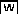-
PDF
- Split View
-
Views
-
Cite
Cite
Xinping Xu, Chunhong Chen, Baofang Fan, Zhixiang Chen, Physical and Functional Interactions between Pathogen-Induced Arabidopsis WRKY18, WRKY40, and WRKY60 Transcription Factors, The Plant Cell, Volume 18, Issue 5, May 2006, Pages 1310–1326, https://doi.org/10.1105/tpc.105.037523
Close - Share Icon Share
Abstract
Limited information is available about the roles of specific WRKY transcription factors in plant defense. We report physical and functional interactions between structurally related and pathogen-induced WRKY18, WRKY40, and WRKY60 transcription factors in Arabidopsis thaliana. The three WRKY proteins formed both homocomplexes and heterocomplexes and DNA binding activities were significantly shifted depending on which WRKY proteins were present in these complexes. Single WRKY mutants exhibited no or small alterations in response to the hemibiotrophic bacterial pathogen Pseudomonas syringae and the necrotrophic fungal pathogen Botrytis cinerea. However, wrky18 wrky40 and wrky18 wrky60 double mutants and the wrky18 wrky40 wrky60 triple mutant were substantially more resistant to P. syringae but more susceptible to B. cinerea than wild-type plants. Thus, the three WRKY proteins have partially redundant roles in plant responses to the two distinct types of pathogens, with WRKY18 playing a more important role than the other two. The contrasting responses of these WRKY mutants to the two pathogens correlated with opposite effects on pathogen-induced expression of salicylic acid–regulated PATHOGENESIS-RELATED1 and jasmonic acid–regulated PDF1.2. While constitutive expression of WRKY18 enhanced resistance to P. syringae, its coexpression with WRKY40 or WRKY60 made plants more susceptible to both P. syringae and B. cinerea. These results indicate that the three WRKY proteins interact both physically and functionally in a complex pattern of overlapping, antagonistic, and distinct roles in plant responses to different types of microbial pathogens.
INTRODUCTION
Plants are constantly exposed to a variety of microbial pathogens and through evolution have developed a battery of active defense mechanisms to protect themselves. In resistance (R) gene–mediated resistance, direct or indirect recognition of race-specific elicitors (avirulence proteins) by the plant R gene product can often lead to rapid activation of hypersensitive response (HR) that confers specific and effective resistance against the races of the pathogen (Dangl and Jones, 2001). Pathogen-induced HR is often associated with activation of salicylic acid (SA)–regulated defense mechanisms in the surrounding or even distal parts of the plants, leading to development of systemic acquired resistance (SAR). HR and associated SA-dependent signaling pathways generally provide defense effective against biotrophic pathogens that feed on living host tissues. Ethylene (ET)- and jasmonate (JA)–mediated signaling pathways, on the other hand, often mediate plant defense against necrotrophic pathogens that promote host cell death at early stages of infection (Glazebrook, 2004). It has been well documented that there is an extensive antagonism between SA-mediated and JA/ET-mediated defense signaling pathways (Kunkel and Brooks, 2002). As a result, blocking SA accumulation in pathogen-infected plants can promote JA signaling (Spoel et al., 2003), and mutations of important regulators of JA signaling, such as MPK4 and COI1, lead to enhanced SA accumulation and signaling in pathogen-infected plants (Petersen et al., 2000; Kloek et al., 2001).
Plant defense responses are associated with the transcriptional activation of a large number of plant host genes after pathogen infection (Rushton and Somssich, 1998). Induction of certain pathogenesis-related (PR) proteins is dependent upon SA-mediated defense signaling pathways and is associated with resistance to biotrophic and hemibiotrophic pathogens. The expression of Arabidopsis thaliana PDF1.2, which encodes a protein with antimicrobial activity, is regulated by JA/ET defense signaling pathways and often correlated with resistance to necrotrophic pathogens (Penninckx et al., 1998; Thomma et al., 1998). Other pathogen-induced genes encode enzymes involved in the biosynthesis of antimicrobial compounds (phytoalexins). Pathogen-induced plant genes also include those encoding regulatory proteins involved in signal transduction of plant defense responses. Given that transcriptional regulation of plant host genes is central to plant defense responses, elucidating the complex regulatory mechanisms controlling the differential expression of plant genes is key to understanding the molecular basis of plant disease resistance.
Transcriptional regulation of gene expression is largely mediated by the specific recognition of cis-acting promoter elements by trans-acting sequence-specific DNA binding transcription factors. Among the several classes of transcription factors associated with plant defense responses are the DNA binding proteins containing WRKY domains (Ulker and Somssich, 2004). The WRKY domains contain a conserved WRKYGQK sequence followed by a Cys2His2 or Cys2HisCys zinc-finger binding motif (Ulker and Somssich, 2004). A number of studies have shown that WRKY proteins have regulatory functions in plant defense responses to pathogen infection. First, pathogen infection or treatment with pathogen elicitors or SA has been shown to induce rapid expression of WRKY genes from a number of plants (Rushton et al., 1996; Chen and Chen, 2000; Hara et al., 2000; Asai et al., 2002; Dong et al., 2003; Kalde et al., 2003; Turck et al., 2004). Secondly, a number of defense-related genes, including the well-studied PR genes, contain W-box elements in their promoter regions (Rushton et al., 1996; Chen and Chen, 2000; Hara et al., 2000; Asai et al., 2002; Dong et al., 2003; Kalde et al., 2003; Turck et al., 2004). A number of studies have shown that these W-box sequences are specifically recognized by WRKY proteins and are necessary for the inducible expression of these genes (Rushton et al., 1996; Chen and Chen, 2000; Hara et al., 2000; Asai et al., 2002; Dong et al., 2003; Kalde et al., 2003; Turck et al., 2004). A microarray study of the gene expression changes in Arabidopsis grown under 14 different SAR-inducing or -repressing conditions has shown that a group of 26 genes, including PR1, is coordinately induced during the establishment of SAR (Maleck et al., 2000). These genes have an average of 4.3 copies per promoter of binding site for WRKY proteins (W-boxes; TTGAC), compared with fewer than two W-boxes per promoter in a randomly selected set of genes (Maleck et al., 2000). More recent studies have provided direct evidence for the involvement of specific WRKY proteins in plant defense responses. For example, two Arabidopsis WRKY genes (WRKY22 and WRKY29) have been shown to be induced by a mitogen-activated protein kinase MAPK pathway involved in plant responses to both bacterial and fungal pathogens, and expression of WRKY29 in transiently transformed leaves led to reduced disease symptoms (Asai et al., 2002). Likewise, constitutive expression of Arabidopsis WRKY18 and WRKY70 led to constitutive or enhanced expression of defense-related genes, including SA-induced PR1, and increased resistance to virulent pathogens (Chen and Chen, 2002; Li et al., 2004). In tobacco (Nicotiana tabacum), virus-induced silencing of three WRKY genes compromises N gene–mediated resistance to Tobacco mosaic virus (Liu et al., 2004). In addition, the resistance gene RRS1 that confers resistance to the bacterial pathogen Ralstonia solanacearum encodes a novel WRKY protein, WRKY52, that combines typical TIR–nucleotide binding site–leucine-rich repeat R protein motifs with a WRKY domain (Lahaye, 2002; Deslandes et al., 2003). These studies indicate that WRKY proteins can function as positive regulators of plant defense responses and disease resistance.
We have previously shown that constitutive expression of Arabidopsis WRKY18 in transgenic plants leads to constitutive PR gene expression and enhanced disease resistance to a virulent strain of Pseudomonas syringae in mature transgenic plants (Chen and Chen, 2002). To study how WRKY18 regulates plant defense responses, we have performed yeast two-hybrid screens to identify proteins that interact with the pathogen-induced WRKY transcription factor. These studies demonstrated that Arabidopsis WRKY18 interacts with itself and with structurally related WRKY40 and WRKY60. Further studies revealed that the physical interactions of these WRKY proteins significantly change their sequence-specific DNA binding activities. To understand the functional significance of the interaction, we have isolated and characterized both loss-of-function mutants and gain-of-function constitutive overexpression transgenic lines for the three WRKY genes. Functional analysis of single, double, and triple combinations of these mutants for response to microbial pathogens indicated that these three pathogen-induced WRKY transcription factors also interact functionally and play complex overlapping, antagonistic, and distinct roles in plant disease resistance.
RESULTS
Structures, Expression, and Subcellular Localization
Sequence Comparison of WRKY18, WRKY40, and WRKY60.
Amino acids identical in three proteins are blue, and residues similar in three proteins are green. The putative Leu zipper motifs with six modules at the N terminus of WRKY18, WRKY40, and WRKY60 are indicated. The fourth residues (Leu, Val, or Met) in the six modules of the Leu zipper motifs are magenta, and the first amino acid residues of these modules are cyan. The WRKY DNA binding motifs are also indicated with the highly conserved WRKYGQK sequences, and the residues forming the C2H2 zinc-fingers are red.
