78. doi: 10.1016/S0924-8579(03)00180-8
30. Bowdish DM, Davidson DJ, Hancock RE. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr Prot Peptide Sci. (2005) 6:35–51. doi: 10.2174/1389203053027494
31. Bowdish DM, Davidson DJ, Scott MG, Hancock RE. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother. (2005) 49:1727–32. doi: 10.1128/AAC.49.5.1727-1732.2005
32. Funderburg N, Lederman MM, Feng Z, Drage MG, Jadlowsky J, Harding CV, et al. Human -defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci USA. (2007) 104:18631–5. doi: 10.1073/pnas.0702130104
33. Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science (2002) 298:1025–9. doi: 10.1126/science.1075565
34. Semple F, MacPherson H, Webb S, Cox SL, Mallin LJ, Tyrrell C, et al. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol. (2011) 41:3291–300. doi: 10.1002/eji.201141648
35. Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science (1999) 286:525–8. doi: 10.1126/science.286.5439.525
36. Kiatsurayanon C, Niyonsaba F, Smithrithee R, Akiyama T, Ushio H, Hara M, et al. Host defense (Antimicrobial) peptide, human beta-defensin-3, improves the function of the epithelial tight-junction barrier in human keratinocytes. J Invest Dermatol. (2014) 134:2163–73. doi: 10.1038/jid.2014.143
37. Otte JM, Werner I, Brand S, Chromik AM, Schmitz F, Kleine M, et al. Human beta defensin 2 promotes intestinal wound healing in vitro. J Cell Biochem. (2008) 104:2286–97. doi: 10.1002/jcb.21787
38. Baroni A, Donnarumma G, Paoletti I, Longanesi-Cattani I, Bifulco K, Tufano MA, et al. Antimicrobial human beta-defensin-2 stimulates migration, proliferation and tube formation of human umbilical vein endothelial cells. Peptides (2009) 30:267–72. doi: 10.1016/j.peptides.2008.11.001
39. Hanaoka Y, Yamaguchi Y, Yamamoto H, Ishii M, Nagase T, Kurihara H, et al. In vitro and in vivo anticancer activity of human beta-defensin-3 and its mouse homolog. Anticancer Res. (2016) 36:5999–6004. doi: 10.21873/anticanres.11188
40. Gerashchenko OL, Zhuravel EV, Skachkova OV, Khranovska NN, Filonenko VV, Pogrebnoy PV, et al. Biologic activities of recombinant human-beta-defensin-4 toward cultured human cancer cells. Exp Oncol. (2013) 35:76–82.
41. Rohrl J, Huber B, Koehl GE, Geissler EK, Hehlgans T. Mouse beta-defensin 14 (Defb14) promotes tumor growth by inducing angiogenesis in a CCR6-dependent manner. J Immunol. (2012) 188:4931–9. doi: 10.4049/jimmunol.1102442
42. Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, et al. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. (2010) 78:2793–800. doi: 10.1128/IAI.00688-09
43. Sass V, Pag U, Tossi A, Bierbaum G, Sahl HG. Mode of action of human beta-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int J Med Microbiol. (2008) 298:619–33. doi: 10.1016/j.ijmm.2008.01.011
44. Jiang Y, Yang D, Li W, Wang B, Jiang Z, Li M. Antiviral activity of recombinant mouse beta-defensin 3 against influenza A virus in vitro and in vivo. Antiv Chem Chemother. (2012) 22:255–62. doi: 10.3851/IMP2077
45. Quinones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS (2003) 17:F39–48. doi: 10.1097/00002030-200311070-00001
46. Vylkova S, Li XS, Berner JC, Edgerton M. Distinct antifungal mechanisms: beta-defensins require Candida albicans Ssa1 protein, while Trk1p mediates activity of cysteine-free cationic peptides. Antimicrob Agents Chemother. (2006) 50:324–31. doi: 10.1128/AAC.50.1.324-331.2006
47. Lande R, Chamilos G, Ganguly D, Demaria O, Frasca L, Durr S, et al. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Eur J Immunol. (2015) 45:203–13. doi: 10.1002/eji.201344277
48. McGlasson SL, Semple F, MacPherson H, Gray M, Davidson DJ, Dorin JR. Human beta-defensin 3 increases the TLR9-dependent response to bacterial DNA. Eur J Immunol. (2017) 47:658–64. doi: 10.1002/eji.201646799
49. Tollner TL, Yudin AI, Treece CA, Overstreet JW, Cherr GN. Macaque sperm coating protein DEFB126 facilitates sperm penetration of cervical mucus. Hum Reprod. (2008) 23:2523–34. doi: 10.1093/humrep/den276
50. Tollner TL, Yudin AI, Tarantal AF, Treece CA, Overstreet JW, Cherr GN. Beta-defensin 126 on the surface of macaque sperm mediates attachment of sperm to oviductal epithelia. Biol Reprod. (2008) 78:400–12. doi: 10.1095/biolreprod.107.064071
51. Fernandez-Fuertes B, Narciandi F, O'Farrelly C, Kelly AK, Fair S, Meade KG, et al. Cauda epididymis-specific beta-defensin 126 promotes sperm motility but not fertilizing ability in cattle. Biol Reprod. (2016) 95:122. doi: 10.1095/biolreprod.116.138792
52. Yudin AI, Generao SE, Tollner TL, Treece CA, Overstreet JW, Cherr GN. Beta-defensin 126 on the cell surface protects sperm from immunorecognition and binding of anti-sperm antibodies. Biol Reprod. (2005) 73:1243–52. doi: 10.1095/biolreprod.105.042432
53. Madison MN, Kleshchenko YY, Nde PN, Simmons KJ, Lima MF, Villalta F. Human defensin alpha-1 causes Trypanosoma cruzi membrane pore formation and induces DNA fragmentation, which leads to trypanosome destruction. Infect Immun. (2007) 75:4780–91. doi: 10.1128/IAI.00557-07
54. Mendez-Samperio P. Role of antimicrobial peptides in host defense against mycobacterial infections. Peptides (2008) 29:1836–41. doi: 10.1016/j.peptides.2008.05.024
55. Chen RB, Zhang K, Zhang H, Gao CY, Li CL. Analysis of the antimicrobial mechanism of porcine beta defensin 2 against E. coli by electron microscopy and differentially expressed genes, Sci Rep. (2018) 8:14711. doi: 10.1038/s41598-018-32822-3
56. Liu R, Zhang Z, Liu H, Hou P, Lang J, Wang S, et al. Human beta-defensin 2 is a novel opener of Ca2+-activated potassium channels and induces vasodilation and hypotension in monkeys. Hypertension (2013) 62:415–25. doi: 10.1161/HYPERTENSIONAHA.111.01076
57. Terkawi MA, Takano R, Furukawa A, Murakoshi F, Kato K. Involvement of beta-defensin 130 (DEFB130) in the macrophage microbicidal mechanisms for killing Plasmodium falciparum. Sci Rep. (2017) 7:41772. doi: 10.1038/srep41772
58. Subramanian H, Gupta K, Lee D, Bayir AK, Ahn H, Ali H. beta-Defensins activate human mast cells via Mas-related gene X2. J Immunol. (2013) 191:345–52. doi: 10.4049/jimmunol.1300023
59. Niyonsaba F, Someya A, Hirata M, Ogawa H, Nagaoka I. Evaluation of the effects of peptide antibiotics human beta-defensins-1/-2 and LL-37 on histamine release and prostaglandin D(2) production from mast cells. Eur J Immunol. (2001) 31:1066–75. doi: 10.1002/1521-4141(200104)31:4<1066::AID-IMMU1066>3.0.CO;2-%23
60. Bhat S, Song YH, Lawyer C, Milner SM. Modulation of the complement system by human beta-defensin 2. J Burns Wounds. (2007) 5:e10.
