Abstract
Acinetobacter pittii 44551 was recovered from a patient with gout combined with tuberculosis and was found to harbor the carbapenemase genes blaNDM-1 and blaOXA-58 on two different plasmids pNDM-44551 and pOXA58-44551, respectively. pNDM-44551 displayed high self-transferability across multiple bacterial species, while pOXA58-44551 was likely co-transferable with pNDM-44551 into A. baumannii receipts. pNDM-44551 was a close variant of the previously characterized pNDM-BJ01 and the blaNDM-1 gene cluster was arranged sequentially as orfA, ISAba14, aphA6, ISAba125, blaNDM-1, bleMBL, ΔtrpF, dsbC, tnpR and zeta. pOXA58-44551 was a repAci9-containing plasmid and blaOXA-58 was embedded in a 372F-ISAba3-like-blaOXA-58-ISAba3 structure. The mobile genetic platforms of blaNDM-1 and blaOXA-58 herein showed some differences from their previously characterized variants. The production of NDM-1 in strain 44551 contributed the majority to its high resistance to carbapenems, while the blaOXA-58 stayed silent most likely due to the lack of an upstream promoter to drive its transcription. Increased surveillance of Acinetobacter co-harboring blaNDM-1 (active) and blaOXA-58 (either active or silent) is urgently needed.
Similar content being viewed by others
Introduction
Acinetobacter spp. are important opportunistic pathogens closely linked to nosocomial infections and the most common species from clinical samples is A. baumannii, followed by A. pittii and A. nosocomialis1. Worldwide increasing emergence of carbapenem-resistant bacteria including Acinetobacter has caused concern over the limited availability of effective antimicrobial agents in the clinic.
The most prevalent mechanism of carbapenem resistance in Acinetobacter is the elevated expression of OXA-type carbapenemase genes, mainly including the horizontally acquired blaOXA-23-like, blaOXA-24-like and blaOXA-58-like genes as well as the intrinsic blaOXA-51-like genes2,3. The blaOXA-58-like and blaOXA-24-like genes are much less frequently detected relative to the blaOXA-23-like genes, but they can confer a high level of carbapenem resistance and cause local outbreaks4,5,6. The carbapenem hydrolytic activity of OXA-58 can be inhibited by NaCl in a Tyr144-Gly-Asn motif-dependent pattern and this property has been used to screen for the expression of blaOXA-587.
At least four types of metallo-β-lactam (MBL) carbapenemase, i.e., IMP, VIM, SIM and NDM, have been identified in Acinetobacter and these MBLs have lower detection rates but higher carbapenemase activities relative to OXAs8,9. Originally found in Klebsiella pneumoniae in 2008, blaNDM has been identified globally in many bacteria and received great attention partly due to its high transferability across different bacterial species, its frequent co-occurrence with other resistance genes and the global spread of blaNDM-carrying strains10,11,12,13,14,15. A large collection of blaNDM-containing plasmids from different bacterial species have been characterized (Table S1 and the references therein). The blaNDM genetic surroundings are commonly composed of ISAba125 (intact or truncated) and bleMBL-ΔtrpF, which are upstream and downstream of blaNDM, respectively, suggesting a similar origin of these blaNDM genes detected (Table S1). Most of the blaNDM genetic platforms from Acinetobacter are a Tn125-related sequence, with a few exceptions containing the truncated forms of this genetic context (Table S1). Tn125 is a blaNDM-containing composite transposon originally described by Pfiefer et al.16 and it has been proposed to be involved in the blaNDM genes dissemination in Acinetobacter17. Almost all the well-characterized blaNDM-containing plasmids from Acinetobacter in China are closely related to the plasmid pNDM-BJ0118 by sharing a novel type IV secretion system backbone (Table S1 and references therein).
Coexistence of blaNDM and blaOXA has been described in Acinetobacter, e.g. blaOXA-23 and blaNDM-1 in A. baumannii from India19 and the Czech Republic20 and blaNDM-1, blaOXA-23 and blaIMP in A. baumannii from China21. However, it remains unclear whether and how these co-existing carbapenemase genes are expressed to contribute to drug resistance. The present study describes the genetic environment, transferability and antibiotic susceptibility of blaNDM-1 and blaOXA-58 harbored on different plasmids in a single clinical A. pittii isolate from China.