A Leu zipper motif is a region of ∼30 to 35 amino acid residues where the sequence can be arranged in modules of seven residues (Landschulz et al., 1988). In each module, the fourth residue is usually Leu, and the first residue is frequently hydrophobic. According to this definition, the potential Leu zipper motifs at the N terminus of WRKY18, WRKY40, and WRKY60 can be divided into six modules (Figure 1). The fourth residues in four of the six modules are Leu, while the other two modules have Val or Met at this position. In addition, the hydrophobic Leu and Ile residues are also found at the first amino acid position of some of these modules. As shown below, the Leu zipper motifs are involved in the physical interaction of these WRKY proteins.
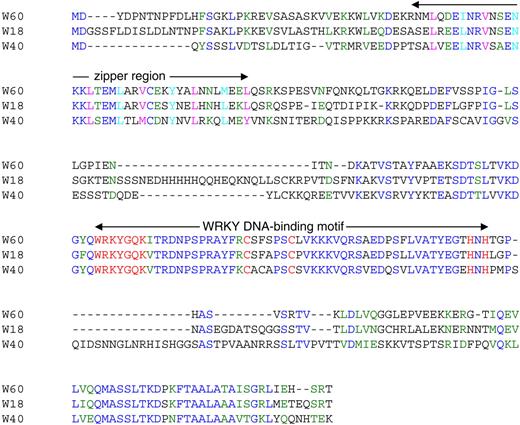
Pathogen-Induced Expression of WRKY18, WRKY40, and WRKY60.
Arabidopsis plants (4 to 5 weeks old) were infiltrated with the virulent strain of P. syringae pv tomato DC3000 (PstDC3000) (OD600 = 0.001 in 10 mM MgCl2) or sprayed with the fungal pathogen B. cinerea (2 × 105 spores/mL in a maltose buffer). Total RNA was isolated from buffer-treated or pathogen-infected leaves that were harvested at indicated time points after treatment/inoculation. Two separate blots were prepared from the same preparation of total RNA, with one probed with the WRKY18 probe and the other probed with the WRKY40 probe. One of the two blots was subsequently stripped and reprobed with the WRKY60 gene probe. The ethidium bromide stain of rRNA from one of the two blots is shown for each lane to allow assessment of equal loading. The experiments were repeated twice with similar results.
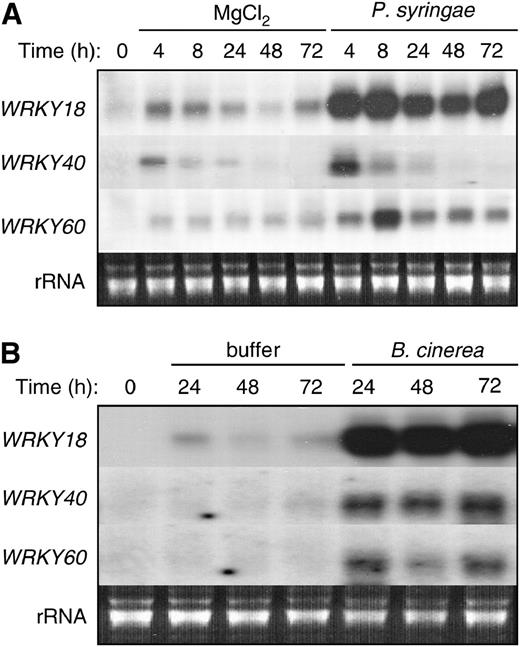
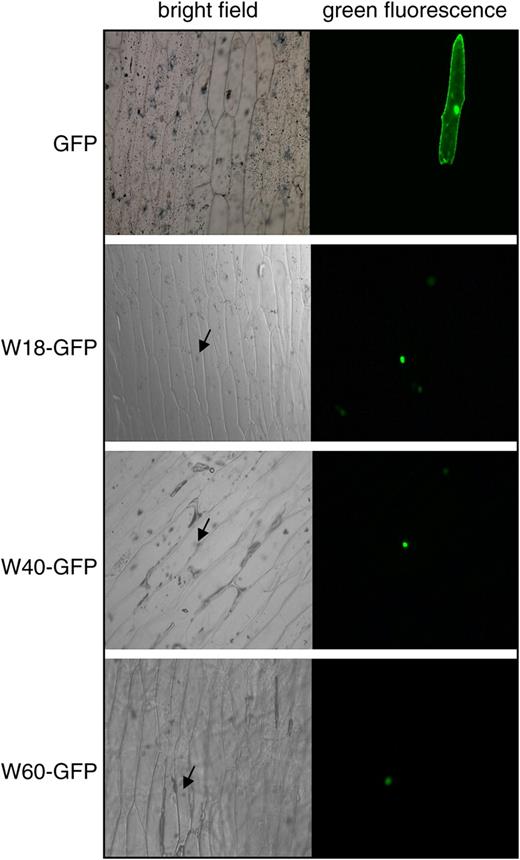
Nuclear Localization of WRKY18, WRKY40, and WRKY60.
WRKY18, WRKY40, and WRKY60 were fused to GFP to yield W18-GFP, WRKY40-GFP, and WRKY60-GFP, respectively. The fusion proteins are localized to the nucleus of onion epidermal cells. GFP alone is localized to both the nucleus and the cytoplasm because of its small size. Bright-field images of the onion epidermal cells are also shown. The arrows point to the nucleus of the cells where GFP fusion proteins are localized.
Mutual and Self-Interactions
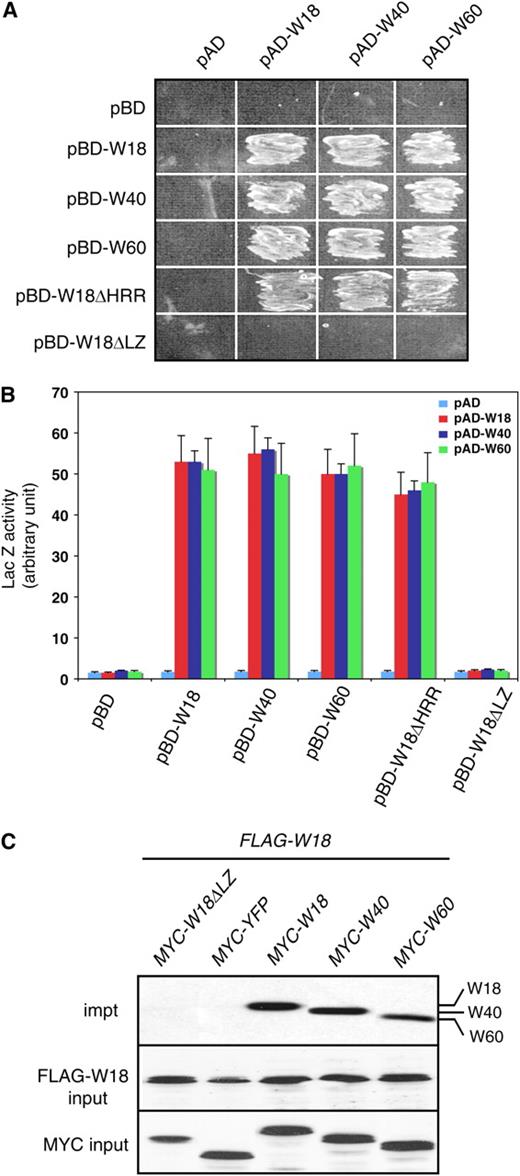
We previously showed that mature transgenic Arabidopsis plants constitutively expressing WRKY18 exhibited enhanced resistance to a virulent strain of P. syringae (Chen and Chen, 2002). To understand how the WRKY protein enhances plant defense responses, we tried to identify its interacting proteins using yeast two-hybrid screens. In these screens, we used a fusion protein of WRKY18 with the Gal4 DNA binding domain as bait. The yeast strain transformed with the bait construct failed to grow in the selective medium (data not shown), indicating that WRKY18 did not confer transcriptional activation activity to the fusion protein in yeast. We screened >2 × 107 independent transformants of a cDNA library generated from SA-treated Arabidopsis plants at a complexity of 2 × 106. The screens yielded two groups of cDNA fragments. The first group, which was isolated twice, encodes WRKY18, suggesting that this DNA binding protein can self-interact. The second group, which was isolated six times, encodes WRKY40.
Mutual and Self-Interaction of WRKY18, WRKY40, and WRKY60.
(A) Protein interactions in the yeast two-hybrid system. The activation domain (AD) fusions were cotransformed with various Gal4 DNA binding domain (BD) vectors into yeast cells and grown in the absence of Leu, Trp, and His.