61. Tomalka J, Azodi E, Narra HP, Patel K, O'Neill S, Cardwell C, et al. beta-Defensin 1 plays a role in acute mucosal defense against Candida albicans. J Immunol. (2015) 194:1788–95. doi: 10.4049/jimmunol.1203239
62. Scott MG, Vreugdenhil AC, Buurman WA, Hancock RE, Gold MR. Cutting edge: cationic antimicrobial peptides block the binding of lipopolysaccharide (LPS) to LPS binding protein. J Immunol. (2000) 164:549–53. doi: 10.4049/jimmunol.164.2.549
63. Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. (2006) 281:2005–11. doi: 10.1074/jbc.M511044200
64. O'Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. (1999) 163:6718–24.
65. Rohrl J, Yang D, Oppenheim JJ, Hehlgans T. Specific binding and chemotactic activity of mBD4 and its functional orthologue hBD2 to CCR6-expressing cells. J Biol Chem. (2010) 285:7028–34. doi: 10.1074/jbc.M109.091090
66. Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. (2006) 4:529–36. doi: 10.1038/nrmicro1441
67. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. (2004) 4:478–85. doi: 10.1038/nri1373
68. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. (2010) 10:159–69. doi: 10.1038/nri2710
69. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science (2000) 288:2222–6. doi: 10.1126/science.288.5474.2222
70. Matamoros S, Gras-Leguen C, Le Vacon F, Potel, G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. (2013) 21:167–73. doi: 10.1016/j.tim.2012.12.001
71. Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. (2015) 194:4081–7. doi: 10.4049/jimmunol.1403169
72. Connor JTO, Clegg TA, More SJ. Efficacy of washing and disinfection in cattle markets in Ireland. Ir Vet J. (2017) 70:6. doi: 10.1186/s13620-017-0081-1
73. Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. (2005) 51:270–4. doi: 10.1007/s00284-005-0020-3
74. Jeon SJ, Cunha F, Vieira-Neto A, Bicalho RC, Lima S, Bicalho ML, et al. Blood as a route of transmission of uterine pathogens from the gut to the uterus in cows. Microbiome (2017) 5:109. doi: 10.1186/s40168-017-0328-9
75. Calkins K, Devaskar SU. Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care (2011) 41:158–76. doi: 10.1016/j.cppeds.2011.01.001
76. Kanellopoulos-Langevin C, Caucheteux SM, Verbeke P, Ojcius DM. Tolerance of the fetus by the maternal immune system: role of inflammatory mediators at the feto-maternal interface. Reprod Biol Endocrinol. (2003) 1:121. doi: 10.1186/1477-7827-1-121
77. Alva-Murillo N, Tellez-Perez AD, Sagrero-Cisneros E, Lopez-Meza JE, Ochoa-Zarzosa A. Expression of antimicrobial peptides by bovine endothelial cells. Cell Immunol. (2012) 280:108–12. doi: 10.1016/j.cellimm.2012.11.016
78. Driver AM, Penagaricano F, Huang W, Ahmad KR, Hackbart KS, Wiltbank MC, et al. RNA-Seq analysis uncovers transcriptomic variations between morphologically similar in vivo- and in vitro-derived bovine blastocysts. BMC Genomics (2012) 13:118. doi: 10.1186/1471-2164-13-118
79. Meyerholz DK, Kawashima K, Gallup JM, Grubor B, Ackermann MR. Expression of select immune genes (surfactant proteins A and D, sheep beta defensin 1, and toll-like receptor 4) by respiratory epithelia is developmentally regulated in the preterm neonatal lamb. Dev Compar Immunol. (2006) 30:1060–9. doi: 10.1016/j.dci.2006.01.001
80. Meade KG, Higgs R, Lloyd AT, Giles S, O'Farrelly C. Differential antimicrobial peptide gene expression patterns during early chicken embryological development. Dev Comp Immunol. (2009) 33:516–24. doi: 10.1016/j.dci.2008.10.003
81. Messens W, Grijspeerdt K, Herman L. Eggshell characteristics and penetration by Salmonella enterica serovar Enteritidis through the production period of a layer flock. Br Poult Sci. (2005) 46:694–700. doi: 10.1080/00071660500395582
82. Buhimschi IA, Jabr M, Buhimschi CS, Petkova AP, Weiner CP, Saed GM. The novel antimicrobial peptide beta3-defensin is produced by the amnion: a possible role of the fetal membranes in innate immunity of the amniotic cavity. Am J Obstet Gynecol. (2004) 191:1678–87. doi: 10.1016/j.ajog.2004.03.081
83. Iavazzo C, Tassis K, Gourgiotis D, Boutsikou M, Baka S, Hassiakos D, et al. The role of human beta defensins 2 and 3 in the second trimester amniotic fluid in predicting preterm labor and premature rupture of membranes. Arch Gynecol Obstet. (2010) 281:793–9. doi: 10.1007/s00404-009-1155-4
84. Szukiewicz D, Alkhalayla H, Pyzlak M, Watroba M, Szewczyk G, Wejman J. Human beta-defensin 1, 2 and 3 production by amniotic epithelial cells with respect to human papillomavirus (HPV) infection, HPV oncogenic potential and the mode of delivery. Microb Pathog. (2016) 97:154–65. doi: 10.1016/j.micpath.2016.06.010
85. Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol. (2011) 38:321–31. doi: 10.1016/j.clp.2011.03.008
86. Lyons NA, Karvountzis S, Knight-Jones TJ. Aspects of bovine caesarean section associated with calf mortality, dam survival and subsequent fertility. Vet J. (2013) 197:342–50. doi: 10.1016/j.tvjl.2013.01.010
87. Singh G, Archana G. Unraveling the mystery of vernix caseosa. Indian J Dermatol. (2008) 53:54–60. doi: 10.4103/0019-5154.41645
88. Tollin M, Bergsson G, Kai-Larsen Y, Lengqvist J, Sjovall J, Griffiths W, et al. Vernix caseosa as a multi-component defence system based on polypeptides, lipids and their interactions. Cell Mol Life Sci. (2005) 62:2390–9. doi: 10.1007/s00018-005-5260-7
89. Baricelli J, Rocafull MA, Vazquez D, Bastidas B, Baez-Ramirez E, Thomas LE. beta-defensin-2 in breast milk displays a broad antimicrobial activity against pathogenic bacteria. J Pediatr. (2015) 91:36–43. doi: 10.1016/j.jped.2014.05.006
90. Lima SF, Teixeira AG, Lima FS, Ganda EK, Higgins CH, Oikonomou G, et al. The bovine colostrum microbiome and its association with clinical mastitis. J Dairy Sci. (2017) 100:3031–42. doi: 10.3168/jds.2016-11604
91. Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics (2007) 119:e724–e732. doi: 10.1542/peds.2006-1649
92. McLean JS. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. (2014) 4:98. doi: 10.3389/fcimb.2014.00098
93. Grassl N, Kulak NA, Pichler G, Geyer PE, Jung J, Schubert S, et al. Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. (2016) 8:44. doi: 10.1186/s13073-016-0293-0
94. Wade WG. The oral microbiome in health and disease. Pharmacol Res. (2013) 69:137–43. doi: 10.1016/j.phrs.2012.11.006
