Abstract
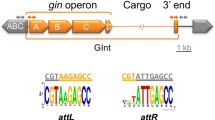
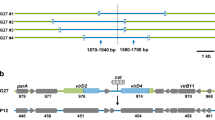
Integrative conjugative elements (ICEs) are a large group of mobile elements found in Gram-positive and Gram-negative bacteria. These genetic elements are replicated by incorporation into the host chromosome, but retain the capability of excision and conjugative transfer. A set of genes in ICEs enables conjugative transfer and control of element removal and integration into the host chromosome. These features indicate that ICEs are directly involved in processes of horizontal transfer of genetic determinants, which increase the adaptive potential of bacterial species, and can also function as universal mobilizing factors for other genetic elements.
Similar content being viewed by others
References
Wozniak, R. and Waldor, M., Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow, Nat. Rev. Microbiol., 2010, Vol. 8, pp. 552–563.
Burrus, V. and Waldor, M., Shaping bacterial genomes with integrative and conjugative elements, Res. Microbiol., 2004, Vol. 155, No. 5, pp. 376–386.
Hastings, P., Rosenberg, S., and Slack, A., Antibioticinduced lateral transfer of antibiotic resistance, Trends Microbiol, 2004, Vol. 12, No. 9, pp. 401–404.
Wozniak, R., Fouts, D., Spagnoletti, M., et al., Comparative ICE genomics: Insights into the evolution of the SXT/R391 family of ICEs, PLoS Genet., 2009, Vol. 5, No. 12, p. e1000786. doi:10.1371/journal.pgen.1000786
Ryan, M., Pembroke, J., and Adley, C., Novel Tn4371ICE like element in Ralstonia pickettii and genome mining for comparative elements, BMC Microbiol., 2009, Vol. 9, p. 242. doi: 10.1186/1471–2180-9–242
Burrus, V., Pavlovic, G., Decaris, B., and Guédon, G., Conjugative transposons The tip of the iceberg, Mol. Microbiol., 2002, Vol. 46, No. 3, pp. 601–610.
Hochhut, B., Lotfi, Y., Mazel, D., et al., Molecular analysis of the antibiotic resistance gene clusters in the Vibrio cholerae O139 and O1 SXT constins, Antimicrob. Agents Chemother., 2001, Vol. 45, No. 11, pp. 2991–3000.
Waldor, M., Tschäpe, H., and Mekalanos, J., A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139, J. Bacteriol., 1996, Vol. 178, No. 14, pp. 4157–4165.
Burrus, V., Marrero, J., and Waldor, M., The current ICE age: Biology and evolution of SXT-related integrating conjugative elements, Plasmid, 2006, Vol. 55, No. 3, pp. 173–183.
Ehara, M., Nguyen, B., Nguyen, D., et al., Drug susceptibility and its genetic basis in epidemic Vibrio cholerae O1 in Vietnam, Epidemiol. Infect., 2004, Vol. 132, No. 4, pp. 595–600.
Iwanaga, M., Toma, C., Miyazato, T., et al., Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos, Antimicrob. Agents Chemother., 2004, Vol. 48, No. 7, pp. 2364–2369.
Thungapathra, M., Amita, A., Sinha, K., et al., Occurrence of antibiotic resistance gene cassettes aac(6')-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India, Antimicrob. Agents Chemother., 2002, Vol. 46, No. 9, pp. 2948–2955.
Ahmed, A., Shinoda, S., and Shimamoto, T., A variant type of Vibrio cholerae SXT element in a multidrugresistant strain of Vibrio fluvialis, FEMS Microbiol. Lett., 2005, Vol. 242, No. 2, pp. 241–247.
Burrus, V., Quezada-Calvillo, R., Marrero, J., et al., SXT-related integrating conjugative element in New World Vibrio cholera, Appl. Environ. Microbiol., 2006, Vol. 72, No. 4, pp. 3054–3057.
Dalsgaard, A., Forslund, A., Sandvang, D., et al., Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons, J. Antimicrob. Chemother., 2001, Vol. 48, pp. 827–838.
Rodríguez-Bianco, A., Lemos, M., and Osorio, C., Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments, Antimicrob. Agents Chemother., 2012, Vol. 56, No. 5, pp. 2619–2626.
Burrus, V., Significance of the SXT/R391 family of integrating conjugative elements in Vibrio cholerae, in Epidemiological and Molecular Aspects on Cholera Infectious Disease, Ramamurthy, T. and Bhattacharya, S.K., Springer, 2011, pp. 161–184.
Osorio, C., Marrero, J., Wozniak, R., et al., Genomic and functional analysis of ICEPdaSpa1, a fish-pathogen-derived SXT related integrating conjugative element that can mobilize a virulence plasmid, J. Bacteriol., 2008, Vol. 190, No. 9, pp. 3353–3361.