(B) Quantitative assays of protein interactions in the yeast two-hybrid systems. The AD fusions were transformed with the various Gal4 DB fusion vectors into yeast cells. Proteins were isolated from the yeast cells and assayed for the β-galactosidase activity using o-nitrophenyl-β-d-galactopyranose as substrate. Data represent means + se (n = 5).
(C) Leaf protein extracts were prepared from transgenic plants coexpressing FLAG-tagged WRKY18 and one of the following MYC-tagged proteins: WRKY18ΔLZ, YFP, WRKY18, WRKY40, or WRKY60. After incubation with a FLAG-specific antibody and precipitation with protein A-agarose, the immunoprecipitated complexes were analyzed with protein gel blotting using anti-MYC-peroxidase (top panel). Protein inputs for FLAG-WRKY18 in the immunoprecipitated complexes (middle panel) and MYC-tagged proteins in the extracts (bottom panel) were detected with anti-FLAG-peroxidase and anti-MYC-peroxidase, respectively.
We next tested the role of two structural motifs of these three WRKY proteins in the protein–protein interactions. As shown in Figure 1, WRKY18 contains two conspicuous structural motifs in addition to the WRKY zinc-finger motif: a His-rich region (HRR) and a Leu zipper (LZ). To test the role of the HRR, we generated a construct encoding an HRR-less WRKY18 (W18ΔHRR) and used it as bait in yeast two-hybrid assays. As shown in Figure 4A, yeast cells transformed with both the bait and the full-length WRKY18, WRKY40, or WRKY60 prey construct were able to grow on the selective medium. Thus, the HRR-less WRKY18 was fully capable of interacting with itself and with WRKY40 and WRKY60, indicating that the HRR was not required for the protein–protein interaction. To analyze the role of LZ in protein–protein interaction, we generated LZ-less WRKY18 (W18ΔLZ) and used it as bait in the yeast two-hybrid assays. Yeast cells transformed with the combination of the LZ-less WRKY18 bait and the WRKY18, WRKY40, or WRKY60 prey construct were unable to grow on the selective medium (Figure 4A) and generated little Gal4-inducible LacZ activity (Figure 4B). In addition, we generated LZ-less WRKY40 and WRKY60 and found that they were unable to interact with themselves or with the other two WRKY proteins (see Supplemental Figure 1 online). These results suggest that the LZ motifs in these WRKY proteins mediate protein–protein interactions with themselves and with each other. Further analysis with site-directed mutations would be required to confirm the role of the LZ motifs in interactions of the WRKY proteins.
We also performed coimmunoprecipitation to determine in vivo interactions of WRKY18 with itself and with WRKY40 and WRKY60. Through genetic crossing or double transformation, we generated transgenic plants harboring a combination of a FLAG-tagged WRKY18 gene driven by the cauliflower mosaic virus (CaMV) 35S promoter and a MYC-tagged WRKY gene driven by a steroid-inducible Gal4 promoter (Aoyama and Chua, 1997). As shown in Figure 4C, the protein complexes immunoprecipitated by the anti-FLAG antibody from protein extracts of transgenic plants coexpressing the FLAG-tagged WRKY18 protein, and a MYC-tagged WRKY protein generated positive interactions to the anti-MYC antisera. By contrast, the immunoprecipitations from protein extracts of transgenic plants expressing the FLAG-tagged WRKY18 protein and a MYC-tagged WRKY18(ΔLZ) or yellow fluorescent protein (YFP) produced no cross-reactivity to the antisera (Figure 4C). These results support in vivo interactions of WRKY18 protein with itself and with WRKY40 and WRKY60.
DNA Binding Activities
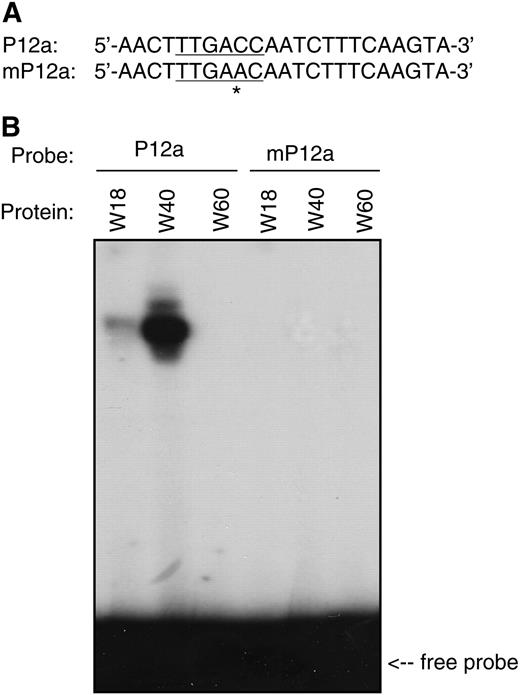
DNA Binding Activity of WRKY18, WRKY40, and WRKY60 Proteins.
(A) Nucleotide sequences of probes used for DNA binding assays. P12a contains a single TTGACC W-box sequence, which is mutated into TTGAAC in mP12a.
(B) Recombinant WRKY18, WRKY40, and WRKY60 were purified from E. coli cells and used for DNA binding assays with P12a and mP12a as probes. The binding reactions (20 μL) contained 2 ng labeled oligo DNA, 5 μg polydeoxyinosinic-deoxycytidylic acid, and 1 μg recombinant protein. The binding assays were repeated twice with independently prepared recombinant proteins with similar results.
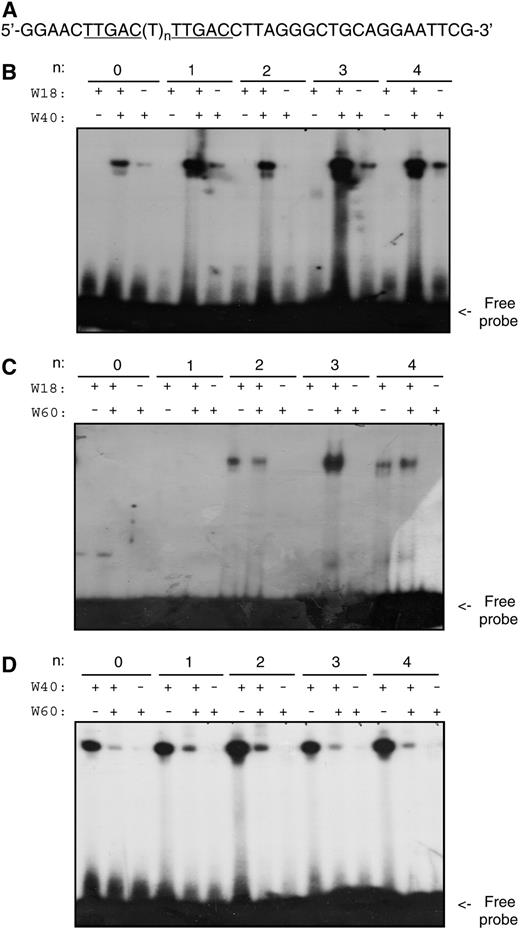
Effects of Protein–Protein Interactions on DNA Binding Activity.
(A) Nucleotide sequences of the probes used for DNA binding assays. The probes contain two TTGAC W-box core sequences separated by a spacer of varying sizes.
(B) DNA binding activity of WRKY18, WRKY40, or a mixture of the two proteins.
(C) DNA binding activity of WRKY18, WRKY60, or a mixture of the two proteins.
(D) DNA binding activity of WRKY40, WRKY60, or a mixture of the two proteins.
The binding reactions and conditions were the same as those in Figure 5 except that the recombinant protein in each binding reaction was reduced to 0.2 μg. The reduced amount of protein was used to facilitate detection of possible effects of protein–protein interactions on their DNA binding activity. The binding assays were repeated twice with independently prepared recombinant proteins with similar results.
WRKY60 alone had little DNA binding activities for W-box sequences. Mixing WRKY60 with WRKY18 did not alter DNA binding activities for a number of DNA sequences containing two W-boxes arranged in various spacings (Figure 6C). However, substantial enhancement of DNA binding activity of WRKY18 was observed by WRKY60 for a DNA molecule with two direct repeats of W-boxes spaced by three nucleotides (Figure 6C). This result suggests that interaction of WRKY18 and WRKY60 selectively enhances their DNA binding activity to DNA molecules containing W-boxes arranged in a specific manner. By contrast, when WRKY40 was mixed with WRKY60, less DNA binding of WRKY40 was observed (Figure 6D). Thus, WRKY60 differentially affected the DNA binding activity of WRKY18 and WRKY40.