95. Hans M, Madaan Hans V. Epithelial antimicrobial peptides: guardian of the oral cavity. Int J Pept. (2014) 2014:370297. doi: 10.1155/2014/370297
96. Bonass WA, High AS, Owen PJ, Devine DA. Expression of beta-defensin genes by human salivary glands. Oral Microbiol Immunol. (1999) 14:371–4. doi: 10.1034/j.1399-302X.1999.140607.x
97. Shinomiya T, Kawaguchi M, Okubo M, Kosuge Y, Yoshikawa M. mRNA expression and localization of LPS-induced beta-defensin isoforms in rat salivary glands. Bull Tokyo Dent Coll. (2014) 55:139–47. doi: 10.2209/tdcpublication.55.139
98. Maekawa M, Beauchemin KA, Christensen DA. Chewing activity, saliva production, and ruminal pH of primiparous and multiparous lactating dairy cows. J Dairy Sci. (2002) 85:1176–82. doi: 10.3168/jds.S0022-0302(02)74180-5
99. Ang CS, Binos S, Knight MI, Moate PJ, Cocks BG, McDonagh MB. Global survey of the bovine salivary proteome: integrating multidimensional prefractionation, targeted, and glycocapture strategies. J Proteome Res. (2011) 10:5059–69. doi: 10.1021/pr200516d
100. Aust V, Knappstein K, Kunz HJ, Kaspar H, Wallmann J, Kaske M. Feeding untreated and pasteurized waste milk and bulk milk to calves: effects on calf performance, health status and antibiotic resistance of faecal bacteria. J Anim Physiol Anim Nutr. (2013) 97:1091–103. doi: 10.1111/jpn.12019
101. Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunol. (2017) 6:e133. doi: 10.1038/cti.2017.6
102. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. (2017) 15:259–70. doi: 10.1038/nrmicro.2017.14
103. Lima SF, Teixeira AG, Higgins CH, Lima FS, Bicalho RC. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci Rep. (2016) 6:29050. doi: 10.1038/srep29050
104. Holman DB, McAllister TA, Topp E, Wright AD, Alexander TW. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet Microbiol. (2015) 180:90–5. doi: 10.1016/j.vetmic.2015.07.031
105. Niederwerder MC. Role of the microbiome in swine respiratory disease. Vet Microbiol (2017) 209:97–106. doi: 10.1016/j.vetmic.2017.02.017
106. Holman DB, Timsit E, Amat S, Abbott DW, Buret AG, Alexander TW. The nasopharyngeal microbiota of beef cattle before and after transport to a feedlot. BMC Microbiol. (2017) 17:70. doi: 10.1186/s12866-017-0978-6
107. Zeineldin M, Lowe J, de Godoy M, Maradiaga N, Ramirez C, Ghanem M, et al. Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet Microbiol. (2017) 208:30–7. doi: 10.1016/j.vetmic.2017.07.006
108. Diamond G, Zasloff M, Eck H, Brasseur M, Maloy WL, Bevins CL. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. (1991) 88:3952–6. doi: 10.1073/pnas.88.9.3952
109. Taha-Abdelaziz K, Perez-Casal J, Schott C, Hsiao J, Attah-Poku S, Slavic D, et al. Bactericidal activity of tracheal antimicrobial peptide against respiratory pathogens of cattle. Vet Immunol Immunopathol. (2013) 152:289–94. doi: 10.1016/j.vetimm.2012.12.016
110. Joo NS, Evans IA, Cho HJ, Park IH, Engelhardt JF, Wine JJ. Proteomic analysis of pure human airway gland mucus reveals a large component of protective proteins. PLoS ONE (2015) 10:e0116756. doi: 10.1371/journal.pone.0116756
111. Hiratsuka T, Mukae H, Iiboshi H, Ashitani J, Nabeshima K, Minematsu T, et al. Increased concentrations of human beta-defensins in plasma and bronchoalveolar lavage fluid of patients with diffuse panbronchiolitis. Thorax (2003) 58:425–30. doi: 10.1136/thorax.58.5.425
112. Rivas-Santiago CE, Rivas-Santiago B, Leon DA, Castaneda-Delgado J, Hernandez Pando R. Induction of beta-defensins by l-isoleucine as novel immunotherapy in experimental murine tuberculosis. Clin Exper Immunol. (2011) 164:80–9. doi: 10.1111/j.1365-2249.2010.04313.x
113. Hagglund S, Blodorn K, Naslund K, Vargmar K, Lind SB, Mi J, et al. Proteome analysis of bronchoalveolar lavage from calves infected with bovine respiratory syncytial virus-Insights in pathogenesis and perspectives for new treatments. PLoS ONE (2017) 12:e0186594. doi: 10.1371/journal.pone.0186594
114. Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol. (2017) 8:738. doi: 10.3389/fmicb.2017.00738
115. Garcia M, Bradford BJ, Nagaraja TG. Invited review: ruminal microbes, microbial products, and systemic inflammation. Profess Animal Scient. (2017) 33:635–650. doi: 10.15232/pas.2017-01663
116. Yanez-Ruiz DR, Abecia L, Newbold CJ. Manipulating rumen microbiome and fermentation through interventions during early life: a review. Front Microbiol. 6:1133. doi: 10.3389/fmicb.2015.01133
117. Kittelmann S, Kirk MR, Jonker A, McCulloch A, Janssen PH. Buccal swabbing as a noninvasive method to determine bacterial, archaeal, and eukaryotic microbial community structures in the rumen. Appl Environ Microbiol. (2015) 81:7470–83. doi: 10.1128/AEM.02385-15
118. Malmuthuge N, Li M, Chen Y, Fries P, Griebel PJ, Baurhoo B, et al. Distinct commensal bacteria associated with ingesta and mucosal epithelium in the gastrointestinal tracts of calves and chickens. FEMS Microbiol Ecol. (2012) 79:337–47. doi: 10.1111/j.1574-6941.2011.01220.x
119. Malmuthuge N, Griebel PJ, Guan le. L Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl Environ Microbiol. (2014) 80:2021–8. doi: 10.1128/AEM.03864-13
120. Liang G, Malmuthuge N, McFadden TB, Bao H, Griebel PJ, Stothard P, et al. Potential regulatory role of microRNAs in the development of bovine gastrointestinal tract during early life. PLoS ONE (2014) 9:e92592. doi: 10.1371/journal.pone.0092592
121. Chen Y, Penner GB, Li M, Oba M, Guan LL. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl Environ Microbiol. (2011) 77:5770–81. doi: 10.1128/AEM.00375-11
122. Menendez A, Ferreira RB, Finlay BB. Defensins keep the peace too. Nat Immunol. (2010) 11:49–50. doi: 10.1038/ni0110-49
123. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. (2010) 11:76–83. doi: 10.1038/ni.1825
124. Veldhuizen EJ, van Dijk A Tersteeg, MH, Kalkhove SI, van der Meulen J, Niewold TA, et al. Expression of beta-defensins pBD-1 and pBD-2 along the small intestinal tract of the pig: lack of upregulation in vivo upon Salmonella typhimurium infection. Mol Immunol. (2007) 44:276–283. doi: 10.1016/j.molimm.2006.03.005
125. Wan ML, Woo CS, Allen KJ, Turner PC, El-Nezami H. Modulation of porcine beta-defensins 1 and 2 upon individual and combined Fusarium toxin exposure in a swine jejunal epithelial cell line. Appl Environ Microbiol. (2013) 79:2225–32. doi: 10.1128/AEM.03277-12
