Genetic manipulation of Staphylococci—breaking through the barrier
- Department of Microbiology, Moyne Institute of Preventive Medicine, School of Genetics and Microbiology, Trinity College Dublin, Dublin, Ireland
Most strains of Staphylococcus aureus and Staphylococcus epidermidis possess a strong restriction barrier that hinders exchange of DNA. Recently, major advances have been made in identifying and characterizing the restriction-modification (RM) systems involved. In particular a novel type IV restriction enzyme that recognizes cytosine methylated DNA has been shown to be the major barrier to transfer of plasmid DNA from Escherichia coli into S. aureus and S. epidermidis. While the conserved type I RM system provides a further barrier. Here we review the recent advances in understanding of restriction systems in staphylococci and highlight how this has been exploited to improve our ability to manipulate genetically previously untransformable strains.
Introduction
The principles behind the use of genetic manipulation to identify virulence factors in pathogenic bacteria were articulated by Stanley Falkow as Molecular Koch's Postulates (Falkow, 1988). The basic premise is that by using precise genetic manipulation, the genes encoding putative virulence factors can be inactivated and the mutants tested for loss of virulence in infection models. It is also necessary to demonstrate that complementation restores virulence to wild-type levels. As pointed out by Falkow, “these postulates place a heavy burden on an investigator. They insist that genetic manipulation of the microorganism is a prerequisite for success, and, of course, for some pathogens, such study is not yet possible.” (Falkow, 1988).
A major barrier to the genetic manipulation of staphylococci and fulfilling Molecular Koch's Postulates is the inability to transform plasmid DNA into the majority of clinical isolates due to a strong restriction-modification (RM) barrier. Consequently studies have focused on a small number of transformable laboratory strains of S. aureus (Voyich et al., 2005; Baba et al., 2008; O'Neill, 2010) and S. epidermidis (Heilmann et al., 1996). In this review we will focus on the recent developments in the understanding of RM systems in staphylococci and will show how these findings, combined with the development of new tools for genetic analysis, have advanced our ability to manipulate staphylococci genetically.
RM Systems of Staphylococci—A Historical Overview
Four different types of RM systems are known but only three are found in staphylococci. Type I RM systems comprise genes that encode a host specificity of DNA (hsd) specificity (S) protein, a modification (M) protein and a restriction (R) endonuclease (Murray, 2000). HsdS functions in an HsdS1 HsdM2 complex which recognizes a specific DNA sequence. The complex methylates hemi-methylated DNA and inhibits cleavage by the endonuclease complex HsdS1 HsdM2 HsdR2 which would otherwise assemble on unmethylated DNA (Murray, 2000). Cleavage of unmodified DNA occurs after HsdR-dependent translocation of the complex along the molecule until it collides with a second complex (or DNA secondary structure), which stimulates the formation of double stranded DNA breaks (Simons and Szczelkun, 2011). Type II RM systems are well known to molecular biologists because the restriction endonucleases are widely used as reagents in molecular biology. The cleavage of DNA is sequence-dependent and can be prevented by the DNA methylation status. In S. aureus the Sau3AI type II RM system is present in a limited subset of strains (Stobberingh et al., 1977). The type IV system is the simplest form of restriction system with a single protein able to detect the methylation status. Examples from E. coli are mcrA, mcrBC which recognize 5-hydroxymethylcytosine and N-4-methylcytosine, respectively, while and mrr recognizes N-6-methyladenine as foreign. DNA containing these motifs are restricted by the corresponding enzyme (Kelleher and Raleigh, 1991).
Over 50 years have passed since RM was first recognized in S. aureus. In early phage typing studies it was observed that some strains were resistant to infection by phage. However, a strain could be infected if a high phage titer was used (Roundtree, 1956) or the recipient was heat-shocked beforehand (Asheshov and Jevons, 1963), suggesting that the barrier to infection could be overwhelmed or by-passed by transient inactivation. Restriction-deficient mutants of the clonal complex (CC—lineages derived from multi locus sequence typing) (Enright et al., 2000) 8 strain 8325 (SA113) (Iordanescu and Surdeanu, 1976) and the CC51 strain 879 (879R4) (Stobberingh and Winkler, 1977) were isolated which could take up foreign DNA and modify it so that it could be transferred to closely related wild-type strains. Both strains are thus r−m+, i.e., defective in restricting foreign DNA but capable of modifying the newly introduced DNA.
In order to facilitate genetic manipulation of S. aureus it is necessary to be able to transform S. aureus with shuttle plasmids that have been constructed in E. coli. Strain 8325-4 (8325 cured of three prophages) was subjected to heavy chemical mutagenesis and then transformed by protoplast transformation with a shuttle plasmid isolated from E. coli in order to isolate a mutant that could accept foreign DNA (Kreiswirth et al., 1983). From these experiments a single transformant was obtained. The plasmid was eliminated and then the S. aureus clone shown to accept the E. coli isolated plasmid at a reasonable frequency. This strain, called RN4220, has been extensively used by staphylococcal researchers ever since. However, it only provides a gateway into a limited set of closely related strains, e.g., in our hands 8325-4 isolated plasmid cannot transform MRSA252 (CC30) and vice versa. Also we cannot transform S. epidermidis isolates tested (RP62a or AMC5) with RN4220 isolated plasmid (unpublished data). The genome sequence of RN4220 revealed a nonsense mutation in the hsdR gene of a type I RM system among the 110 single nucleotide polymorphisms by which it differs from the parental strain (Nair et al., 2011). It had been previously shown by Waldron and Lindsay (Waldron and Lindsay, 2006) that complementation of RN4220 mutant hsdR allele with wild-type hsdR expressed from a low copy number plasmid prevented transformation by electroporation with a shuttle plasmid isolated from E. coli K-12, inhibited transduction and reduced the frequency of conjugation of a plasmid from Enterococcus faecalis. However Veiga and Pinho (2009) were unable to confirm the role of HsdR as the barrier to uptake of foreign DNA when they deleted hsdR in 8325-4 and COL. Mild heat shock (56°C for 2 min) prior to electroporation allowed transformation of 8325-4ΔhsdR but not the parental 8325-4. These results suggested that an additional heat-sensitive restriction system prevented transformation with plasmid DNA from E. coli K-12 (Veiga and Pinho, 2009). Interestingly, the majority of sequenced S. aureus isolates contain 2 sets of hsdMS genes located on the alpha and beta pathogenicity islands (Waldron and Lindsay, 2006), with hsdR located at a third site on the chromosome. This is in direct contrast to S. epidermidis and S. lugdunensis where the type I RM genes are clustered together (unpublished observation). In some MRSA strains, a third complete hsdMSR has also been identified in the staphylococcal cassette chromosome mec element (SCCmec) III, with hsdMR found in the SCCmec VII (Malachowa and DeLeo, 2010). The functionality of the modification and specificity genes in staphylococci has not been published. In S. aureus the sequences of the hsdM genes are highly conserved, while the two hsdS genes are divergent (Waldron and Lindsay, 2006). hsdS sequence variation is localized to the two target recognition domains (TRDs) within the gene, with hsdS gene content shown to be lineage specific, e.g., CC30 cluster together as do CC8 strains (Cockfield et al., 2007; Lindsay, 2010).