Pembroke, J. and Piterina, A., A novel ICE in the genome of Shewanella putrefaciens W3–18-1: Comparison with the SXT/R391 ICE-like elements, FEMS Microbiol. Lett., 2006, Vol. 264, No. 1, pp. 80–88.
Burrus, V. and Waldor, M., Shaping bacterial genomes with integrative and conjugative elements, Res. Microbiol, 2004, Vol. 155, No. 5, pp. 376–386.
Seth-Smith, H., Fookes, M., Okoro, C., et al., Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae, J. Bacteriol., 2012, Vol. 194, No. 6, pp. 1494–1504.
Ghinet, M., Bordeleau, E., Beaudin, J., et al., Uncovering the prevalence and diversity of integrating conjugative elements in Actinobacteria, Mob. Genet. Elements, 2012, Vol. 2, No. 2, pp. 119–124.
Beaber, J. and Waldor, M., Identification of operators and promoters that control SXT conjugative transfer, J. Bacteriol., 2004, Vol. 186, No. 17, pp. 5945–5949.
Yokota, T. and Kuwahara, S., Temperature-sensitive R plasmid obtained from naturally isolated drug-resistant Vibrio cholerae (biotype El Tor), Antimicrob. Agents Chemother., 1977, Vol. 11, No. 1, pp. 13–20.
Beaber, J., Burrus, V., Hochhut, B., et al., Comparison of SXT and R391, two conjugative integrating elements definition of a genetic backbone for the mobilization of resistance determinants, Cell. Mol. Life Sci., 2002, Vol. 59, No. 12, pp. 2065–2070.
Hochhut, B. and Waldor, M., Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC, Mol. Microbiol., 1999, Vol. 32, No. 1, pp. 99–110.
Marrero, J. and Waldor, M., Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer, Dev. Cell, 2005, Vol. 8, No. 6, pp. 963–970.
Ramamurthy, T., Garg, S., Sharma, R., et al., Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India, Lancet, 1993, Vol. 341, No. 8846, pp. 703–704.
Beaber, J., Hochhut, B., and Waldor, M., SOS response promotes horizontal dissemination of antibiotic resistance genes, Nature, 2004, Vol. 427, No. 6969, pp. 72–74.
Beaber, J., Hochhut, B., and Waldor, M., Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholera, J. Bacteriol., 2002, Vol. 184, No. 15, pp. 4259–4269.
Bordeleau, E., Brouillette, E., Robichaud, N., and Burrus, V., Beyond antibiotic resistance: Integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-diGMP signalling in Vibrio cholerae, Environ. Microbiol., 2010, Vol. 12, No. 2, pp. 510–523.
Sherburne, C., Lawley, T., Gilmour, M., et al., The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer, Nucleic Acids Res., 2000, Vol. 28, No. 10, pp. 2177–2186.
Garriss, G., Waldor, M., and Burrus, V., Mobile antibiotic resistance encoding elements promote their own diversity, PLoS Genet., 2009, Vol. 5, No. 12, p. e1000775. doi: 10.1371/journal.pgen.1000775
Roberts, A. and Mullany, P., A modular master on the move: The Tn916 family of mobile genetic elements, Trends Microbiol., 2009, Vol. 17, No. 6, pp. 251–258.
Gaillard, M., Vallaeys, T., Vorhölter, F., et al., The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties, J. Bacteriol., 2006, Vol. 188, No. 5, pp. 1999–2013.
Moon, K., Sonnenburg, J., and Salyers, A., Unexpected effect of a Bacteroides conjugative transposon, CTnDOT, on chromosomal gene expression in its bacterial host, Mol. Microbiol., 2007, Vol. 64, No. 6, pp. 1562–1571.
Smyth, D. and Robinson, D., Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus, J. Bacteriol., 2009, Vol. 191, No. 19, pp. 5964–5975.
Frost, L., Leplae, R., Summers, A., and Toussaint, A., Mobile genetic elements: The agents of open source evolution, Nat. Rev. Microbiol., 2005, Vol. 3, No. 9, pp. 722–732.
Mazel, D., Dychinco, B., Webb, V., and Davies, J., Antibiotic resistance in the ECOR collection: Integrals and identification of a novel aad gene, Antimicrob. Agents Chemother., 2000, Vol. 44, No. 6, pp. 1568–1574.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © I.B. Zakharova, D.V. Viktorov, 2015, published in Molekulyarnaya Genetika, Mikrobiologiya i Virusologiya, 2015, No. 3, pp. 9–16.
About this article
Cite this article
Zakharova, I.B., Viktorov, D.V. Integrative conjugative elements (ICEs) of microorganisms. Mol. Genet. Microbiol. Virol. 30, 114–123 (2015). https://doi.org/10.3103/S0891416815030076
Received:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0891416815030076