126. Bentley-Hewitt KL, Blatchford PA, Parkar SG, Ansell J, Pernthaner A. Digested and fermented green kiwifruit increases human beta-defensin 1 and 2 production in vitro. Plant Foods Hum Nutr. (2012) 67:208–14. doi: 10.1007/s11130-012-0305-1
127. Baldwin RLT, Wu S, Li W, Li C, Bequette BJ, Li RW. Quantification of Transcriptome Responses of the Rumen Epithelium to Butyrate Infusion using RNA-seq Technology. Gene Regul Syst Biol. (2012) 6:67–80. doi: 10.4137/GRSB.S9687
128. Huttner KM, Brezinski-Caliguri DJ, Mahoney MM, Diamond G. Antimicrobial peptide expression is developmentally regulated in the ovine gastrointestinal tract. J Nutr. (1998) 128(2 Suppl.):297S−9S. doi: 10.1093/jn/128.2.297S
129. Sommer F, Nookaew I, Sommer N, Fogelstrand P, Backhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. (2015) 16:62. doi: 10.1186/s13059-015-0614-4
130. Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature (2003) 422:522–6. doi: 10.1038/nature01520
131. Sunkara LT, Achanta M, Schreiber NB, Bommineni YR, Dai G, Jiang W, et al. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS ONE (2011) 6:e27225. doi: 10.1371/journal.pone.0027225
132. Cobo ER, Chadee K. Antimicrobial human beta-defensins in the colon and their role in infectious and non-infectious diseases. Pathogens (2013) 2:177–92. doi: 10.3390/pathogens2010177
133. Tarver AP, Clark DP, Diamond G, Russell JP, Erdjument-Bromage H, Tempst P, et al. Enteric beta-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect Immun. (1998) 66:1045–56.
134. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome (2016) 4:1. doi: 10.1186/s40168-015-0145-y
135. Falentin H, Rault L, Nicolas A, Bouchard DS, Lassalas J, Lamberton P, et al. Bovine teat microbiome analysis revealed reduced alpha diversity and significant changes in taxonomic profiles in quarters with a history of mastitis. Front Microbiol. (2016) 7:480. doi: 10.3389/fmicb.2016.00480
136. Roosen S, Exner K, Paul S, Schroder JM, Kalm E, Looft C. Bovine beta-defensins: identification and characterization of novel bovine beta-defensin genes and their expression in mammary gland tissue. Mamm Genome (2004) 15:834–42. doi: 10.1007/s00335-004-2387-z
137. Isobe N, Nakamura J, Nakano H, Yoshimura Y. Existence of functional lingual antimicrobial peptide in bovine milk. J Dairy Sci. (2009) 92:2691–5. doi: 10.3168/jds.2008-1940
138. Whelehan CJ, Meade KG, Eckersall PD, Young FJ, O'Farrelly C. Experimental Staphylococcus aureus infection of the mammary gland induces region-specific changes in innate immune gene expression. Veter Immunol Immunopathol. (2011) 140:181–9. doi: 10.1016/j.vetimm.2010.11.013
139. Yarbrough VL, Winkle S, Herbst-Kralovetz MM. Antimicrobial peptides in the female reproductive tract: a critical component of the mucosal immune barrier with physiological and clinical implications. Hum Reprod Update (2015) 21:353–77. doi: 10.1093/humupd/dmu065
140. Frew L, Stock SJ. Antimicrobial peptides and pregnancy. Reproduction (2011) 141:725–35. doi: 10.1530/REP-10-0537
141. Aloe S, Weber F, Behr B, Sauter-Louis C, Zerbe H. Modulatory effects of bovine seminal plasma on uterine inflammatory processes. Reproduct Domest Anim Zuchthyg. (2012) 47:12–9. doi: 10.1111/j.1439-0531.2011.01792.x
142. Alan E, Liman N. Immunohistochemical localization of beta defensins in the endometrium of rat uterus during the postpartum involution period. Vet Res Commun. (2012) 36:173–85. doi: 10.1007/s11259-012-9529-7
143. Hickey DK, Fahey JV, Wira CR. Mouse estrous cycle regulation of vaginal versus uterine cytokines, chemokines, alpha-/beta-defensins and TLRs. Innate Immun. (2013) 19:121–31. doi: 10.1177/1753425912454026
144. Fahey JV, Wright JA, Shen L, Smith JM, Ghosh M, Rossoll RM, et al. Estradiol selectively regulates innate immune function by polarized human uterine epithelial cells in culture. Mucosal Immunol. (2008) 1:317–25. doi: 10.1038/mi.2008.20
145. Wen S, Cao G, Bao T, Cheng L, Li H, Du C, et al. Modulation of ovine SBD-1 expression by 17beta-estradiol in ovine oviduct epithelial cells. BMC Vet Res. (2012) 8:143. doi: 10.1186/1746-6148-8-143
146. Patel MV, Fahey JV, Rossoll RM, Wira CR. Innate immunity in the vagina (part I): estradiol inhibits HBD2 and elafin secretion by human vaginal epithelial cells. Am J Reprod Immunol. (2013) 69:463–74. doi: 10.1111/aji.12078
147. Han JH, Kim MS, Lee MY, Kim TH, Lee MK, Kim HR, et al. Modulation of human beta-defensin-2 expression by 17beta-estradiol and progesterone in vaginal epithelial cells. Cytokine (2010) 49:209–14. doi: 10.1016/j.cyto.2009.09.005
148. Moore SG, Ericsson AC, Poock SE, Melendez P, Lucy MC. Hot topic: 16S rRNA gene sequencing reveals the microbiome of the virgin and pregnant bovine uterus. J Dairy Sci. (2017) 100:4953–60. doi: 10.3168/jds.2017-12592
149. Hansen PJ. Physiology and endocrinology symposium: maternal immunological adjustments to pregnancy and parturition in ruminants and possible implications for postpartum uterine health: is there a prepartum-postpartum nexus? J Anim Sci. (2013) 91:1639–49. doi: 10.2527/jas.2012-5934
150. King AE, Paltoo A, Kelly RW, Sallenave JM, Bocking AD, Challis JR. Expression of natural antimicrobials by human placenta and fetal membranes. Placenta (2007) 28:161–9. doi: 10.1016/j.placenta.2006.01.006
151. King AE, Fleming DC, Critchley HO, Kelly RW. Regulation of natural antibiotic expression by inflammatory mediators and mimics of infection in human endometrial epithelial cells. Mol Hum Reprod. (2002) 8:341–9. doi: 10.1093/molehr/8.4.341
152. King AE, Fleming DC, Critchley HO, Kelly RW. Differential expression of the natural antimicrobials, beta-defensins 3 and 4, in human endometrium. J Reprod Immunol. (2003) 59:1–16. doi: 10.1016/S0165-0378(02)00083-9
153. Fan SR, Liu XP, Liao QP. Human defensins and cytokines in vaginal lavage fluid of women with bacterial vaginosis. Int J Gynaecol Obstet. (2008) 103:50–4. doi: 10.1016/j.ijgo.2008.05.020
154. Vodstrcil LA, Twin J, Garland SM, Fairley CK, Hocking JS, Law MG, et al. The influence of sexual activity on the vaginal microbiota and Gardnerella vaginalis clade diversity in young women. PLoS ONE 12:e0171856. doi: 10.1371/journal.pone.0171856
155. Becknell B, Spencer JD, Carpenter AR, Chen X, Singh A, Ploeger S, et al. Expression and antimicrobial function of beta-defensin 1 in the lower urinary tract. PLoS ONE 8:e77714. doi: 10.1371/journal.pone.0077714