A major advance in the understanding of staphylococcal RM occurred with the discovery of a novel type IV restriction enzyme, which was shown to be the dominant barrier to prevent the uptake of foreign DNA by S. aureus (Corvaglia et al., 2010). Mutants of hsdR in UAMS-1 (CC30) and SA564 (CC5) were not or poorly transformable (respectively) with plasmid DNA isolated from E. coli K-12 (Corvaglia et al., 2010). UV mutagenesis of SA564hsdR− and subsequent transformation of the pooled survivors with a shuttle plasmid from E. coli K-12 resulted in 18 transformants. The genome of the strain that exhibited the highest transformation efficiency was sequenced along with the parental SA564. A frameshift mutation was identified in an ORF that has 98% identity to Sao_2790 of 8325. This gene was subsequently designated sauUSI (Xu et al., 2011). Disruption of sauUSI in SA564 and UAMS-1 yielded a strain that was transformable with the E. coli K-12-derived plasmid. Analysis of sauUSI in RN4220 identified a nonsense mutation in the middle of the gene and complementation using a multicopy plasmid carrying wild-type sauUSI reduced transformation into RN4220 100-fold (Corvaglia et al., 2010).
We have restored the sauUSI gene in the chromosome of RN4220 to wild-type by allelic exchange which resulted in a 10−4-fold reduction in the transformation frequency in RN4220sauUSI+ compared to RN4220 (Monk et al., 2012). The SauUSI protein has a very limited similarity to HsdR except for a DNA helicase domain. Deletion of the type I RM specificity genes hsdS1 and hsdS2 in SA564 did not yield a transformable strain indicating that SauUSI acts independently of the type I RM system (Corvaglia et al., 2010). The gene upstream of sauUSI in strain Newman (called Sae_2385) encodes a protein with homology to a nudix hydrolase that could potentially be involved in removal of toxic nucleotide derivatives. However deletion of this gene did not enhance transformation indicating that it is not important for SauUSI activity (Monk et al., 2012). SauUSI is highly conserved in S. aureus. However the CC5 strains N315 and Mu50 contain an allele of sauUSI with a nonsense mutation within the middle of the gene. Loss of SauUSI has made the strains permissive to transformation with plasmid DNA isolated from Enterococcus faecalis strain JH2-2 (Corvaglia et al., 2010), which could have implications for the enhanced the spread of antibiotic resistance between these organisms in the hospital environment (Zhu et al., 2008). Homologues of sauUSI occur in S. epidermidis and S. pseudintermedius and also in some enterococci, bacilli and lactobacilli.
The biochemical properties of SauUSI were recently characterized (Xu et al., 2011). The enzyme was shown to be a type IV endonuclease. The motif recognized by SauUSI was identified as methylation of cytosine bases in the motif C/GmCNGC/G. The E. coli K-12 strains that are widely used for cloning such as DH5∝, TOP10, XL1-Blue and DH10B methylate both adenine (dam) and cytosine (dcm) residues. Plasmids isolated from these strains are readily degraded by SauUSI. In order to bypass the type IV restriction barrier in S. aureus the plasmid must be isolated from an E. coli strain that is defective in cytosine methylation. DNA methylation at cytosine residues is not only limited to E. coli. Some type II RM systems use cytosine methylation of target sites to prevent the activity of the cognate restriction enzyme, with these including S. aureus lineage CC398 (Bosch et al., 2010), some Listeria monocytogenes (Yildirim et al., 2010) and Lactococcus lactis strains (O'Driscoll et al., 2005).
The loss of Dam methylation in E. coli leads to deregulated mismatch repair and an elevated frequency of transition mutations (Wion and Casadesus, 2006) which means that a dam mutant is unsuitable for cloning. However a dcm mutant of E. coli does not have an enhanced mutation rate (Palmer and Marinus, 1994). A dcm mutant of a high efficiency cloning strain of E. coli would be a useful host for constructing recombinant plasmids prior to direct transformation into a wild-type SauUSI proficient strain of S. aureus.
Electroporation of Staphylococci—Bypassing the Restriction Barrier
While S. aureus contains homologues of genes involved in natural competence, and induction of some of the com-like genes has been shown upon the over expression of the comX homolog sigH (Morikawa et al., 2003), there is no experimental evidence that facilitated uptake of foreign DNA can occur. Electroporation is the method of choice for introducing plasmid DNA into S. aureus.
The most widely used protocol involves the growth of cells into early logarithmic phase followed by washing with a hypertonic buffer (e.g., 500 mM sucrose) to remove salts and to stabilize the cells (Augustin and Gotz, 1990; Kraemer and Iandolo, 1990; Oskouian and Stewart, 1990; Schenk and Laddaga, 1992; Lee, 1995). The cells are concentrated to 1–3 × 1010 CFU/ml and purified plasmid DNA added. A defined electric pulse is discharged through the cells to facilitate the uptake of the DNA. The cells are then grown in broth for a short period of time to allow recovery and for growth to begin prior to plating on media containing an antibiotic that selects for the plasmid-containing transformants. Lofblom et al. (2007) described extensive optimization of electroporation for Staphylococcus carnosus. Application of the final protocol with minor modifications to S. aureus strain Newman gave a 50-fold improvement over the sucrose-wash protocol (Monk et al., 2012) with transformants being obtained directly with DNA isolated from E. coli K-12 strains at a low frequency (between 101 and 102 CFU/5 μg plasmid DNA). In strain Newman the restriction barriers cause a 10−4 reduction in the transformation efficiency when comparing uptake of plasmid DNA isolated from wild-type Newman with that isolated from E. coli K-12. Thus improving the efficiency of electroporation allowed the type IV and type I RM systems to be bypassed.