Knockout Mutants
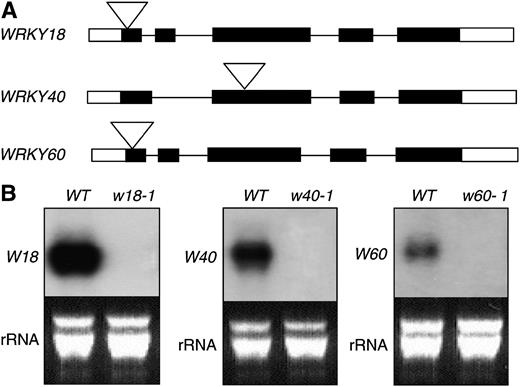
Loss-of-Function Mutants for the WRKY Genes.
(A) Diagram of WRKY18, WRKY40, and WRKY60 and their T-DNA insertion or transposon tagging mutants.
(B) RNA gel blot analysis of wrky18-1, wrky40-1, and wrky60-1 mutants. Wild-type and mutant plants were infiltrated with PstDC3000 (OD600 = 0.01). Two hours after the inoculation, the inoculated leaves were harvested and total RNA was isolated. After separation on the gel and blotting to a nylon membrane, the blot was probed with gene-specific DNA fragments. The RNA gel blotting was repeated at least three times with independently isolated RNA with similar results.
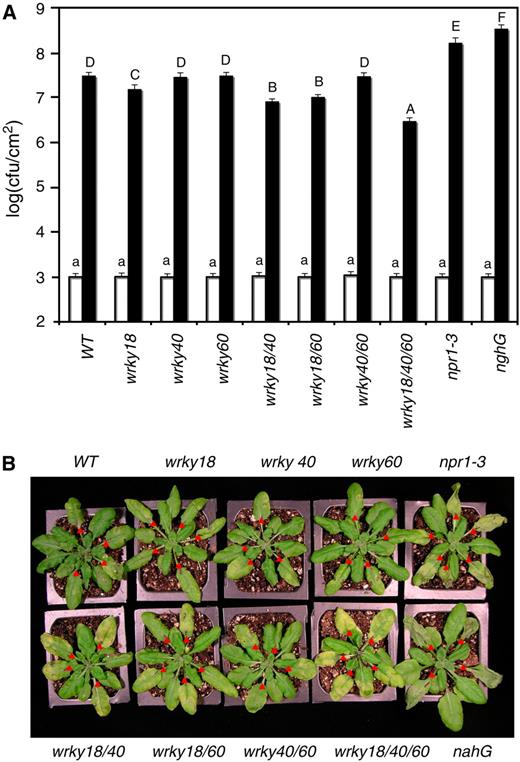
Altered Response of WRKY Mutants to P. syringae.
(A) Wild-type, single, double, and triple mutants were inoculated through infiltration with a suspension of PstDC3000 (OD600 = 0.001 in 10 mM MgCl2). Samples were taken at 0 d (open bars) or 3 d (closed bars) after inoculation (DAI) to determine the growth of the bacterial pathogen. The means and standard errors were calculated from 10 plants for each treatment. According to Duncan's multiple range test (P = 0.05), means of colony-forming units (cfu) at 0 DAI do not differ significantly if they are indicated with the same lowercase letter; means of cfu at 3 DAI do not differ significantly if they are indicated with the same capital letter.
(B) Wild-type and wrky mutants were inoculated through infiltration with a suspension of PstDC3000 (OD600 = 0.001 in 10 mM MgCl2). Pictures of representative inoculated leaves were taken 3 DAI. Infiltrated leaves are indicated with red triangles.
These experiments were repeated twice with similar results.
Some of the mutants developed more chlorotic symptoms than wild-type plants after infection of the bacterial pathogen (Figure 8B). Surprisingly, the extent of chlorosis on the inoculated leaves was negatively correlated with bacterial growth. Thus, the wild type, wrky40, wrky60, and wrky40/60 mutants supported the highest bacterial growth and had the least chlorosis (Figure 8B). The wrky18 wrky40 and wrky18 wrky60 double mutants had modest levels of reduction in the bacterial growth and were accompanied with modest levels of chlorosis (Figure 8B). The wrky18 wrky40 wrky60 triple mutant had the largest reduction in bacterial growth and developed the most extensive chlorosis (Figure 8B).
To determine how the reduction in the bacterial growth was associated with enhanced chlorosis in the mutants, we compared them with the npr1-3 mutant (Cao et al., 1997) and transgenic nahG plants (Delaney et al., 1994). As shown in Figure 8A, the npr1-3 and transgenic nahG plants had increased growth (∼7- to 12-fold) of the bacterial pathogen relative to wild-type plants. When compared with the wrky18 wrky40 wrky60 triple mutant, the npr1-3 and transgenic nahG plants had ∼70- to 100-fold increase in the bacterial growth. The npr1-3 and transgenic nahG plants also developed severe disease symptoms when compared with the wild-type plants (Figure 8B). However, there was a marked difference between the symptoms on the npr1 and transgenic nahG plants and those on the WRKY mutants with reduced bacterial growth (Figure 8B). In the transgenic nahG plants, which had the highest bacterial growth (Figure 8A), the inoculated leaves first developed water-soaking symptoms followed by necrotic and wilting leaves with little chlorosis after the infection (Figure 8B). The npr1 mutant plants also developed extensively necrotic and slightly chlorotic symptoms, and the wild-type plants had mixed chlorotic and necrotic symptoms (Figure 8B). Inoculated leaves of the WRKY double and triple mutants with reduced bacterial growth, particularly the wrky18 wrky40 wrky60 triple mutant, had extensive yellowing with little necrotic collapsing or wilting (Figure 8B).
Altered Response of WRKY Mutants to B. cinerea.
(A) Wild-type, single, double, and triple mutants were inoculated by spraying spore suspension at a density of 2 × 104 spores/mL and kept at high humidity. The pictures of the plants were taken 4 DAI.
(B) Total RNA was isolated from inoculated plants 3 DAI and probed with a Botrytis β-tubulin gene probe to determine the accumulation of the Botrytis β-tubulin mRNA for measurement of the biomass of the fungal pathogen on infected plants.
The experiments were repeated twice with similar results.
To confirm that the altered disease resistance of the mutants was due to disruption of the WRKY genes, we performed genetic complementation of the wrky18 wrky40 wrky60 triple mutant because of its strongest phenotypes in disease assays. Full-length cDNA clones for the three WRKY genes were placed behind their native promoters and transformed into the mutant. T1 transformants were identified through Basta selection and analyzed for expression of the WRKY transgenes and responses to P. syringae and B. cinerea. Transformation of the mutant with WRKY18 resulted in enhanced growth of P. syringae but reduced susceptibility to B. cinerea to the levels close to those in wild-type plants, while transformation of the same mutant with WRKY40 and WRKY60 led to partial restoration of the altered phenotypes in the pathogen assays (see Supplemental Figure 2 online). These results indicate that the altered disease resistance of the wrky18 wrky40 wrky60 mutant was attributed to mutations of the WRKY genes.
Constitutive Expression in Transgenic Plants
Construction of Overexpression Lines.
(A) RNA gel analysis of WRKY40 and WRKY60 expression in transgenic plants. RNA samples were prepared from leaves of 32-d-old wild-type and transgenic plants and probed with WRKY gene-specific DNA fragments. Transgenic WRKY40 line 7 and transgenic WRKY60 line 1 contained a single T-DNA insertion in their genomes and exhibited stable expression of their respective transgenes. Their F3 homozygous progeny plants were used in all the experiments in this study.
(B) Morphology of 5-week-old wild-type and transgenic plants overexpressing one or two WRKY genes.
(C) Total RNA was isolated from 5-week-old transgenic plants expressing WRKY18 alone or coexpressing WRKY18 and WRKY40 or WRKY18 and WRKY60. Two separate blots were prepared from the same preparation of total RNA, with one probed with the PR1 probe and the other probed with the WRKY18 probe. The two blots were subsequently stripped and reprobed with the WRKY40 and WRKY60 gene probes, respectively.
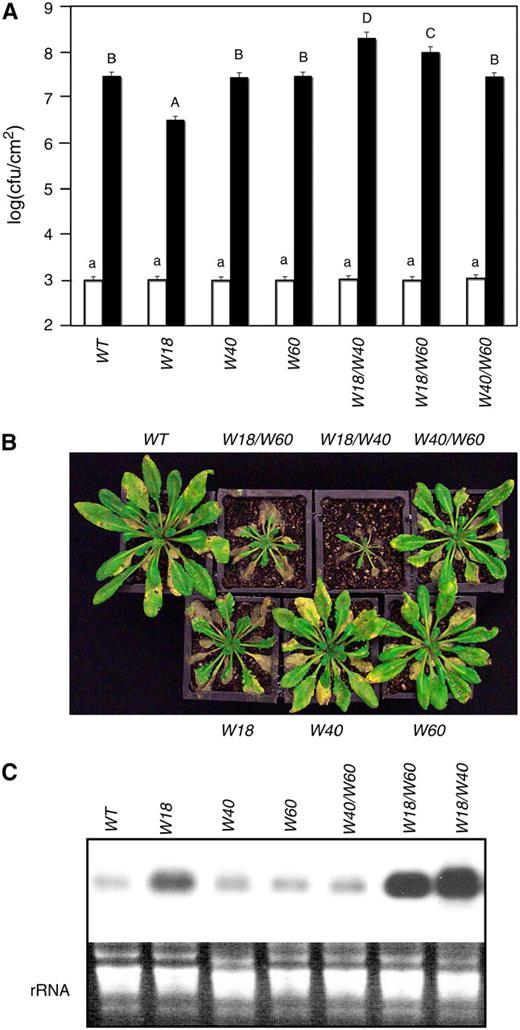
Altered Responses of Overexpression Lines to P. syringae and B. cinerea.