156. Meng W, Du R, Wang Y, Chen Z, Ding Y. Human beta-defensin messenger RNA is overexpressed in the cervical epithelia of patients with nongonococcal cervicitis. J Low Genit Tract Dis. (2013) 17:440–5. doi: 10.1097/LGT.0b013e318281f1a0
157. Hein M, Valore EV, Helmig RB, Uldbjerg N, Ganz T. Antimicrobial factors in the cervical mucus plug. Am J Obstet Gynecol. (2002) 187:137–44. doi: 10.1067/mob.2002.123034
158. Davies D, Meade KG, Herath S, Eckersall PD, Gonzalez D, White JO, et al. Toll-like receptor and antimicrobial peptide expression in the bovine endometrium. Reprod Biol Endocrinol. (2008) 6:53. doi: 10.1186/1477-7827-6-53
159. Foley C, Chapwanya A, Callanan JJ, Whiston R, Miranda-CasoLuengo R, Lu J, Meade KG, et al. Integrated analysis of the local and systemic changes preceding the development of post-partum cytological endometritis. BMC Genomics (2015) 16:811. doi: 10.1186/s12864-015-1967-5
160. Klamminger S, Prunner I, Giuliodori MJ, Drillich M. Uterine infection with bovine herpesvirus type 4 in dairy cows. Reproduct Domest Anim. (2017) 52:115–21. doi: 10.1111/rda.12865
161. Ghosh M, Fahey JV, Shen Z, Lahey T, Cu-Uvin S, Wu Z, et al. Anti-HIV activity in cervical-vaginal secretions from HIV-positive and -negative women correlate with innate antimicrobial levels and IgG antibodies. PLoS ONE (2010) 5:e11366. doi: 10.1371/journal.pone.0011366
162. Racicot K, Cardenas I, Wunsche V, Aldo P, Guller S, Means RE, et al. Viral infection of the pregnant cervix predisposes to ascending bacterial infection. J Immunol. (2013) 191:934–41. doi: 10.4049/jimmunol.1300661
163. Reid G, Brigidi P, Burton JP, Contractor N, Duncan S, Fargier E, et al. Microbes central to human reproduction. Am J Reprod Immunol. (2015) 73:1–11. doi: 10.1111/aji.12319
164. Ono C, Yoshida M, Kawana N, Miyado K, Umezawa A. Staphylococcus epidermidis is involved in a mechanism for female reproduction in mice. Regener Therapy (2015) 1:11–7. doi: 10.1016/j.reth.2014.12.003
165. Hall TJ, McQuillan C, Finlay EK, O'Farrelly C, Fair S, Meade KG. Comparative genomic identification and validation of beta-defensin genes in the Ovis aries genome. BMC Genomics (2017) 18:278. doi: 10.1186/s12864-017-3666-x
166. Javurek AB, Spollen WG, Ali AM, Johnson SA, Lubahn DB, Bivens NJ, et al. Discovery of a novel seminal fluid microbiome and influence of estrogen receptor alpha genetic status. Sci Rep. (2016) 6:23027. doi: 10.1038/srep23027
167. Narciandi F, Lloyd AT, Chapwanya A, O' Farrelly C, Meade KG. Reproductive tissue-specific expression profiling and genetic variation across a 19 gene bovine beta-defensin cluster. Immunogenetics (2011) 63:641–51. doi: 10.1007/s00251-011-0551-7
168. Browne JA, Yang R, Leir SH, Eggener SE, Harris A. Expression profiles of human epididymis epithelial cells reveal the functional diversity of caput, corpus and cauda regions. Mol Hum Reprod. (2016) 22:69–82. doi: 10.1093/molehr/gav066
169. Tollner TL, Yudin AI, Treece CA, Overstreet JW, Cherr GN. Macaque sperm release ESP13.2 and PSP94 during capacitation: the absence of ESP13.2 is linked to sperm-zona recognition and binding. Mol Reproduct Dev. (2004) 69:325–37. doi: 10.1002/mrd.20132
170. Zhao Y, Diao H, Ni Z, Hu S, Yu H, Zhang Y. The epididymis-specific antimicrobial peptide beta-defensin 15 is required for sperm motility and male fertility in the rat (Rattus norvegicus). Cell Mol Life Sci. (2011) 68:697–708. doi: 10.1007/s00018-010-0478-4
171. Diao R, Fok KL, Chen H, Yu MK, Duan Y, Chung CM, et al. Deficient human beta-defensin 1 underlies male infertility associated with poor sperm motility and genital tract infection. Sci Transl Med. (2014) 6:249ra108. doi: 10.1126/scitranslmed.3009071
172. Whiston R, Finlay EK, McCabe MS, Cormican P, Flynn P, Cromie A, et al. A dual targeted beta-defensin and exome sequencing approach to identify, validate and functionally characterise genes associated with bull fertility. Sci Rep. (2017) 7:12287. doi: 10.1038/s41598-017-12498-x
173. Liu CM, Osborne BJ, Hungate BA, Shahabi K, Huibner S, Lester R, et al. The semen microbiome and its relationship with local immunology and viral load in HIV infection. PLoS Pathog. (2014) 10:e1004262. doi: 10.1371/journal.ppat.1004262
174. Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T, et al. Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS ONE (2014) 9:e110152. doi: 10.1371/journal.pone.0110152
175. Mandar R. Microbiota of male genital tract: impact on the health of man and his partner. Pharmacol Res. (2013) 69:32–41. doi: 10.1016/j.phrs.2012.10.019
176. Dejucq N, Jegou B. Viruses in the mammalian male genital tract and their effects on the reproductive system. Microbiol Mol Biol Rev. (2001) 65:208–31. doi: 10.1128/MMBR.65.2.208-231.2001
177. Watanabe K, Gilchrist CA, Uddin MJ, Burgess SL, Abhyankar MM, Moonah SN, et al. Microbiome-mediated neutrophil recruitment via CXCR2 and protection from amebic colitis. PLoS Pathog. (2017) 13:e1006513. doi: 10.1371/journal.ppat.1006513
178. Dong H, Lv Y, Zhao D, Barrow P, Zhou X. Defensins: the case for their use against mycobacterial infections. J Immunol Res. (2016) 2016:7515687. doi: 10.1155/2016/7515687
179. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature (2012) 489:220–30. doi: 10.1038/nature11550
180. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. (2017) 17:219–32. doi: 10.1038/nri.2017.7
181. Lone AG, Selinger LB, Uwiera RR, Xu Y, Inglis GD. Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PLoS ONE (2013) 8:e75325. doi: 10.1371/journal.pone.0075325
182. Sankaran-Walters S, Hart R, Dills C. Guardians of the gut: enteric defensins. Front Microbiol. (2017) 8:647. doi: 10.3389/fmicb.2017.00647
183. Fecteau ME, Pitta DW, Vecchiarelli B, Indugu N, Kumar S, Gallagher SC, et al. Dysbiosis of the fecal microbiota in cattle infected with mycobacterium avium subsp. paratuberculosis PLoS ONE. 11:e0160353. doi: 10.1371/journal.pone.0160353
184. Walters CE, Ingham E, Eady EA, Cove JH, Kearney JN, Cunliffe WJ. In vitro modulation of keratinocyte-derived interleukin-1 alpha (IL-1 alpha) and peripheral blood mononuclear cell-derived IL-1 beta release in response to cutaneous commensal microorganisms. Infect Immun. (1995) 63:1223–8.