DC10B—A Universal Host for Constructing Plasmid for Introduction into Staphylococci
To improve the transformation of S. aureus we created an unmarked dcm deletion mutation in the high efficiency cloning strain E. coli DH10B to generate strain DC10B (Monk et al., 2012). The absence of cytosine methylation allows plasmid DNA to bypass the type IV restriction barrier. Transformation with plasmid DNA isolated from DC10B occurred in 15 strains from different CCs that we have so far tested. The CCs were chosen to represent a diverse selection of S. aureus lineages and to encompass the major MRSA CCs. The only strain we were unable to transform was from CC97 (Monk et al., 2012). Using DC10B we have been able to transform and isolate chromosomal mutations in strains that were previously refractory to genetic manipulation, for example the CC30 strains Cowan and MRSA252 (Monk et al., 2012). The improved transformation protocol has enabled us to transform several strains of S. lugdunensis (Heilbronner and Foster, unpublished) and combined with DC10B has allowed the direct transformation of S. epidermidis. For both coagulase negative species, a reduced efficiency was found compared to S. aureus Newman with a maximum of 103 CFU/5 μg plasmid DNA. Deletion of the sauUSI homologue (termed mcrR for methylated cytosine recognition and restriction) in S. epidermidis isolate RP62a (Gill et al., 2005) produced a strain that could accept plasmid DNA from a Dcm+ E. coli K-12 host. This directly demonstrates the importance of the type IV restriction barrier in this species (Monk et al., 2012). In conclusion using plasmid DNA isolated from the DC10B strain of E. coli and or an enhanced electroporation protocol will dramatically improve our ability to conduct genetic studies in many different staphylococci.
E. coli Strains that Modify Plasmid DNA for Staphylococci
While bypassing the type IV barrier allows us to transform DNA directly into wild-type staphylococci, the efficiency of plasmid transfer is still low and for some applications borderline for selection (e.g., transferring pVW01ts into S. epidermidis RP62a or direct integration of plasmids at phage att sites mediated by integrase [see below]). Bypassing the type I RM barrier would require the decoration of plasmid with the methylation pattern determined by the hsdMS genes in the strain to be transformed. There is a paucity of information on the properties of the type I RM systems in S. aureus (Waldron and Lindsay, 2006; Sung and Lindsay, 2007). They appear to be involved in the limiting uptake of phage DNA from unrelated staphylococci (Veiga and Pinho, 2009), play an additive role with SauUSI in restricting foreign DNA (Monk et al., 2012) and impede the transfer of DNA between staphylococci (Corvaglia et al., 2010; Lindsay, 2010). In the simplest system where only one hsdRMS operon is present (e.g., S. epidermidis RP62a) expression of the hsdMS genes in E. coli DC10B should further improve the efficiency of plasmid transfer. There is a 60-fold reduction in transformation of RP62a with plasmid isolated from DC10B compared to RP62a isolated plasmid, suggesting the presence of a second active RM system (unpublished data). The term plasmid artificial modification (PAM) has been coined to describe pre-methylation of plasmid DNA in an E. coli strain which expresses the target strain's modification and specificity genes (Suzuki and Yasui, 2011).
Two groups have described the use of this technology for bifidobacteria. O'Connell Motherway et al. (2009) isolated the modification genes of a two different type II RM systems from Bifidobacterium breve UCC2003 and expressed them either from a plasmid or from a chromosomal locus in E. coli. An increase in transformation by 1000-fold was observed for the plasmid-borne methylation genes, while a 50-fold improvement was observed for the chromosomally encoded genes compared to DNA from the parental E. coli strain. Two type II RM methylase genes from Bifidobacterium adolescentis were cloned into an E. coli plasmid giving a ∼10,000-fold increase in transformation frequency (Yasui et al., 2009). There is only one example of PAM being applied to a type I RM system (Yasui et al., 2009). The expression of the hsdMS genes of Lactococcus lactis IO-1 from a plasmid in E. coli BL21 (DE3) yielded a seven fold improvement in transformation.
We have constructed a strain of E. coli that expresses the functional set of hsdMS genes from MRSA252 from an intergenic location in the chromosome (manuscript in preparation, Monk and Foster) (Figure 1). Plasmid DNA isolated from SA30B (DC10B::hsdMSMRSA252) was transformed into MRSA252 at the same frequency as the plasmid isolated from MRSA252 with a 1000-fold improvement in transformation efficiency compared to plasmid isolated from DC10B. We are currently introducing the functional hsdMS genes from strains of different CC's of S. aureus and from different staphylococcal species into DC10B. These hosts will be invaluable for generating plasmids for genetic manipulation of staphylococcal strains that are currently refractory to transformation and will permit fulfillment of Molecular Koch's Postulates in diverse hosts.
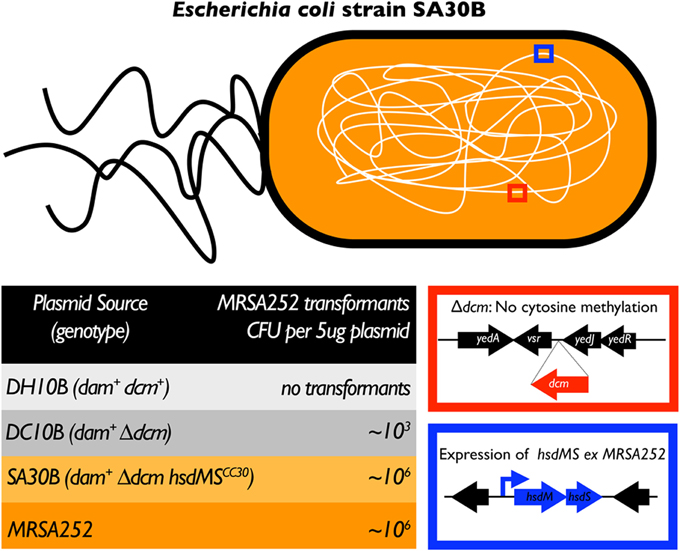
Figure 1. Plasmid artificial modification—application to staphylococci. In a cloning strain of E. coli (DH10B), the chromosome was manipulated by recombineering to develop a host for the maximal efficiency of plasmid transfer into the target staphylococcal strain. To bypass the type IV restriction system, the dcm gene was deleted resulting in a loss of cytosine methylation (red box), creating strain DC10B. Secondly, the hsdMS genes of the target staphylococcal strain, under the control of a strong promoter (blue box) were introduced onto the chromosome of DC10B at a neutral site forming, for example. E. coli strain SA30B—with hsdMS genes derived from MRSA252 (clonal complex 30). A shuttle plasmid was isolated from each E. coli strain described above and S. aureus MRSA252. Five micrograms of shuttle plasmid was transformed into MRSA252 with the CFU enumerated after growth on antibiotic-containing selective agar (bottom left table).