Results and Discussion
blaNDM-1 and blaOXA-58 on different plasmids in a single A. pittii isolate
A total of 31 carbapenem-resistant Acinetobacter isolates were collected at the Meizhou People's Hospital, Guangdong Province, China, from June 2010 to December 2012. They were classified into A. pittii (n = 1, designated 44551), A. nosocomialis (n = 2) and A. baumannii (n = 28). PCR screening for the MBL and OXA carbapenemase genes in these strains indicated the presence of blaNDM-1 and blaOXA-58 in strain 44551 and the habitation of blaOXA-23 in 21 A. baumannii isolates (67.7%), while all the other PCR reactions gave negative results. DNA sequencing further confirmed the presence of blaNDM-1 and blaOXA-58 in 44551. PCR detection of the extended spectrum β-lactamase genes blaTEM, blaCTX-M, blaPER, blaSHV, blaDHA and blaCMY22,23,24,25 in 44551 showed negative results. The observation of blaOXA-23 as the most prevalent carbapenemase gene in A. baumannii is consistent with previous findings26.
A. pittii 44551 was isolated from the sputum of a 62-year-old male hospitalized for gouty arthritis with a skin soft tissue infection combined with pulmonary tuberculosis (TB) in August 2011. After the initiation of anti-tuberculosis treatment with HREZ (isoniazid, rifampicin, ethambutol hydrochloride and pyrazinamide), the patient's sputum samples were screened weekly for Mycobacterium tuberculosis. The patient's pulmonary infection symptoms, including fever, weakness and coughing, were relieved in response to anti-TB therapy and his sputum acid-fast stain gave negative results after two weeks of treatment. After one month of hospitalization, the patient improved perceptibly with stable vital signs. He was discharged with instructions to continue taking 3HREZ/9-12HRZ as prescribed and to undergo regular follow-up. Unfortunately, the patient did not return for regular checkup and has been out of touch with our clinic.
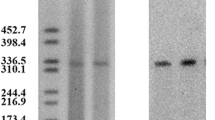
S1-nuclease pulsed-field gel electrophoresis (S1-PFGE) assay (Fig. 1a) showed that A. pittii 44551 harbored four different plasmids at ~20 kb, ~40 kb, ~50 kb and ~90 kb. Subsequent Southern hybridization (Fig. 1b) with a probe specific to blaNDM-1 or blaOXA-58 revealed that these two genes were located on the ~40 kb (designated pNDM-44551) and ~50 kb (pOXA58-44551) plasmids, respectively. Using the PCR-based replicon typing scheme of A. baumannii27, repAci9 was detected in pOXA58-44551 but no classified type of replicase could be identified from pNDM-44551.
S1-PFGE and Southern blot assay of strain 44551 and its conjugants.
Shown are EtBr-stained PFGE gel of S1-digested of genomic DNA samples (a) and subsequent Southern blot hybridization with the DIG-labeled probe specific to blaNDM-1 (b) or blaOXA-58 (c). The white arrow and star indicate the locations of blaOXA-58- and blaNDM-1-positive signals, respectively. Lane 1: 44551; 2: J53; 3: J53-44551; 4: EC600; 5: EC600-44551; 6: MZPB; 7: MZPB-44551; 8: MZPB-44551OXA58+; M: marker.
Mating out experiments showed that plasmid pNDM-44551 could be transferred from 44551 into E. coli J53 and EC600 and A. baumannii MZPB at similar frequencies of 2.36 × 10−2, 2.81 × 10−2 and 3.54 × 10−2 per donor cell, respectively. The initial cultures of all the chosen J53 and EC600 conjugant clones and most MZPB clones were blaNDM-1-positive and blaOXA-58-negative as revealed by PCR/sequencing. A single resulting conjugant from each of these J53, EC600 and MZPB conjugants, designated J53-44551 EC600-44551 and MZPB-44551, respectively, was selected for S1-PFGE/Southern hybridization (Fig. 1a and 1b). Besides detection of ~40 kb pNDM-44551 in all the tested conjugants, an additional blaNDM-1-positive signal at ~80 kb was found in J53-44551 and EC600-44551, suggesting that a recombination event occurred for pNDM-44551 upon conjugal transfer from Acinetobacter to Enterobacteriaceae. The above observations denoted high self-transferability of pNDM-44551 across multiple bacterial species.
A weak PCR signal of blaOXA-58 was observed in a small portion of the blaNDM-1-positive MZPB-44551 clones at a 1:10 ratio, as well as in their passage cultures, which was further confirmed by sequencing. Meanwhile, Random Amplified Polymorphic DNA showed that these blaOXA-58- and blaNDM-1-positive conjugants were MZPB-originated species. The PCR results of one of the above conjugants, designated MZPB-44551OXA58+, were shown in Fig. S1a and S1b. Nonetheless, the pOXA58-44551 signal was invisible by S1-PFGE/Southern hybridization in MZPB-44551OXA58+ (Fig. 1a), which was attributable to the very low copy number of pOXA58-44551. In addition, no PCR signal of blaOXA-58 was detected in all the blaNDM-1-haboring J53 or EC600 conjugants tested, which is consistent with the fact that blaOXA-58 has only been found in Acinetobacter28. In all, it seemed that pOXA58-44551 could co-transfer with pNDM-44551 only into the Acinetobacter recipient.