(A) Wild-type and transgenic overexpression plants were inoculated with PstDC3000 (OD600 = 0.001). Samples were taken at 0 (open bars) and 3 (closed bars) DAI to determine the growth of the bacterial pathogen. The means and standard errors were calculated from 6 to 10 plants. According to Duncan's multiple range test (P = 0.05), means of cfu at 0 DAI do not differ significantly if they are indicated with the same lowercase letter; means of cfu at 3 DAI do not differ significantly if they are indicated with the same capital letter.
(B) Wild-type and transgenic overexpression lines constitutively expressing one or two WRKY genes were inoculated by spraying spore suspension at a density of 105 spores/mL and kept at high humidity. The pictures of the plants were taken 4 DAI.
(C) Total RNA was isolated from inoculated plants 3 DAI and probed with a Botrytis β-tubulin gene probe to determine the accumulation of the Botrytis β-tubulin mRNA for measurement of the biomass of the fungal pathogen on infected plants.
The experiments were repeated twice with similar results.
We also analyzed the responses of the transgenic overexpression plants to B. cinerea. Based on symptom development (Figure 11B) and quantification of biomass of the fungal pathogen (Figure 11C), constitutive expression or coexpression of WRKY40 or WRKY60 had no significant effect on plant response to the pathogen. Constitutive expression of WRKY18, on the other hand, resulted in a substantial increase in the susceptibility to the fungal pathogen (Figures 11B and 11C). Coexpression of WRKY40 or WRKY60 with WRKY18 resulted in further enhanced susceptibility to the fungal pathogen (Figures 11B and 11C). Thus, while constitutive overexpression and knockout mutations of some of these WRKY genes had opposite effects on response to P. syringae, both overexpression and disruption of some of the WRKY genes resulted in enhanced susceptibility to B. cinerea.
Defense-Related Gene Expression
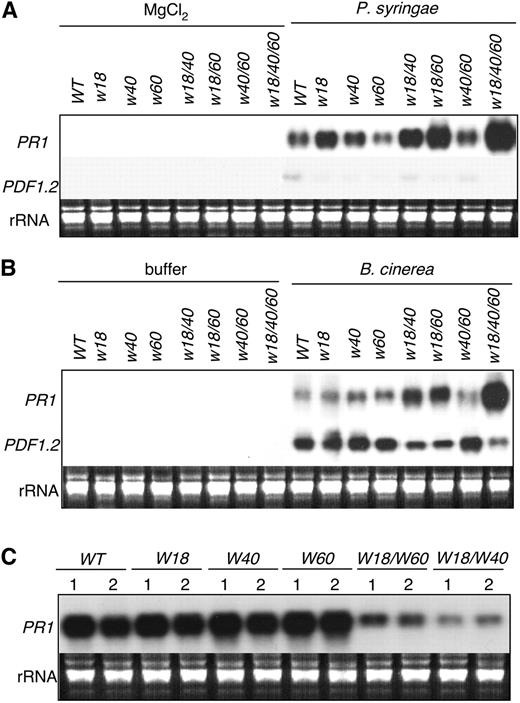
Defense-Related Gene Expression.
(A) Plants of wild-type, single, double, and triple mutants were infiltrated with 10 mM MgCl2 or PstDC3000 (OD600 = 0.001 in 10 mM MgCl2). Total RNA was isolated from infiltrated leaves harvested at 24 h after infiltration and probed with a PR1 probe. The blot was stripped and reprobed with a PDF1.2 probe.
(B) Plants of wild-type, single, double, and triple mutants were sprayed with a maltose buffer or with B. cinerea spore suspension at a density of 105 spores/mL and kept at high humidity. Total RNA was isolated from leaves harvested 24 h after spraying and probed with a PR1 probe. The blot was stripped and reprobed with a PDF1.2 probe.
(C) Three leaves from wild-type and transgenic plants constitutively expressing WRKY18, WRKY40, and WRKY60 alone or coexpressing WRKY18 and WRKY60 or WRKY18 and WRKY40 were inoculated with PstDC3000 (OD600 = 0.001). Total RNA was isolated from the inoculated leaves harvested at 48 h after inoculation, separated on an agarose (1.2%)–formaldehyde gel, and probed with a PR1 gene fragment.
These RNA gel blot experiments were repeated twice using independently isolated RNA with similar results.
Enhanced resistance to P. syringae in transgenic plants constitutively expressing WRKY18 is associated with constitutive expression of PR1 genes (Figure 10C). Coexpression of WRKY40 or WRKY60 rendered transgenic WRKY18 plants more susceptible to the bacterial pathogen and, at the same time, abolished the constitutive PR1 expression in the transgenic plants (Figure 10C). Pathogen infection did not further enhance PR1 expression in the transgenic WRKY18 plants (Figure 12C), as previously reported (Chen and Chen, 2002). The levels of PR1 transcripts in transgenic plants expressing WRKY40 or WRKY60 were similar to those observed in the wild-type plants after infection by P. syringae (Figure 12C), consistent with their unaltered resistance to the bacterial pathogen (Figure 11A). On the other hand, pathogen-induced PR1 expression was substantially reduced in the transgenic plants coexpressing WRKY18 and WRKY60 or WRKY18 and WRKY40 (Figure 12C), consistent with their enhanced susceptibility to the bacterial pathogen (Figure 11A).
DISCUSSION
Roles in Defense Responses
Arabidopsis WRKY18, WRKY40, and WRKY60 are pathogen-induced and encode three structurally related WRKY proteins (Dong et al., 2003). To determine their roles in plant defense against microbial pathogens, we have analyzed both their knockout mutants and constitutive overexpression lines to the bacterial pathogen P. syringae and the fungal pathogen B. cinerea. P. syringae is biotrophic in the early stages of infection but can become necrotrophic at late stages of infection and should probably be considered a hemibiotroph. Arabidopsis mutants defective in SA biosynthesis or signaling, including eds1 (Aarts et al., 1998), pad4 (Zhou et al., 1998), eds5 (Rogers and Ausubel, 1997), sid2 (Nawrath and Metraux, 1999), and npr1 (Glazebrook et al., 1996), allow increased growth of P. syringae, indicating that SA-mediated signaling mechanisms play a vital role in limiting P. syringae growth. Among the three single knockout mutants analyzed, only wrky18 exhibited a small reduction in bacterial growth, while wrky40 and wrky60 displayed no significant phenotype after infection of the bacterial pathogen (Figure 8A). Two double knockout mutants, wrky18 wrky40 and wrky18 wrky60, on the other hand, supported modestly reduced bacterial growth, while the wrky18 wrky40 wrky60 triple knockout mutant had the most reduced bacterial growth when compared with wild-type plants (Figure 8A). In transgenic overexpression plants, although constitutive expression of WRKY18 led to constitutive expression of the SA-related PR1 gene and enhanced resistance to P. syringae, its coexpression with WRKY40 or WRKY60 resulted in increased growth of the bacterial pathogen (Figure 11A). Based on these results, WRKY18, WRKY40, and WRKY60 function redundantly and cooperatively as negative regulators in Arabidopsis defense that limits the growth of the bacterial pathogen.
B. cinerea is a necrotrophic fungal pathogen that promotes host cell death at very early stages in infection. Arabidopsis resistance to B. cinerea depends on JA and ET signaling pathways since mutations that block JA signaling, including coi1 and jar1, or ET signaling, including ein2, result in enhanced susceptibility (Glazebrook, 2004). Our analyses revealed that loss-of-function mutants for the three WRKY genes exhibited altered phenotypes in response to the necrotrophic fungal pathogen, and these altered phenotypes were opposite to those observed with the bacterial pathogen P. syringae. Among the three single knockout mutants, only the wrky18 mutant exhibited slightly reduced growth of P. syringae (Figure 8) and was also slightly more susceptible to B. cinerea than the wild-type plants (Figure 9). Double knockout mutants wrky18 wrky40 and wrky18 wrky60 but not wrky40 wrky60 exhibited modestly reduced growth of P. syringae (Figure 8) and were modestly more susceptible to the fungal pathogen than wild-type plants (Figure 9). The wrky18 wrky40 wrky60 triple knockout mutant had the most reduced bacterial growth (Figure 8) and was most susceptible to B. cinerea among all these mutants tested (Figure 9).