Efficient Allelic Exchange
Several plasmids have been developed to facilitate the construction of mutations in the chromosome of staphylococci by allelic exchange. The preferred method employs a temperature sensitive (ts) plasmid, which replicates by the rolling circle mechanism. A ts version of pE194 is the most widely used replicon for allelic exchange in staphylococci (Gryczan et al., 1982). The procedure for creating a mutation is a two-step process (Figure 2). First, the plasmid carrying the mutational cassette is transformed into the target strain at a temperature that is permissive for replication. Then integration by a single crossover (SCO) event at either the upstream or downstream region of homology is selected by growing at the non-permissive temperature for replication while selecting for antibiotic resistance encoded by the plasmid. This forms an integrant and creates a duplication of the locus to be mutated with one copy being wild-type and the other copy carrying the mutation. Excision of the plasmid by a SCO event is stimulated by decreasing the temperature to one permissive for plasmid replication, in the absence of antibiotic selection. This triggers recombination and loss of the plasmid. If the second crossover event occurs at the region of homology used for integration the strain remains wild-type. If recombination occurs at the opposite region of homology the mutant allele is left in the chromosome. Excision at the same site as that used for integration should occur in theory at the same frequency as at the heterologous site giving a mutation frequency of 50%. In practice mutation frequencies as low as 1% (or lower) may occur (Biswas et al., 1993; Bae and Schneewind, 2006). This can make identification of a mutant that lacks a selectable marker a laborious and time consuming process. A number of improvements to allelic exchange have been devised for staphylococci and are detailed below.
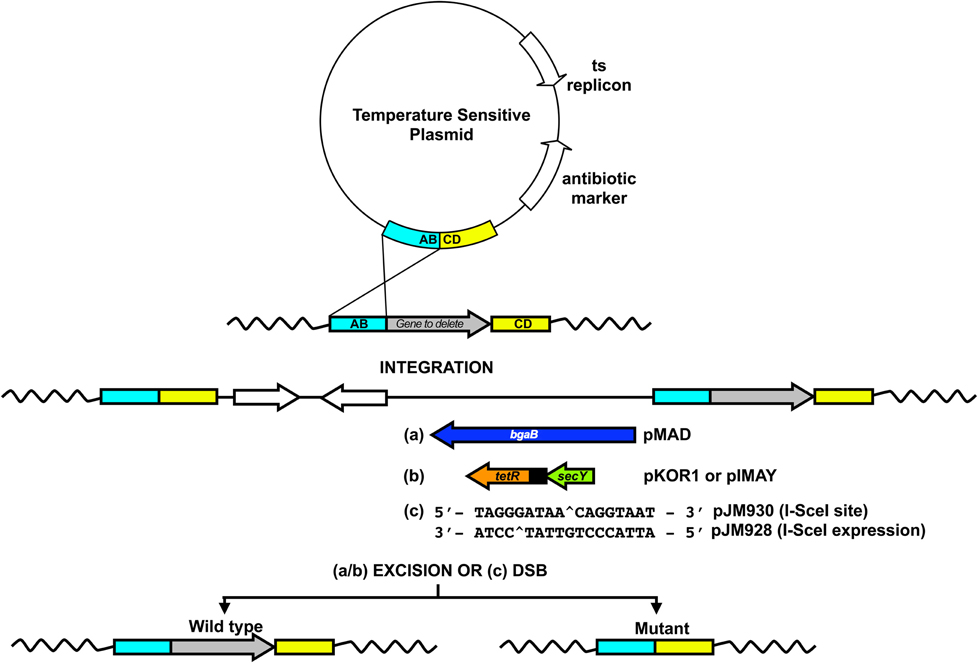
Figure 2. Allelic exchange in staphylococci. The two-step approach (integration/excision) is shown above for the creation of directed mutations. A deletion construct is assembled in the multiple cloning site of a temperature sensitive plasmid and then transformed into the target strain at a temperature permissive for replication. A temperature shift, to one non-permissive for plasmid replication (in the presence of selection for antibiotic resistance encoded by the plasmid), stimulates integration through either the up (AB—shown here) or downstream (CD) region of cloned homologous DNA. Decreasing the temperature and removing antibiotic selection stimulates rolling circle replication which leads to vector excision. As described in the text, the different allelic exchange plasmids developed for staphylococci contain additional features which aid in discrimination of colonies lacking the plasmid either post excision, e.g., (a) pMAD: constitutive bgaB for the hydrolysis of the colourmetric substrate—detected as white colonies on X-gal containing agar, (b) pKOR1/pIMAY: ATc inducible secY antisense to repress growth of plasmid-containing strains or (c) I-SceI induced double strained DNA breaks (DSB) which promote homologous recombination.
pMAD/pORI280
A temperature sensitive shuttle plasmid was created by joining pE194ts to pBR322 with the subsequent addition of a gene encoding a constitutively expressed thermostable ß-galactosidase (Arnaud et al., 2004). While plasmid excision cannot be selected, colonies that lack the plasmid can be identified on plates containing X-gal where they form white colonies. A similar concept has been applied in lactococci with the pORI280 two-plasmid system (Leenhouts et al., 1996). A suicide plasmid missing two of the replication genes and encoding ß-galactosidase is used for allelic exchange, with a second ts plasmid, pVE6007 (with a pWV01ts replicon) supplying the missing repAD encoded replication functions in trans. By growth at a temperature that is restrictive for replication pVE6007 is lost and the integrants with pORI280 in the chromosome selected with erythromycin. Resolution of integrants occurs after growth in the absence of antibiotic. We have been unable to manipulate S. aureus using the pORI280 two-plasmid system even though two groups have previously reported success (Pinho and Errington, 2004; Daly et al., 2010).
pKOR1
Bae and Schneewind (2006) revolutionized the isolation of mutations in S. aureus by introducing counter selection into the procedure for allelic exchange. A secY antisense transcript which hybridizes to mRNA encoding part of the essential Sec protein secretion system acts as an inhibitor of growth impairing colony formation on agar. Combining tetracycline inducible secY expression and Gateway cloning into the pTS1 ts shuttle plasmid (pE194ts replicon) (Greene et al., 1995) generated the vector pKOR1 (Bae and Schneewind, 2006). The initial stages of allelic exchange are the same as for any ts plasmid. Cells where the plasmid has been lost by reverse SCO can be selected directly on agar following induction by anhydrotetracycline of secY antisense giving cells lacking pKOR1 a growth advantage. However, conditions used for allelic exchange with pE194ts replicons can produce secondary mutations in sae, a locus which encodes a two component signal transduction system (Sun et al., 2010). The elevated temperature of growth (43°C), aeration and low levels of antibiotic resistance expressed by the plasmid's chloramphenicol resistance determinant, particularly when at a single copy in the chromosome, can promote the selection of sae mutations. sae mutations can influence the expression of other genes and alter virulence (Herbert et al., 2010).