185. Guo S, Guo Y, Ergun A, Lu L, Walker WA, Ganguli K. Secreted metabolites of bifidobacterium infantis and lactobacillus acidophilus protect immature human enterocytes from IL-1beta-induced inflammation: a transcription profiling analysis. PLoS ONE (2015) 10:e0124549. doi: 10.1371/journal.pone.0124549
186. Menard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut (2004) 53:821–8. doi: 10.1136/gut.2003.026252
187. Seo SU, Kamada N, Munoz-Planillo R, Kim YG, Kim D, Koizumi Y, et al. Distinct commensals induce interleukin-1beta via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity (2015) 42:744–55. doi: 10.1016/j.immuni.2015.03.004
188. Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. (2012) 13:449–56. doi: 10.1038/ni.2263
189. Perregaux DG, Bhavsar K, Contillo L, Shi J, Gabel CA. Antimicrobial peptides initiate IL-1 beta posttranslational processing: a novel role beyond innate immunity. J Immunol. (2002) 168:3024–32. doi: 10.4049/jimmunol.168.6.3024
190. Coretti L, Natale A, Cuomo M, Florio E, Keller S, Lembo F, et al. The interplay between defensins and microbiota in Crohn's Disease. Mediat Inflamm. (2017) 2017:8392523. doi: 10.1155/2017/8392523
191. Chamaillard M, Dessein R. Defensins couple dysbiosis to primary immunodeficiency in Crohn's disease. World J Gastroenterol. (2011) 17:567–71. doi: 10.3748/wjg.v17.i5.567
192. Schulzke JD, Ploeger S, Amasheh M, Fromm A, Zeissig S, Troeger H, et al. Epithelial tight junctions in intestinal inflammation. Ann N Y Acad Sci. (2009) 1165:294–300. doi: 10.1111/j.1749-6632.2009.04062.x
193. Guttman JA, Finlay BB. Tight junctions as targets of infectious agents. Biochim Biophys Acta. (2009) 1788:832–41. doi: 10.1016/j.bbamem.2008.10.028
194. Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS ONE (2008) 3:e2369. doi: 10.1371/journal.pone.0002369
195. Shukla PK, Meena AS, Rao V, Rao RG, Balazs L, Rao R. Human defensin-5 blocks ethanol and colitis-induced dysbiosis, tight junction disruption and inflammation in mouse intestine. Sci Rep. (2018) 8:16241. doi: 10.1038/s41598-018-34263-4
196. Dupont A, Heinbockel L, Brandenburg K, Hornef MW. Antimicrobial peptides and the enteric mucus layer act in concert to protect the intestinal mucosa. Gut Microbes (2014) 5:761–5. doi: 10.4161/19490976.2014.972238
197. Semple F, Webb S, Li HN, Patel HB, Perretti M, Jackson IJ, et al. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol. (2010) 40:1073–8. doi: 10.1002/eji.200940041
198. Han F, Zhang H, Xia X, Xiong H, Song D, Zong X, et al. Porcine beta-defensin 2 attenuates inflammation and mucosal lesions in dextran sodium sulfate-induced colitis. J Immunol. (2015) 194:1882–93. doi: 10.4049/jimmunol.1402300
199. Semple F, Dorin JR. beta-Defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun. (2012) 4:337–48. doi: 10.1159/000336619
200. Aldhous MC, Noble CL, Satsangi J. Dysregulation of human beta-defensin-2 protein in inflammatory bowel disease. PLoS ONE (2009) 4:e6285. doi: 10.1371/journal.pone.0006285
201. Boniotto M, Jordan WJ, Eskdale J, Tossi A, Antcheva N, Crovella S, et al. Human beta-defensin 2 induces a vigorous cytokine response in peripheral blood mononuclear cells. Antimicrob Agents Chemother. (2006) 50:1433–41. doi: 10.1128/AAC.50.4.1433-1441.2006
202. Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. (2015) 16:191. doi: 10.1186/s13059-015-0759-1
203. Jacobs J, Braun J. Host genes and their effect on the intestinal microbiome garden. Genome Med. (2014) 6:119. doi: 10.1186/s13073-014-0119-x
204. Wojdak-Maksymiec K, Kmiec M, Zukiewicz A. Associations between defensin polymorphism and somatic cell count in milk and milk utility traits in Jersey dairy cows. J Veter Med A Physiol Pathol Clin Med. (2006) 53:495–500. doi: 10.1111/j.1439-0442.2006.00899.x
205. Bagnicka E, Strzalkowska N, Flisikowski K, Szreder T, Jozwik A, Prusak B, et al. The polymorphism in the beta4-defensin gene and its association with production and somatic cell count in Holstein-Friesian cows. J Anim Breed Genet Zeitschrift fur Tierzuchtung und Zuchtungsbiologie. (2007) 124:150–6. doi: 10.1111/j.1439-0388.2007.00649.x
206. Machado LR, Ottolini B. An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Front Immunol. (2015) 6:115. doi: 10.3389/fimmu.2015.00115
207. Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nature Genetics (2008) 40:23–5. doi: 10.1038/ng.2007.48
208. Hollox EJ. Beta-defensins and Crohn's disease: confusion from counting copies. Am J Gastroenterol. (2010) 105:360–2. doi: 10.1038/ajg.2009.573
209. Hollox EJ. Copy number variation of beta-defensins and relevance to disease. Cytogenet Genom Res. (2008) 123:148–55. doi: 10.1159/000184702
210. Bickhart DM, Hou Y, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. (2012) 22:778–90. doi: 10.1101/gr.133967.111
211. Boussaha M, Esquerre D, Barbieri J, Djari A, Pinton A, Letaief R, et al. Genome-wide study of structural variants in bovine holstein, montbeliarde and normande dairy breeds. PLoS ONE (2015) 10:e0135931. doi: 10.1371/journal.pone.0135931
212. Semple CA, Taylor K, Eastwood H, Barran PE, Dorin JR. Beta-defensin evolution: selection complexity and clues for residues of functional importance. Biochem Soc Transact. (2006) 34(Pt 2):257–62. doi: 10.1042/BST0340257
213. Semple CA, Gautier P, Taylor K, Dorin JR. The changing of the guard: Molecular diversity and rapid evolution of beta-defensins. Mol Divers. (2006) 10:575–84. doi: 10.1007/s11030-006-9031-7
214. Semple CA, Rolfe M, Dorin JR. Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. (2003) 4:R31. doi: 10.1186/gb-2003-4-5-r31
215. Luenser K, Ludwig A. Variability and evolution of bovine beta-defensin genes. Genes Immun. (2005) 6:115–22. doi: 10.1038/sj.gene.6364153
216. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science (2000) 290:1151–5. doi: 10.1126/science.290.5494.1151