Based on analyses of pathogen-induced expression of SA-induced PR1 and JA-regulated PDF1.2 in these mutants (Figures 12A and 12B), the opposite effects of mutations of these WRKY genes on plant responses to the two pathogens probably reflected the antagonism between SA- and JA-mediated defense signaling pathways (Kunkel and Brooks, 2002; Li et al., 2004; Takahashi et al., 2004). After infection by P. syringae, enhanced induction of SA-regulated PR1 expression was observed in some of these WRKY mutants, including wrky18 wrky40, wrky18 wrky60, and wrky18 wrky40 wrky60 (Figure 12A), and was correlated with increased resistance to the bacterial pathogen (Figure 8). After infection by B. cinerea, on the other hand, reduced expression of JA-regulated PDF1.2 was observed in the wrky18 wrky40, wrky18 wrky60, and wrky18 wrky40 wrky60 mutants (Figure 12B) and was correlated with enhanced susceptibility of these mutants to the necrotrophic fungal pathogen (Figure 9). Furthermore, the induction level of JA-regulated PDF1.2 by the fungal pathogen was inversely correlated with that of SA-regulated PR1 among the wild type and WRKY mutants (Figure 12B). These results suggest that these three WRKY proteins may function redundantly as negative regulators in SA-dependent pathways but play a positive role in JA-mediated pathways. Recently, Arabidopsis WRKY70 has also been shown to be involved in the crosstalk between SA and JA signaling. However, studies with both overexpression and antisense lines indicated that WRKY70 plays a positive role in SA signaling and functions as a negative regulator of JA-inducible genes (Li et al., 2004). In a recent study using yeast two-hybrid screening, Arabidopsis mitogen-activated protein kinase 4 (MPK4), a repressor of SA-dependent resistance (Petersen et al., 2000), was found to interact with a MPK4 substrate MKS1 that in turn interacts with Arabidopsis WRKY25 and WRKY33 (Andreasson et al., 2005). In addition, WRKY25 and WRKY33 were shown to be in vitro substrates of MPK4, and a wrky33 knockout mutant was found to express enhanced levels of the PR1 gene under a short-day growth condition (Andreasson et al., 2005). These results suggest that WRKY25 and WRKY33 may function as downstream components of the MPK4-mediated signaling pathway and also act as repressors of SA-dependent disease resistance. Thus, different WRKY proteins play distinct roles in various signaling pathways of plant defense responses.
A recent study using the β-glucuronidase reporter gene driven by the Arabidopsis WRKY18 gene promoter has shown that this gene is expressed in vascular bundles throughout the plant but not in very young leaves, suggesting that WRKY18 may play a role in vascular development (Ohashi-Ito et al., 2005). However, no obvious phenotype in vascular development was observed in the knockout mutants or overexpression plants for WRKY18. Since WRKY18 is induced by pathogens and defense-inducing molecules such as SA, its constitutive expression in vascular bundles in the absence of pathogen may be caused by certain defense-mimic molecules or conditions such as lignification and reactive oxygen species that are associated with vascular tissues. The preferential expression of WRKY18 in the vascular bundles might contribute to plant defense as these tissues may be important for systemic defense signaling.
Analysis of the response of the overexpression and knockout lines for the three WRKY genes to P. syringae and B. cinerea have also revealed intriguing phenotypes that might reflect the complex nature of plant defense mechanisms. First, both mutations of these WRKY genes and constitutive expression of WRKY18 and its coexpression with WRKY40 or WRKY60 resulted in enhanced susceptibility to B. cinerea (Figures 9 and 11). Thus, the roles of these WRKY genes in plant responses to the necrotrophic fungal pathogen may be more complex. It appeared that resistance to the fungal pathogen was associated with certain expression levels of the three WRKY genes. It has been proposed that the cell death induced by B. cinerea is a form of HR and is important for the virulence of the fungal pathogen (Govrin and Levine, 2000). HR is a form of programmed cell death that can be activated or positively influenced by a variety of signaling mechanisms, including those that normally antagonize each other. In plants, for example, SA and ET can activate signaling pathways antagonistic to each other, but they both are also known to induce or enhance pathogen-induced cell death (Shirasu et al., 1997; Devadas et al., 2002; Samuel et al., 2005). Therefore, it is possible that mutations of the WRKY genes may lead to activation or suppression of signaling mechanisms different from those in the transgenic overexpression lines, but these different signaling events may then converge to enhance cell death upon infection by the necrotrophic pathogen and promote its virulence.
Second, we observed reduced necrosis but enhanced chlorosis in the WRKY mutants with reduced bacterial growth after infection by P. syringae (Figure 8B). This was in contrast with the extensive necrosis but limited chlorosis in the npr1 mutant and transgenic nahG plants with increased growth of the bacterial pathogen (Figure 8B). P. syringae is a biotroph during early stages of infection but after a period of multiplication in the plant becomes necrotrophic (Collmer and Bauer, 1994). Population density–dependent sensing is important for the expression of virulence in plant pathogens (Pierson et al., 1998) and may also be important for the transition from biotrophic to necrotrophic phases of P. syringae. In the npr1 mutants, suppressed SA-mediated defense mechanisms support enhanced growth of the bacterial pathogen and may promote initiation of quorum sensing and switch to the necrotrophic phase. Likewise, expression of nahG in transgenic plants results in pleiotropic changes in defense pathways, including those mediated by elevated SA accumulation (Heck et al., 2003; van Wees and Glazebrook, 2003), and may also promote switch of the bacterial pathogen to necrotrophic phase by supporting enhanced bacterial growth. In the WRKY double and triple mutants with reduced bacterial growth, necrosis was limited probably because the bacterial density remained low and the initiation of quorum sensing and switch to the necrotrophic phase might be inhibited.
Physical and Functional Interactions
Despite similar structures and expression patterns, WRKY18, WRKY40, and WRKY60 were found to be distinct in a number of molecular and functional characteristics. For example, the three WRKY proteins differed in in vitro binding activity for DNA molecules containing the TTGACC/T W-box sequences: WRKY18 and WRKY40 had binding activity, while WRKY60 had little binding activity for the same DNA probes containing the W-box sequences (Figures 5 and 6). The sequence-specific DNA binding activity of WRKY18 and WRKY40 was correlated with altered growth phenotypes (e.g., reduced growth and enhanced serration of leaves) in the transgenic overexpression plants constitutively expressing WRKY18 or WRKY40 (Figure 10B). In addition, mature transgenic 35S:WRKY18 plants constitutively expressed the PR1 gene and became more resistant to P. syringae but more susceptible to B. cinerea (Figures 10 and 11). No such alteration in defense responses was observed in transgenic plants overexpressing WRKY40 or WRKY60 (Figures 10 and 11). Likewise, the wrky18 single mutant has a weak, but detectable, phenotype in response to the bacterial and fungal pathogens, while no such phenotype was observed in the wrky40 or wrky60 mutant (Figures 8 and 9). These observations suggest that while the DNA binding activity of the WRKY proteins may be important for altered growth and defense phenotypes, there are additional properties that may differ among these WRKY proteins and that can influence their roles in plant growth and defense.
Using yeast two-hybrid assays and immunoprecipitation, we have shown that these three WRKY proteins interact with themselves and with each other to form both homocomplexes and heterocomplexes (Figure 4). Pathogen-induced expression of all three WRKY genes was detected in leaves (Figure 2), and it is likely that they can interact with each other in the leaf tissues, although additional experiments would be required to confirm that they are expressed in the same cells. Physical interactions have been widely reported between structurally related DNA binding transcription factors with comparable affinities to the same DNA sequences in vitro, and such interactions are often required for achieving the necessary selectivity and specificity of DNA binding in vivo. For example, the animal steroid hormone receptors bind exclusively as homodimers to response elements where the half-sites are organized in palindromic orientation (Freedman and Luisi, 1993). Other nuclear receptors, such as those for vitamin D, thyroid hormone, or trans-retinoic acid, form heterodimers with the cis-retinoic acid receptor and recognize response elements with half-sites arranged as direct repeats with different heterodimers recognizing response elements with different spacer lengths (Freedman and Luisi, 1993; Towers et al., 1993). Our analysis of in vitro DNA binding with mixed WRKY proteins indicated that such protein–protein interactions did not result in the high selectivity or specificity of DNA binding observed in other DNA binding proteins. For example, WRKY18 and WRKY40, alone or in combination, could bind DNA molecules containing two W-box sequences separated by various numbers of nucleotides (Figure 6B). On the other hand, distinct effects of protein–protein interactions on the DNA binding activity and specificity of these WRKY proteins were observed. Interactions between WRKY18 and WRKY40 had a positive effect on their DNA binding activity (Figure 6B). WRKY60, a protein with little in vitro DNA binding activity for W-box sequences, differentially influenced the DNA binding activity of the two other WRKY proteins. When mixed with WRKY18, WRKY60 enhanced the binding to a DNA molecule containing two specifically arranged W-box repeats (Figure 6). When mixed with WRKY40, WRKY60 reduced the DNA binding activity of WRKY40 (Figure 6). The altered DNA binding activities due to formation of heterocomplexes of two WRKY proteins could result from cooperative binding of the proteins or from a conformational change of one WRKY protein by the other interacting WRKY protein. These protein–protein interactions might even affect other properties important for their regulatory functions in transcription.