pIMAY
To alleviate problems associated with using pKOR1 we have constructed a plasmid vector for allele exchange that has a strongly expressed drug resistance marker and a ts replicon that allows selection of integrants at 37°C (Monk et al., 2012). The pVWO1ts replicon on pIMAY is functional in staphylococci (S. aureus, S. epidermidis and S. lugdunenesis have so far been tested) at the permissive temperature (30°C) but the plasmid cannot replicate at the restrictive temperature of 37°C. The replicon used to propagate the plasmid in E. coli is low copy number which should improve the stability of cloned staphylococcal DNA. The chloramphenicol resistance (cat) gene is expressed from a strong promoter which allows efficient selection as a single copy when integrated into the staphylococcal chromosome. The plasmid carries the inducible secY antisense counterselection determinant of pKOR1. Furthermore, we have recently applied a sequence- and ligation-independent cloning (Li and Elledge, 2007) to pIMAY which increases the cloning efficiency (greater than 90% of colonies screened contain inserts) and reduces the costs and time involved in production of deletion constructs. From the start of cloning to mutant confirmation can be conducted in under two weeks (Monk et al., 2012).
I-SceI
Counter selection with secY enriches for cells which have lost the integrated plasmid following the second SCO event. However, enhancing the rate of plasmid excision requires initiating DNA replication at the integrated plasmid's origin. An alternative approach utilizes a rare cutting restriction enzyme SceI which recognizes an 18 bp sequence yet to be found in bacteria (Posfai et al., 1999). It was first shown in mammalian cells that the induction of double stranded DNA breaks (DSBs) following expression of SceI induced homologous recombination (Choulika et al., 1995). After codon optimization of sceI the enzyme was applied to several Gram-negative and Gram-positive bacteria (Janes and Stibitz, 2006; Szurmant et al., 2007; Flannagan et al., 2008; Blank et al., 2011; Martinez-Garcia and de Lorenzo, 2011). Pagels et al. (2010) developed the SceI system for use in S. aureus. The allelic exchange plasmid with the pE194ts replicon carries the SceI restriction cleavage site (pJM930). After integration into the chromosome by SCO at the locus to be deleted, a second plasmid (pJM928) with a compatible pT181ts replicon constitutively expressing SceI was delivered by transduction. The enzyme introduces a DSB within the integrated plasmid. This induces homologous recombination which promotes plasmid excision, which in other bacteria has increased the frequency of wild-type to mutant up to 50% (Posfai et al., 1999; Janes and Stibitz, 2006). Cells where the plasmid has been excised will not have a DNA break in the chromosome while the excised plasmid carrying the cleavage site will be destroyed. This approach should dramatically improve the efficiency of generating mutations. Ideally an inducible sceI gene should be part of the mutational plasmid to avoid the need to use a second plasmid, but this will require a very strong repressor to prevent SceI expression during plasmid propagation and integration.
Temperature—An Important Consideration
A recent publication has identified sequential non-selective passage at a reduced temperature (25°C) as a method to improve the frequency of co-integrate resolution (Kato and Sugai, 2011). Their results also highlighted the requirement for the isolation of both SCOs, either through the up or downstream region of homology. We have found that resuspension of a colony and direct plating rather than broth growth at the non-permissive temperature for plasmid replication improves the isolation of both SCOs (Monk et al., 2012). Combining the DC10B strain with pIMAY or other ts plasmids will enhance isolation of mutations in laboratory strains and diverse clinical isolates.
Validation of Mutations by Complementation
After construction of a mutant, any change in phenotype should be corroborated by complementation in order to prevent attributing properties to the missing gene that are actually due to secondary mutations. Currently four approaches to complementation can be applied.
Shuttle Plasmids
A number of shuttle plasmids that replicate both in E. coli and staphylococci have been constructed that utilize several different plasmid replicons (pE194, pC194, pT181, pKS1). The effect of plasmid copy number should be considered when attempting to complement a mutation. A gene-dosage effect may lead to high level expression which could potentially be toxic. This could be reduced by using an inducible gene expression system (see below). Plasmid-based complementation can be established quickly compared to other methods. However it can be difficult to use in animal infection experiments where the plasmid is often lost in the absence of antibiotic selection in vivo (Cho et al., 2011). It is possible to maintain selection by administering antibiotics into drinking water but this is not ideal (de Azavedo et al., 1985; Bubeck Wardenburg et al., 2006). For an extensive review of plasmids used in staphylococcal research see McNamara (2008).
Inducible Gene Expression
Promoters that can be activated by the inducers IPTG (Zhang et al., 2000), xylose (Peschel et al., 1996), cadmium (Charpentier et al., 2004) and anhydrotetracycline (ATc) (Bateman et al., 2001) have been used in staphylococci. The ATc inducible vector pRMC2 was derived from pALC2073 (Bateman et al., 2001) by increasing the level of expression of the TetR repressor to reduce leakiness (Corrigan and Foster, 2009). Recently the laboratory of R. Bertram has constructed and validated a series of improved ATc inducible expression vectors that were derived from pRMC2. By inserting a second tetO binding site for TetR downstream of the −10 box of the Pxyl/tet promoter, creating pRAB11, a greater level of repression compared to that of pRMC2 was observed (Helle et al., 2011). Mutations in the –10 and –35 boxes of the Pxyl/tet promoter in pRAB11 resulted in reduced expression, but achieved a higher level of repression. Finally, a hybrid tetR gene (tetR-BD) improved both the level of expression when induced and the level of repression when uninduced compared to pRAB11 while introduction revtetR (reverse tetR—contains 3 amino acid changes which reverse the activity of TetR) yielded a construct comparable to pRAB11, except exhibiting repression in the presence of ATc but induction in the absence (Helle et al., 2011). These vectors should be of great value for experiments requiring controlled expression of a cloned gene in vitro.
Phage Integrase Vectors
Integrating vectors that utilize the att sites and integrases of lysogenic phages have been developed to eliminate the problems associated with complementation by extrachromosomally replicating plasmids. These allow the integration of a plasmid at a specific phage attachment site in the chromosome directed by a phage integrase (van Mellaert et al., 1998; Haldimann and Wanner, 2001; Lauer et al., 2002; Huff et al., 2010). The integrated plasmid lacks a replicon that functions in S. aureus and is stable even in the absence of selective antibiotic. The presence of only a single copy of the plasmid eliminates gene-dosage effects.