217. Hardwick RJ, Machado LR, Zuccherato LW, Antolinos S, Xue Y, Shawa N, et al. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Human Mutation. (2011) 32:743–50. doi: 10.1002/humu.21491
218. Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. (2009) 324:522–8. doi: 10.1126/science.1169588
219. Fouts DE, Szpakowski S, Purushe J, Torralba M, Waterman RC, MacNeil MD, et al. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS ONE (2012) 7:e48289. doi: 10.1371/journal.pone.0048289
220. Johnson GP, Lloyd AT, O'Farrelly C, Meade KG, Fair S. (2015). Comparative genomic identification and expression profiling of a novel B-defensin gene cluster in the equine reproductive tract. Reprod Fertil Dev. 28:1499–508. doi: 10.1071/RD14345
221. Guyonnet B, Marot G, Dacheux JL, Mercat MJ, Schwob S, Jaffrezic F, et al. The adult boar testicular and epididymal transcriptomes. BMC Genomics. 10:369. doi: 10.1186/1471-2164-10-369
222. Brennan PL. Genital evolution: cock-a-doodle-don't. Curr Biol. (2013) 23:R523–5. doi: 10.1016/j.cub.2013.04.035
223. Huang Y, Li Y, Burt DW, Chen H, Zhang Y, Qian W, Li N, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet. (2013) 45:776–83. doi: 10.1038/ng.2657
224. Kern S, Robertson SA, Mau VJ, Maddocks S. Cytokine secretion by macrophages in the rat testis. Biol Reprod. (1995) 53:1407–16. doi: 10.1095/biolreprod53.6.1407
225. Zhao S, Zhu W, Xue S, Han D. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol. (2014) 11:428–37. doi: 10.1038/cmi.2014.38
226. Bagu ET, Gordon JR, Rawlings NC. Post-natal changes in testicular concentrations of interleukin-1 alpha and beta and interleukin-6 during sexual maturation in bulls. Reprod Domest Anim. (2010) 45:336–41. doi: 10.1111/j.1439-0531.2008.01318.x
227. Jiang L, Zheng T, Huang J, Mo J, Zhou H, Liu M, et al. Association of semen cytokines with reactive oxygen species and histone transition abnormalities. J Assist Reprod Genet. (2016) 33:1239–46. doi: 10.1007/s10815-016-0756-7
228. Vera O, Vasqucz LA, Munoz MG. Semen quality and presence of cytokines in seminal fluid of bull ejaculates. Theriogenology (2003) 60:553–8. doi: 10.1016/S0093-691X(03)00031-1
229. Eggert-Kruse W, Boit R, Rohr G, Aufenanger J, Hund M, Strowitzki T. Relationship of seminal plasma interleukin (IL)−8 and IL-6 with semen quality. Hum Reprod. (2001) 16:517–28. doi: 10.1093/humrep/16.3.517
230. Com E, Bourgeon F, Evrard B, Ganz T, Colleu D, Jegou B, et al. Expression of antimicrobial defensins in the male reproductive tract of rats, mice, and humans. Biol Reprod. (2003) 68:95–104. doi: 10.1095/biolreprod.102.005389
231. Kolar SS, McDermott AM. Role of host-defence peptides in eye diseases. Cell Mol Life Sci. (2011) 68:2201–13. doi: 10.1007/s00018-011-0713-7
232. Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. (2009) 130:7–15. doi: 10.1016/j.clim.2008.08.015
233. Kallsen K, Andresen E, Heine H. Histone deacetylase (HDAC) 1 controls the expression of beta defensin 1 in human lung epithelial cells. PLoS ONE 7:e50000. doi: 10.1371/journal.pone.0050000
234. Andresen E, Gunther G, Bullwinkel J, Lange C, Heine H. Increased expression of beta-defensin 1 (DEFB1) in chronic obstructive pulmonary disease. PLoS ONE 6:e21898. doi: 10.1371/journal.pone.0021898
235. Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell (2016) 166:1231–1246 e13. doi: 10.1016/j.cell.2016.07.043
236. Camp JG, Frank CL, Lickwar CR, Guturu H, Rube T, Wenger AM, et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. (2014) 24:1504–16. doi: 10.1101/gr.165845.113
237. Adams LG, Khare S, Lawhon SD, Rossetti CA, Lewin HA, Lipton MS, et al. Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: Linking systems biology with vaccine development. Vaccine (2011) 29:7197–206. doi: 10.1016/j.vaccine.2011.05.080
238. Rivas-Santiago B, Cervantes-Villagrana A, Sada E, Hernandez-Pando R. Expression of beta defensin 2 in experimental pulmonary tuberculosis: tentative approach for vaccine development. Arch Med Res. (2012) 43:324–8. doi: 10.1016/j.arcmed.2012.06.005
239. Kohlgraf KG, Pingel LC, Dietrich DE, Brogden KA. Defensins as anti-inflammatory compounds and mucosal adjuvants. Fut Microbiol. (2010) 5:99–113. doi: 10.2217/fmb.09.104
240. Mei HF, Jin XB, Zhu JY, Zeng AH, Wu Q, Lu XM, et al. beta-defensin 2 as an adjuvant promotes anti-melanoma immune responses and inhibits the growth of implanted murine melanoma in vivo. PLoS ONE (2012) 7:e31328. doi: 10.1371/journal.pone.0031328
241. Rohrl J, Yang D, Oppenheim JJ, Hehlgans T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J Immunol. (2010) 184:6688–94. doi: 10.4049/jimmunol.0903984
242. Tewary P, de la Rosa G, Sharma N, Rodriguez LG, Tarasov SG, Howard OM, et al. beta-Defensin 2 and 3 promote the uptake of self or CpG DNA, enhance IFN-alpha production by human plasmacytoid dendritic cells, and promote inflammation. J Immunol. (2013) 191:865–874. doi: 10.4049/jimmunol.1201648
243. Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. (2008) 8:435–46. doi: 10.1038/nri2335
244. Uhlig HH, Powrie F. Dendritic cells and the intestinal bacterial flora: a role for localized mucosal immune responses. J Clin Invest. (2003) 112:648–51. doi: 10.1172/JCI19545
245. Lynn DJ, Pulendran B. The potential of the microbiota to influence vaccine responses. J Leukoc Biol. (2017) 103:225–31. doi: 10.1189/jlb.5MR0617-216R
246. Han SH, Kim YE, Park JA, Park JB, Kim YS, Lee Y, et al. Expression of human beta-defensin-2 gene induced by CpG-DNA in human B cells. Biochem Biophys Res Commun. (2009) 389:443–8. doi: 10.1016/j.bbrc.2009.08.162
247. Ju SM, Goh AR, Kwon DJ, Youn GS, Kwon HJ, Bae YS, et al. Extracellular HIV-1 Tat induces human beta-defensin-2 production via NF-kappaB/AP-1 dependent pathways in human B cells. Mol Cells (2012) 33:335–41. doi: 10.1007/s10059-012-2287-0