The physical interactions among these three WRKY proteins could explain some of the functional interactions observed in our analysis of their roles in plant defense responses. For example, there was a striking difference in the phenotype between transgenic plants expressing a single WRKY gene and those coexpressing two WRKY genes. In mature transgenic plants expressing WRKY18, there was constitutive expression of PR1 and enhanced resistance to P. syringae (Figures 10C and 11A). When the plants were crossed with plants expressing WRKY40 or WRKY60, the progeny plants were more stunted and the constitutive expression of PR1 and enhanced resistance to the bacterial pathogen was abolished or even reversed (Figures 10C and 11A). This observation indicated that at least in these transgenic plants, WRKY40 and WRKY60 antagonized WRKY18 in the activation of defense responses to the bacterial pathogen. However, since the transgenic plants expressing WRKY40 or WRKY60 alone did not have phenotypes opposite to those observed in the transgenic WRKY18 plants (Figures 10C and 11A), it appeared that the phenotypes of transgenic WRKY18/40 and WRKY18/60 plants were the result of not only simple antagonistic actions but also functional cooperation of these WRKY proteins. Functional interactions were also obvious from the loss-of-function mutants in that the mutation of WRKY40 or WRKY60 was less consequential when it was single or in combination (Figures 8 and 9). However, when either mutation was combined with the wrky18 mutation, an enhanced phenotype was observed (Figures 8 and 9). The functional interactions among the three WRKY proteins in influencing disease resistance are consistent with the changes in DNA binding activity. WRKY18 has a DNA binding activity (Figure 5) and can itself influence plant defense and disease resistance based on the altered phenotypes of its transgenic overexpression plants and the T-DNA insertion mutant (Figures 8, 9, and 11). Interaction of WRKY18 with WRKY40 and/or WRKY60 results in enhanced DNA binding activity and specificity (Figure 6) that may contribute to their increased roles in plant defense and disease resistance as exhibited from the stronger phenotypes of T-DNA double and triple mutants than those of the single mutants (Figures 8 and 9). The changes in DNA binding activity as a result of physical interactions may also explain the apparent antagonism of WRKY18 by WRKY40 and WRKY60 in transgenic overexpression plants. The enhanced PR1 gene expression and resistance to P. syringae in the transgenic WRKY18 expression plants (Figure 11) is apparently not consistent with the role of WRKY18 in plant defense deduced from the similar phenotypes of its T-DNA insertion mutant (Figure 8). It is, therefore, possible that the observed phenotypes in the transgenic WRKY18 overexpression plants might result from pleiotropic effects of WRKY18 overexpression that could lead to nonspecific activation or repression of genes that are not normally regulated by WRKY18. Coexpression of WRKY40 or WRKY60 may enhance binding specificity of WRKY18 and reduce its nonspecific effects on gene expression. On the other hand, WRKY40 interaction with WRKY60 resulted in decreased DNA binding activity (Figure 6), and no positive functional interactions were observed between the two WRKY genes since their coexpression or double knockout did not affect disease resistance phenotypes (Figures 8, 9, and 11). When the cognate target genes of these WRKY proteins are identified in future studies, they will serve as more sensitive and reliable markers for determining the phenotypes of the WRKY single, double, and triple mutants. These molecular phenotypes can then be used for more rigorous analysis of the functional significance of the physical interactions between the WRKY proteins. These studies should provide important insights into the mode of action of the large number of plant WRKY proteins in plant defense and other biological processes.
METHODS
Plant Materials
The [α-32P]dATP (>3000 Ci/mmol) was obtained from New England Nuclear; other common chemicals were purchased from Sigma-Aldrich. Arabidopsis thaliana plants were grown in a growth chamber at 22°C with 150 μE m−2 s−1 light in a photoperiod of 12 h light and 12 h dark.
cDNA Library Screening
The cDNA library was prepared from Arabidopsis plants harvested for 4 h after spraying with 2 mM SA. The library (in ZAP Express λ-vector from Stratagene) of 106 phages was screened using 32P-labeled PCR-amplified products with primers designed from the genomic sequences of the WRKY genes. The hybridization was performed in the buffer of 5× SSPE, 0.5% SDS, 5× Denhardt's solution, and 100 μg/mL salmon sperm DNA for 16 h at 55°C. The filters were washed three to four times for 30 min each with 0.5× SSPE and 0.5% SDS at 55°C. The phagemid for each isolated clone was obtained through in vivo excision according to the manufacturer's instructions. DNA sequencing was performed by the dideoxynucleotide chain-termination method at the Genomics Center of Purdue University.
RNA Gel Blot Analysis
For RNA gel blot analysis of plant gene expression, Arabidopsis plants were treated and two fully expanded leaves from each plant were harvested at each time point for total RNA isolation using TRIZOL reagent (BRL Life Technologies). The RNA was separated on agarose (1.2%)–formaldehyde gels and blotted onto nylon membrane. Hybridization was performed using random-primed 32P-labeled DNA probes in PerfectHyb plus hybridization buffer (Sigma-Aldrich) overnight at 68°C. The membranes were washed at 68°C once in 2× SSC and 0.1% SDS for 5 min, twice in 0.5× SSC and 0.1% SDS, and twice in 0.1× SSC and 0.1% SDS.
Subcellular Localization
Onion (Allium cepa) epidermal cell layers were peeled and placed inside up on the Murashige and Skoog (MS) plates. Plasmid DNAs of appropriate fusion genes (0.5 μg) were introduced to the onion cells using a pneumatic particle gun (PDS 1000; DuPont). The condition of bombardment was vacuum of 28-inch Hg, helium pressure of 1100 or 1300 psi, and 6 cm of target distance using 1.1 μm of tungsten microcarriers. After bombardment, tissues were incubated on the MS plates for 24 h at 22°C. Samples were observed directly or transferred to glass slides.
Yeast Two-Hybrid Screens
WRKY18-interacting proteins were identified using a Gal4-based two-hybrid system as described by the manufacturer (Stratagene). The Arabidopsis HybridZAP-2.1 two-hybrid cDNA library was prepared from Arabidopsis plants harvested for 4 h after spraying with 2 mM SA. The DNA fragments corresponding to different WRKY proteins were amplified by PCR and confirmed by sequencing. Overlapping PCR as previously described (Chen et al., 2003) was used to generate WRKY deletion constructs according to the following WRKY protein amino acids: HRR of WRKY18, 126 to 134; LZ of WRKY18, 44 to 80; LZ of WRKY40, 30 to 64; LZ of WRKY60, 42 to 76. The fragments were inserted into the pBD-Gal4Cam plasmid to generate bait plasmids. The HybridZAP cDNA library and corresponding bait plasmids were used to transform yeast strain YRG-2. Yeast transformants were plated onto the selection medium lacking Trp, Leu, and His. β-Galactosidase activity was assayed using o-nitrophenyl-β-d-galactopyranose as substrate. Plasmid DNA was recovered from positive yeast colonies, transformed into Escherichia coli strain DH5α, and isolated for DNA sequencing.
Immunoprecipitation
To generate the FLAG-tagged WRKY18, a WRKY18 cDNA fragment was generated by PCR amplification and subsequently subcloned into a tagging plasmid behind the FLAG tag sequence. The tagged gene was subcloned into pTA7002 behind the steroid-inducible Gal4 promoter (Aoyama and Chua, 1997) and transformed into Arabidopsis plants. To analyze expression of the tagged transgene, transformants were treated with 5 μM dexamethasone, a steroid, and leaves were harvested 24 h later for RNA gel blot and protein gel blot analyses. MYC-tagged WRKY18, WRKY40, WRKY60, WRKY18(ΔLZ), and YFP constructs were generated using a similar approach, subcloned into the plant transformation vector pOCA30, and transformed into Arabidopsis plants. Transformants expressing the tagged transgenes were identified by protein gel blotting using anti-MYC-peroxidase (Invitrogen). Plants expressing FLAG-tagged WRKY18 and a MYC-tagged protein were generated through genetic crossing or double transformation and identified through protein gel blotting using epitope-recognizing antibodies after treatment with 5 μM dexamethasone.