The pLL39 phage integrase vector encodes both ϕL54a and ϕ11 attP sites and in the presence of the appropriate phage integrase can insert into the matching chromosomal attB site (Luong and Lee, 2007). This vector is a refinement of the previously developed and successfully applied pCL83/pCL84 vectors (Lee et al., 1991). To stimulate pLL39 integration, the integrase is encoded on a second extrachromosomally replicating plasmid (Luong and Lee, 2007). Similar two plasmid systems have been created based on the phage related S. aureus pathogenicity island (SaPI1) using the SaPI1 int and attS to direct integration (Charpentier et al., 2004). However until now both pLL39 and the SaPI1 based vector system could only be integrated into the chromosome of RN4220 and must then be moved by generalized transduction into the target strain. Transduction from RN4220 can be problematic because DNA that is located adjacent to the integrated plasmid will also be introduced by the double reciprocal recombination event. This could introduce mutations into the recipient. Also transfer is limited to strains that are closely related to RN4220, similar to plasmid transformation discussed earlier, due to RM. It is also possible that the integration of the plasmid itself could cause a change in virulence of the host. It is of note that the ϕL54a attB site is located within the geh gene which encodes an extracellular lipase (Lee and Iandolo, 1985). Although this could be controlled for by comparing the complemented mutant strain with the mutant carrying the empty vector, the ideal integrating vector would insert into an attB site that is located in an intergenic region. Lei et al. (2012) recently constructed a phage integrase vector, which recognizes an engineered core attB sequence based on the ϕL54a. This synthetic attB sequence was inserted onto the chromosome of RN4220 at a region characterized with negligible transcriptional activity and allowed the transformation with the attP compatible vector (pLL102). The region is highly conserved among S. aureus isolates which should allow the transfer either by phage transduction or by direct electroporation.
We are currently testing two new small (∼3 kb) phage integrase vectors that incorporate phage ϕ11 or ϕ80 attP sites as well as the appropriate integrase gene. The plasmids can be propagated in E. coli DC10B and transformed directly into the wild type S. aureus strain where they will integrate under the direction of the encoded integrase. This eliminates the need for a second plasmid encoding the integrase and improves the efficiency of the process (Monk, Tan, Shah and Foster, unpublished).
Gene Reconstitution
Sometimes genes cloned into multicopy plasmids can be toxic to E. coli or the target strain (Pilgrim et al., 2003; Alonzo et al., 2011), expression from a phage integrase vector may occur at a different level compared to wild-type despite the presence of the native promoter (Lauer et al., 2002) or polar effects may need to be ruled out. Reconstitution of the mutated gene to wild-type (Donegan and Cheung, 2009; Diep et al., 2010; Monk et al., 2012) by reverse allelic exchange can be employed. The wild-type gene and flanking DNA are cloned into a ts allelic exchange-promoting plasmid such as pIMAY. A codon change that creates a novel restriction site without altering the amino acid sequence of the encoded protein (http://emboss.bioinformatics.nl/cgi-bin/emboss/silent)is introduced to facilitate identification of the restored gene. This is time consuming but results in a complemented strain that differs from wild-type only by the introduced restriction site.
Conclusions and Future Perspectives
In this article we have described a genetic toolbox that is applicable to many species of Staphylococcus and we have reviewed how recent developments in understanding of RM systems have greatly improved the ability to manipulate these bacteria genetically. Bypassing the host encoded RM systems with plasmid DNA isolated from E. coli DC10B expressing staphylococcal HsdMS proteins will enable the rapid construction of mutant strains by allelic exchange and for their complementation.
S. epidermidis strains are much more difficult to work with than S. aureus because they are transformable by electroporation at a much lower frequency. Wall teichoic acid has recently been identified as a barrier to transformation in S. epidermidis (Holland et al., 2011). Optimization of conditions for generating competent cells for electroporation (Lofblom et al., 2007) will be required to increase the frequency of transformation. Development of a single plasmid phage integrase vector for S. epidermidis would require enhanced transformation.
To take advantage of the high transformation efficiencies that can be achieved in S. aureus, we propose to apply single stranded DNA recombineering where point mutations, small insertions or deletions can be rapidly constructed in chromosomal genes (Swingle et al., 2010). This technology has been used E. coli, Shigella flexneri, Salmonella typhimurium, Pseudomonas syringae (Swingle et al., 2010), Mycobacteria (van Kessel and Hatfull, 2008) and more recently to lactic acid bacteria (van Pijkeren and Britton, 2012). Expression of the recT-encoded single strand DNA annealing protein enhances the incorporation of synthetic oligonucleotides designed to bypass the mismatch repair pathway. We have identified several recT homologues that are functional in S. aureus and are currently optimizing the procedure (Monk, van Pijkeren, Britton and Foster, unpublished).
Through mining staphylococcal genome sequences, we have observed that both the type I and/or type IV systems are highly conserved in staphylococci. However, some strains (e.g., S. aureus RF122, Cowan, JKD6159 and S. pseudintermedius ED99) encode additional type I or II RM systems, which can impair transformation (unpublished data). The type IV restriction system can be by-passed using plasmid that lacks cytosine methylation while a type I or a type II system bypass would require the plasmid obtain the methylation profile of the specific RM system.
In summary the improvements in vectors and transformation described above has facilitated genetic manipulation in many strains of S. aureus and other staphylococcal species which were previously not amenable to transformation with plasmid DNA (Monk et al., 2012). Also the time taken to construct strains and to perform genetic manipulation has been reduced significantly. This will facilitate more sophisticated genetic manipulation in clinically relevant strains and will contribute directly to rapid advances in knowledge about this important group of organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alonzo, F. R., McMullen, P., and Freitag, N. (2011). Actin polymerization drives septation of Listeria monocytogenes namA hydrolase mutants, demonstrating host correction of a bacterial defect. Infect. Immun. 79, 1458–1470.
Arnaud, M., Chastanet, A., and Debarbouille, M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bac-teria. Appl. Environ. Microbiol. 70, 6887–6891.
Asheshov, E., and Jevons, M. (1963). The effect of heat on the ability of a host strain to support the growth of a Staphylococcus phage. J. Gen. Microbiol. 31, 97–107.
Augustin, J., and Gotz, F. (1990). Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54, 203–207.
Baba, T., Bae, T., Schneewind, O., Takeuchi, F., and Hiramatsu, K. (2008). Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190, 300–310.