248. Allaker RP. Host defence peptides-a bridge between the innate and adaptive immune responses. Trans R Soc Trop Med Hyg. (2008) 102:3–4. doi: 10.1016/j.trstmh.2007.07.005
249. Hand TW. The role of the microbiota in shaping infectious immunity. Trends Immunol. (2016) 37:647–58. doi: 10.1016/j.it.2016.08.007
250. Steimle A, Autenrieth IB, Frick JS. Structure and function: lipid a modifications in commensals and pathogens. Int J Med Microbiol. (2016) 306:290–301. doi: 10.1016/j.ijmm.2016.03.001
251. Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science (2015) 347:170–5. doi: 10.1126/science.1260580
252. Rasmussen MA, Carlson SA, Franklin SK, McCuddin ZP, Wu MT, Sharma VK. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect Immun. (2005) 73:4668–75. doi: 10.1128/IAI.73.8.4668-4675.2005
253. Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome (2017) 5:48. doi: 10.1186/s40168-017-0268-4
254. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell (2015) 161:264–76. doi: 10.1016/j.cell.2015.02.047
255. Shamova OV, Lesnikova MP, Kokriakov VN, Shkhinek EK, Korneva EA. The action of defensins on the corticosterone level of the blood and on the immune response in stress. Biull Eksp Biol Med. (1993) 115:646–9. doi: 10.1007/BF00791176
256. Mitchell GB, Al-Haddawi MH, Clark ME, Beveridge JD, Caswell JL. Effect of corticosteroids and neuropeptides on the expression of defensins in bovine tracheal epithelial cells. Infect Immun. (2007) 75:1325–34. doi: 10.1128/IAI.00686-06
257. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. (2014) 14:405–16. doi: 10.1038/nri3684
258. Cadwell K. Expanding the role of the virome: commensalism in the gut. J Virol. (2015) 89:1951–3. doi: 10.1128/JVI.02966-14
259. Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. (2013) 14:654–9. doi: 10.1038/ni.2614
260. Williams WM, Castellani RJ, Weinberg A, Perry G, Smith MA. Do beta-defensins and other antimicrobial peptides play a role in neuroimmune function and neurodegeneration? Sci World J. (2012) 2012:905785. doi: 10.1100/2012/905785
261. Yoon YJ, Park JW, Lee EJ. Presence of hBD-1 and hBD-2 in human cerumen and external auditory canal skin. Acta Otolaryngol. (2008) 128:871–5. doi: 10.1080/00016480701785020
262. Jin JY, Zhou L, Wang Y, Li Z, Zhao JG, Zhang QY, et al. Antibacterial and antiviral roles of a fish beta-defensin expressed both in pituitary and testis. PLoS ONE (2010) 5:e12883. doi: 10.1371/journal.pone.0012883
263. Morrison GM, Semple CA, Kilanowski FM, Hill RE, Dorin JR. Signal sequence conservation and mature peptide divergence within subgroups of the murine beta-defensin gene family. Mol Biol Evolut. (2003) 20:460–70. doi: 10.1093/molbev/msg060
264. Froy O, Hananel A, Chapnik N, Madar Z. Differential expression of rat beta-defensins. IUBMB Life (2005) 57:41–3. doi: 10.1080/15216540500088912
265. Toutain PL, Ferran AA, Bousquet-Melou A, Pelligand L, Lees P. Veterinary medicine needs new green antimicrobial drugs. Front Microbiol. (2016) 7:1196. doi: 10.3389/fmicb.2016.01196
266. Nijnik A, Hancock R. Host defence peptides: antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg Health Threats J. (2009) 2:e1. doi: 10.3402/ehtj.v2i0.7078
267. Wang S, Zeng X, Yang Q, Qiao S. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci. (2016) 17:E603. doi: 10.3390/ijms17050603
268. Robinson K, Deng Z, Hou Y, Zhang G. Regulation of the intestinal barrier function by host defense peptides. Front Vet Sci. (2015) 2:57. doi: 10.3389/fvets.2015.00057
269. Liu Q, Yao S, Chen Y, Gao S, Yang Y, Deng J, et al. Use of antimicrobial peptides as a feed additive for juvenile goats. Sci Rep. (2017) 7:12254. doi: 10.1038/s41598-017-12394-4
270. Pryce JE, Parker Gaddis KL, Koeck A, Bastin C, Abdelsayed M, Gengler N, et al. Invited review: opportunities for genetic improvement of metabolic diseases. J Dairy Sci. (2016) 99:6855–73. doi: 10.3168/jds.2016-10854
271. McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome (2013) 1:17. doi: 10.1186/2049-2618-1-17
272. Oyinloye BE, Adenowo AF, Kappo AP. Reactive oxygen species, apoptosis, antimicrobial peptides and human inflammatory diseases. Pharmaceuticals (2015) 8:151–75. doi: 10.3390/ph8020151
273. Spiljar M, Merkler D, Trajkovski M. The immune system bridges the gut microbiota with systemic energy homeostasis: focus on TLRs, Mucosal Barrier, and SCFAs. Front Immunol. (2017) 8:1353. doi: 10.3389/fimmu.2017.01353
274. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front Immunol. (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
275. Zhou YS, Webb S, Lettice L, Tardif S, Kilanowski F, Tyrrell C, et al. Partial deletion of chromosome 8 beta-defensin cluster confers sperm dysfunction and infertility in male mice. PLoS Genet. (2013) 9:e1003826. doi: 10.1371/journal.pgen.1003826
276. Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. (1999) 96:9833–8. doi: 10.1073/pnas.96.17.9833
277. Gilmore MS, Ferretti JJ. Microbiology. The thin line between gut commensal and pathogen. Science (2003) 299:1999–2002. doi: 10.1126/science.1083534
Keywords: defensin, immune privilege, innate, microbiome, basal, constitutive
Citation: Meade KG and O'Farrelly C (2019) β-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 9:3072. doi: 10.3389/fimmu.2018.03072
Received: 12 September 2018; Accepted: 11 December 2018;
Published: 25 January 2019.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Christine McDonald, Lerner Research Institute, United StatesLarry J. Dishaw, University of South Florida St. Petersburg, United States
Copyright © 2019 Meade and O'Farrelly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kieran G. Meade, kieran.meade@teagasc.ie
 Kieran G. Meade
Kieran G. Meade Cliona O'Farrelly
Cliona O'Farrelly