Protein extracts (100 μg) from control plants and transgenic plants expressing FLAG-tagged WRKY18 and a MYC-tagged protein were incubated with the anti-FLAG epitope antibody at 4°C for 5 h on a shaker. Protein A-agarose (50 μL packed volume) was added and incubated for another 2 h. Agarose-protein complexes were collected by brief centrifugation. After washing three times with a wash buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 0.5% Triton X-100, and 10 μg/mL phenylmethylsulfonyl fluoride), the protein complexes were boiled for 5 min in 1× SDS-PAGE sample buffer. After centrifugation, the supernatant was separated by SDS-PAGE, and MYC-tagged WRKY protein in the immunoprecipitated FLAG-tagged WRKY18 complexes were detected with protein gel blot analysis using anti-MYC-peroxidase (Invitrogen).
Recombinant Protein and DNA Binding
Preparation of recombinant WRKY proteins and DNA binding assays were performed as previously described (Yu et al., 2001).
Identification and Genotyping of Knockout Mutants
The wrky18-1 (Salk_093916) and wrky60-1 (Salk_120706) T-DNA insertion mutants were identified from the Salk Arabidopsis T-DNA insertion population (Alonso et al., 2003) and are in the Columbia (Col) ecotype. The wrky40 mutant was a Ds transposon insertion mutant obtained from Cold Spring Harbor Laboratory gene trap lines (Sundaresan et al., 1995) and is in the Landsberg erecta (Ler) background. Confirmation of the insertions was done by performing PCR using a combination of a border primer of T-DNA (5′-GCTTGCTGCAACTCTCTCAG-3′) or Ds transposon (5′-ACCCGACCGGATCGTATCGGT-3′) and a gene-specific primer (W18, 5′-TCATAGAAATTGAAGGGATACAAAAA-3′; W40, 5′-CGGAGATGCTAACTTTGATGTG-3′; W60, 5′-GCTGAAGCCGAGACTTCTCTT-3′). Another PCR was performed to identify plants homozygous for the insertions using the above gene-specific primers and respective reverse primers (W18R, 5′-TGATTTTTCATTTTCGTTAAAGC-3′; W40R, 5′-GGAGCACAAGCACATTTGAA-3′; W60R, 5′-ATTTTCCACCCAAATCGTCA-3′). To remove additional insertions or mutations from the mutant, backcrosses to wild-type plants were performed, and plants homozygous for the insertion were again identified. Since the wrky40 mutant was in the Ler ecotype, it was backcrossed to Col wild-type plants three times, and the progeny plants from each cross were genotyped with a collection of 22 simple sequence length polymorphism markers spaced evenly over the entire genome in order to identify plants that contained the most Col genetic background. Double and triple mutants were generated from genetic crosses of single mutants and identified through PCR genotyping.
To generate WRKY genes for genetic complementation, their cDNA clones were placed behind their native promoter fragments (∼1.2 kb) that were amplified by PCR. The genes were subcloned into a modified plant transformation vector pKMB (Mylne and Botella, 1998) and directly transformed into the wrky18 wrky40 wrky60 triple mutants. T1 transformants were identified based on their resistance to spraying with 1.384% (v/v) Finale (Farnam Companies) in soil and used for analysis of disease resistance.
Construction of Transgenic Plants
The transgenic line constitutively expressing WRKY18 (W18Δ-1) has been previously described (Chen and Chen, 2002) and was used in this study. The cDNA fragments that contain the full coding sequences and 3′ untranslated regions of WRKY40 or WRKY60 were excised from the cloning plasmid and subcloned into the Agrobacterium tumefaciens transformation vector pOCA30 (Chen and Chen, 2002) in the sense orientation behind the CaMV 35S promoter. Arabidopsis transformation was performed by the floral dip procedure (Clough and Bent, 1998). The seeds were collected from the infiltrated plants and selected in MS medium containing 50 μg/mL kanamycin. Kanamycin-resistant plants were transferred to soil 9 d later and grown in a growth chamber.
Pathogen Infection
Pathogen inoculations were performed by infiltration of leaves of 6 to 10 plants for each treatment with the PstDC3000 strain that contains the pVSP61 kanamycin-resistant empty plasmid vector (OD600 = 0.001 in 10 mM MgCl2). Inoculated leaves were harvested 3 DAI and homogenized in 10 mM MgCl2. Diluted leaf extracts were plated on King's B medium supplemented with rifampicin (100 μg/mL) and kanamycin (25 μg/mL) and incubated at 25°C for 2 d before counting the colony-forming units.
Botrytis cinerea was grown on 2xV8 agar as described previously (Mengiste et al., 2003). To infect plants, conidia were collected from 10-d-old culture, and the spore density was adjusted in Sabouraud Maltose Broth and sprayed using a Preval sprayer. Inoculated plants were maintained at high humidity with a transparent cover in a growth chamber, and symptom development was observed 3 to 4 d after the inoculation. Biomass of the fungal pathogen was quantified by RNA gel blotting of total RNA isolated from inoculated plants. A Botrytis β-tubulin gene fragment amplified from the Botrytis genome DNA was used as a probe as described previously (Mengiste et al., 2003).
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: WRKY18, At4g31800; WRKY40, At1g80840; WRKY60, At2g25000; PR1, At2g14610; PDF1.2, At5g44420.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitative Assays of the Effect of the Leu Zipper Motif on WRKY Protein Interactions.
Supplemental Figure 2. Genetic Complementation of the wrky18 wrky 40 wrky60 Mutant.
ACKNOWLEDGMENTS
We thank the ABRC at the Ohio State University (Columbus, OH) and R. Martienssen from the Cold Spring Harbor Laboratory (Cold Spring Harbor, NY) for the Arabidopsis mutants. We also thank W. Gassmann (University of Missouri, Columbia, MO) for providing the Pseudomonas strain and T. Mengiste (Purdue University) for providing the Botrytis culture. We are grateful to J. Banks (Purdue University) for critically reading the manuscript. This work was supported in part by National Science Foundation Grant MCB-0209819. This is journal paper 2006-17841 of the Purdue University Agricultural Research Program.
REFERENCES
Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (
Alonso, J.M., et al. (
Andreasson, E., et al. (
Aoyama, T., and Chua, N.-H. (
Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (
Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (
Chen, C., and Chen, Z. (
Chen, C., and Chen, Z. (
Chen, K., Du, L., and Chen, Z. (
Clough, S.J., and Bent, A.F. (
Collmer, A., and Bauer, D.W. (
Dangl, J.L., and Jones, J.D. (
Delaney, T.P.U.S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (
Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (
Devadas, S.K., Enyedi, A., and Raina, R. (
Dong, J., Chen, C., and Chen, Z. (
Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (
Freedman, L.P., and Luisi, B.F. (
Glazebrook, J. (
Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (
Govrin, E.M., and Levine, A. (
Hara, K., Yagi, M., Kusano, T., and Sano, H. (
Heck, S., Grau, T., Buchala, A., Metraux, J.P., and Nawrath, C. (
Kalde, M., Barth, M., Somssich, I.E., and Lippok, B. (
Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (
Kunkel, B.N., and Brooks, D.M. (
Lahaye, T. (
Landschulz, W.H., Johnson, P.F., and McKnight, S.L. (
Li, J., Brader, G., and Palva, E.T. (
Liu, Y., Schiff, M., and Dinesh-Kumar, S.P. (
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (
Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (
Mylne, J., and Botella, J.R. (
Nawrath, C., and Metraux, J.P. (
Ohashi-Ito, K., Kubo, M., Demura, T., and Fukuda, H. (
Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (
Petersen, M., et al. (
Pierson III, L.S., Wood, D.W., and Pierson, E.A. (
Rogers, E.E., and Ausubel, F.M. (
Rushton, P.J., and Somssich, I.E. (
Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (
Samuel, M.A., Walia, A., Mansfield, S.D., and Ellis, B.E. (
Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (
Spoel, S.H., et al. (
Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (
Takahashi, H., Kanayama, Y., Zheng, M.S., Kusano, T., Hase, S., Ikegami, M., and Shah, J. (
Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (
Towers, T.L., Luisi, B.F., Asianov, A., and Freedman, L.P. (
Turck, F., Zhou, A., and Somssich, I.E. (
Ulker, B., and Somssich, I.E. (
van Wees, S.C., and Glazebrook, J. (
Yu, D., Chen, C., and Chen, Z. (
Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (
Author notes
Current address: Key Laboratory of Gene Engineering of the Ministry of Education, Biotechnology Research Center, Zhongshan University, Guangzhou 510275, People's Republic of China.
Current address: AgResearch Limited, Tennent Drive, Private Bag 11008, Palmerston North, New Zealand.
To whom correspondence should be addressed. Email zhixiang@purdue.edu; fax 765-494-5896.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhixiang Chen (zhixiang@purdue.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037523.