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plas-mid 55, 58–63.
Bateman, B., Donegan, N., Jarry, T., Palma, M., and Cheung, A. (2001). Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69, 7851–7857.
Biswas, I., Gruss, A., Ehrlich, S., and Maguin, E. (1993). High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175, 3628–3635.
Blank, K., Hensel, M., and Gerlach, R. (2011). Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One 6:e15763. doi: 10.1371/journal.pone.0015763
Bosch, T., De Neeling, A. J., Schouls, L. M., Van Der Zwaluw, K. W., Kluytmans, J. A., Grundmann, H., and Huijsdens, X. W. (2010). PFGE diversity within the methicillin-resistant Staphylococcus aureus clonal lineage ST398. BMC Microbiol. 10, 40.
Bubeck Wardenburg, J., Williams, W., and Missiakas, D. (2006). Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 103, 13831–13836.
Charpentier, E., Anton, A., Barry, P., Alfonso, B., Fang, Y., and Novick, R. (2004). Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70, 6076–6085.
Cho, J., Zussman, J., Donegan, N., Ramos, R., Garcia, N., Uslan, D., Iwakura, Y., Simon, S., Cheung, A., Modlin, R., Kim, J., and Miller, L. (2011). Noninvasive in vivo imaging to evaluate immune responses and antimicrobial therapy against Staphylococcus aureus and USA300 MRSA skin infections. J. Invest. Dermatol. 131, 907–915.
Choulika, A., Perrin, A., Dujon, B., and Nicolas, J. (1995). Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 1968–1973.
Cockfield, J. D., Pathak, S., Edgeworth, J. D., and Lindsay, J. A. (2007). Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J. Med. Microbiol. 56, 614–619.
Corrigan, R., and Foster, T. (2009). An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61, 126–129.
Corvaglia, A., Francois, P., Hernandez, D., Perron, K., Linder, P., and Schrenzel, J. (2010). A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. U.S.A. 107, 11954–11958.
Daly, K., Upton, M., Sandiford, S., Draper, L., Wescombe, P., Jack, R., O'Connor, P., Rossney, A., Gotz, F., Hill, C., Cotter, P., Ross, R., and Tagg, J. (2010). Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J. Bacteriol. 192, 1131–1142.
de Azavedo, J., Foster, T., Hartigan, P., Arbuthnott, J., O'Reilly, M., Kreiswirth, B., and Novick, R. (1985). Expression of the cloned toxic shock syndrome toxin 1 gene (tst) in vivo with a rabbit uterine model. Infect. Immun. 50, 304–309.
Diep, B. A., Chan, L., Tattevin, P., Kajikawa, O., Martin, T. R., Basuino, L., Mai, T. T., Marbach, H., Braughton, K. R., Whitney, A. R., Gardner, D. J., Fan, X., Tseng, C. W., Liu, G. Y., Badiou, C., Etienne, J., Lina, G., Matthay, M. A., DeLeo, F. R., and Chambers, H. F. (2010). Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. U.S.A. 107, 5587–5592.
Donegan, N., and Cheung, A. (2009). Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 191, 2795–2805.
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Falkow, S. (1988). Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10 (Suppl. 2), S274–S276.
Flannagan, R., Linn, T., and Valvano, M. (2008). A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10, 1652–1660.
Gill, S., Fouts, D., Archer, G., Mongodin, E., Deboy, R., Ravel, J., Paulsen, I., Kolonay, J., Brinkac, L., Beanan, M., Dodson, R., Daugherty, S., Madupu, R., Angiuoli, S., Durkin, A., Haft, D., Vamathevan, J., Khouri, H., Utterback, T., Lee, C., Dimitrov, G., Jiang, L., Qin, H., Weidman, J., Tran, K., Kang, K., Hance, I., Nelson, K., and Fraser, C. (2005). Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438.
Greene, C., McDevitt, D., Francois, P., Vaudaux, P., Lew, D., and Foster, T. (1995). Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17, 1143–1152.
Gryczan, T., Hahn, J., Contente, S., and Dubnau, D. (1982). Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J. Bacteriol. 152, 722–735.
Haldimann, A., and Wanner, B. (2001). Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183, 6384–6393.
Heilmann, C., Gerke, C., Perdreau-Remington, F., and Gotz, F. (1996). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282.
Helle, L., Kull, M., Mayer, S., Marincola, G., Zelder, M., Goerke, C., Wolz, C., and Bertram, R. (2011). Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157, 3314–3323.
Herbert, S., Ziebandt, A., Ohlsen, K., Schafer, T., Hecker, M., Albrecht, D., Novick, R., and Gotz, F. (2010). Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78, 2877–2889.
Holland, L., Conlon, B., and O'Gara, J. (2011). Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157, 408–418.
Huff, J., Czyz, A., Landick, R., and Niederweis, M. (2010). Taking phage integration to the next level as a genetic tool for mycobacteria. Gene 468, 8–19.
Iordanescu, S., and Surdeanu, M. (1976). Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96, 277–281.
Janes, B., and Stibitz, S. (2006). Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74, 1949–1953.
Kato, F., and Sugai, M. (2011). A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods 87, 76–81.
Kelleher, J., and Raleigh, E. (1991). A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J. Bacteriol. 173, 5220–5223.
Kraemer, G., and Iandolo, J. (1990). High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21, 373–376.
Kreiswirth, B., Lofdahl, S., Betley, M., O'Reilly, M., Schlievert, P., Bergdoll, M., and Novick, R. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712.
Lauer, P., Chow, M., Loessner, M., Portnoy, D., and Calendar, R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186.
Lee, C., and Iandolo, J. (1985). Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J. Bacteriol. 164, 288–293.
Lee, C. Y., Buranen, S. L., and Ye, Z. H. (1991). Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103, 101–105.
Leenhouts, K., Buist, G., Bolhuis, A. Ten Berge, A., Kiel, J., Mierau, I., Dabrowska, M., Venema, G., and Kok, J. (1996). A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253, 217–224.
Lei, M. G., Cue, D., Alba, J., Junecko, J., Graham, J. W., and Lee, C. Y. (2012). A single copy integration vector that integrates at an engineered site on the Staphylococcus aureus chromosome. BMC Res. Notes 5, 5.
Li, M., and Elledge, S. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256.
Lindsay, J. A. (2010). Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 98–103.
Lofblom, J., Kronqvist, N., Uhlen, M., Stahl, S., and Wernerus, H. (2007). Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 102, 736–747.
Luong, T., and Lee, C. (2007). Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70, 186–190.
Malachowa, N., and DeLeo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071.
Martinez-Garcia, E., and de Lorenzo, V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716.
McNamara, P. (2008). “Genetic manipulation of Staphylococcus aureus,” in Staphylococcus: Molecular Genetics, ed J. Lindsay (Norfolk, UK: Caister Academic Press), 89–130.
Monk, I. R., ShahI. M., Xu, M., Tan, M-W., and Foster, T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio. 3, e00277–e00211.
Morikawa, K., Inose, Y., Okamura, H., Maruyama, A., Hayashi, H., Takeyasu, K., and Ohta, T. (2003). A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8, 699–712.
Murray, N. (2000). Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64, 412–434.
Nair, D., Memmi, G., Hernandez, D., Bard, J., Beaume, M., Gill, S., Francois, P., and Cheung, A. (2011). Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193, 2332–2335.
O'Connell Motherway, M., O'Driscoll, J., Fitzgerald, G. F., and Van Sinderen, D. (2009). Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2, 321–332.
O'Driscoll, J., Fitzgerald, G. F., and Van Sinderen, D. (2005). A dichotomous epigenetic mechanism governs expression of the LlaJI restriction/modification system. Mol. Microbiol. 57, 1532–1544.
O'Neill, A. (2010). Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 51, 358–361.
Oskouian, B., and Stewart, G. (1990). Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J. Bacteriol. 172, 3804–3812.
Pagels, M., Fuchs, S., Pane-Farre, J., Kohler, C., Menschner, L., Hecker, M., McNamara, P., Bauer, M., Von Wachenfeldt, C., Liebeke, M., Lalk, M., Sander, G. Von Eiff, C., Proctor, R., and Engelmann, S. (2010). Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161.
Palmer, B., and Marinus, M. (1994). The dam and dcm strains of Escherichia coli–a review. Gene 143, 1–12.
Peschel, A., Ottenwalder, B., and Gotz, F. (1996). Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137, 279–284.
Pilgrim, S., Kolb-Maurer, A., Gentschev, I., Goebel, W., and Kuhn, M. (2003). Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 71, 3473–3484.
Pinho, M., and Errington, J. (2004). A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol. Lett. 240, 145–149.
Posfai, G., Kolisnychenko, V., Bereczki, Z., and Blattner, F. (1999). Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27, 4409–4415.
Roundtree, P. (1956). Variations in a related series of Staphylococcal bacteriophages. J. Gen. Microbiol. 15, 266–279.
Schenk, S., and Laddaga, R. (1992). Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73, 133–138.
Simons, M., and Szczelkun, M. (2011). Recycling of protein subunits during DNA translocation and cleavage by Type I restriction-modification enzymes. Nucleic Acids Res. 39, 7656–7666.
Stobberingh, E., Schiphof, R., and Sussenbach, J. (1977). Occurrence of a class II restriction endonuclease in Staphylococcus aureus. J. Bacteriol. 131, 645–649.
Stobberingh, E., and Winkler, K. (1977). Restriction-deficient mutants of Staphylococcus aureus. J. Gen. Microbiol. 99, 359–367.
Sun, F., Cho, H., Jeong, D., Li, C., He, C., and Bae, T. (2010). Aureusimines in Staphylococcus aureus are not involved in virulence. PLoS One 5:e15703. doi: 10.1371/journal.pone.0015703
Sung, J. M., and Lindsay, J. A. (2007). Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 51, 2189–2191.
Suzuki, T., and Yasui, K. (2011). Plasmid artificial modification: a novel method for efficient DNA transfer into bacteria. Methods Mol. Biol. 765, 309–326.
Swingle, B., Markel, E., Costantino, N., Bubunenko, M., Cartinhour, S., and Court, D. (2010). Oligonucleotide recombination in Gram-negative bacteria. Mol. Microbiol. 75, 138–148.
Szurmant, H., Mohan, M., Imus, P., and Hoch, J. (2007). YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189, 3280–3289.
van Kessel, J., and Hatfull, G. (2008). Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol. Microbiol. 67, 1094–1107.
van Mellaert, L., Mei, L., Lammertyn, E., Schacht, S., and Anne, J. (1998). Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of a VWB-based integrative vector. Microbiology 144, 3351–3358.
van Pijkeren, J. P., and Britton, R. A. (2012). High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. doi: 10.1093/nar/gks147. [Epub ahead of print].
Veiga, H., and Pinho, M. (2009). Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl. Environ. Microbiol. 75, 3034–3038.
Voyich, J., Braughton, K., Sturdevant, D., Whitney, A., Said-Salim, B., Porcella, S., Long, R., Dorward, D., Gardner, D., Kreiswirth, B., Musser, J., and DeLeo, F. (2005). Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175, 3907–3919.
Waldron, D. E., and Lindsay, J. A. (2006). Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188, 5578–5585.
Wion, D., and Casadesus, J. (2006). N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192.
Xu, S., Corvaglia, A., Chan, S., Zheng, Y., and Linder, P. (2011). A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39, 5597–5610.
Yasui, K., Kano, Y., Tanaka, K., Watanabe, K., Shimizu-Kadota, M., Yoshikawa, H., and Suzuki, T. (2009). Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 37, e3.
Yildirim, S., Elhanafi, D., Lin, W., Hitchins, A. D., Siletzky, R. M., and Kathariou, S. (2010). Conservation of genomic localization and sequence content of Sau3AI-like restriction-modification gene cassettes among Listeria monocytogenes epidemic clone I and selected strains of serotype 1/2a. Appl. Environ. Microbiol. 76, 5577–5584.
Zhang, L., Fan, F., Palmer, L., Lonetto, M., Petit, C., Voelker, L., St John, A., Bankosky, B., Rosenberg, M., and McDevitt, D. (2000). Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255, 297–305.
Keywords: transformation, mutation, staphylococcus, allelic exchange, electroporation, aureus, epidermidis, restriction-modification
Citation: Monk IR and Foster TJ (2012) Genetic manipulation of Staphylococci—breaking through the barrier. Front. Cell. Inf. Microbio. 2:49. doi: 10.3389/fcimb.2012.00049
Received: 14 January 2012; Accepted: 20 March 2012;
Published online: 12 April 2012.
Edited by:
David Heinrichs, University of Western Ontario, CanadaReviewed by:
Jodi A. Lindsay, St. George's, University of London, UKChia Lee, University of Arkansas for Medical Sciences, USA
Copyright: © 2012 Monk and Foster. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ian R. Monk, Department of Microbiology, Moyne Institute of Preventive Medicine, School of Genetics and Microbiology, Trinity College Dublin, Dublin 2, Ireland. e-mail: monki@tcd.ie