Plasmid pBBRIMCS3-NDM-1, containing the ISAba125-blaNDM-1 fragment cloned from pNDM-44551, was transformed into E. coli DH5α and the resulting transformant clone was named DH5α-NDM1. All clones of 44551, DH5α-NDM1, J53-44551 and EC600-44551 showed high resistance to the penicillin/cephalosporin drugs tested. 44551 and DH5α-NDM1 also displayed high resistance to IPM and MEM, but J53-44551 and EC600-44551 remained susceptible to these two carbapenem drugs (Table 1). Therefore, we speculated that more functional NDM-1 proteins might be present in 44551 and DH5α-NDM1 than in J53-44551 and EC600-44551, although there was no discernible difference in the pNDM-44551 copy number among these strains (Fig. 1a and 1b). This speculation was supported by the northern blot and quantitative reverse-transcription PCR (RT-qPCR) assays, which showed that the blaNDM-1 mRNA abundance in 44551 or DH5α-NDM1 was much higher than that in J53-44551 or EC600-44551 (Fig. S2).
It is worth noting that MZPB-44551 showed distinct antimicrobial susceptibility profile from 44551 across almost all the tested β-lactams (Table 1), while the pNDM-44551 copy number (Fig. 1a) or the blaNDM-1 mRNA abundance (Fig. S2) was comparable between these two strains. Based on the spontaneous PB-resistance of MZPB under PB selection, it is reasonable to speculate that this distinction may be mainly due to different species backgrounds.
Genetic surroundings of blaNDM-1
pNDM-BJ01 was recovered from A. lwoffii in China in 2012 and harbored four regions encoding for conjugate transfer, plasmid replication and stability, a type IV secretion system and the blaNDM-1 gene cluster18. Primer walking combined PCR/sequencing indicated that pNDM-44551 contained 41 of the 46 CDSs annotated in pNDM-BJ01 with >99% sequence identity and the missing five CDSs were located in a tandem manner within the blaNDM-1 gene cluster.
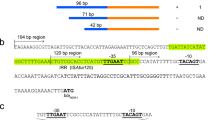
The blaNDM-1 gene cluster of pNDM-44551 was arranged sequentially as orfA and orfB of ISAba14, aphA6, ISAba125, blaNDM-1, bleMBL, ΔtrpF, dsbC, tnpR and zeta from left to right; a five-gene region (cutA, ΔgroS, groEL, insE and ISAba125) located between dsbc and tnpR of pNDM-BJ01 was absent from pNDM-44551; the cleavage of the above DNA region led to a 97 bp 3′-remnant (GC content ≈ 60%) of the cutA coding region (CDS) to be spliced directly with a 372 bp 3′-flanking sequence (GC content ≈ 40%) of tnpR at the junction site AGGGAT-ATATAG, generating a ‘new' dsbC-tnpR intergenic region in pNDM-44551 (Fig. 2a).
Schematic alignment of blaNDM-1 genetic surroundings.
Genes and their transcriptional orientations are represented by differently patterned horizontal arrows. orfA and orfB: ISAba14 transposase; aphA6: aminoglycoside 3′-phosphotransferase; blaNDM-1: New Delhi metallo-β-lactamase 1; bleMBL: bleomycin resistance protein; ΔtrpF: truncated phosphoribosylanthranilate isomerase; dsbC: tat twin-arginine translocation pathway signal sequence domain protein; cutA: periplasmic divalent cation tolerance protein; ΔgroS and groEL: chaperonin subunits; insE: ISCR3 transposase. (a) The blaNDM-1 gene clusters from pNDM-44551 and pNDM-BJ01. The identical DNA regions are shaded in gray, the GC skew is shown with a sliding window of 100 nucleotides and the primers used for PCR/sequencing are marked with thin black arrows as well as corresponding primer names. (b) Schematic showing the sequence differences of aphA6 CDSs and their 3′ flanking regions from pNDM-BJ01, pNDM-44551 and Tnaph6.
The aphA6 gene was usually found adjacently upstream of the ISAba125-blaNDM-1 structure in the pNDM-BJ01-like plasmids (Table S1). It has been postulated that a Tn125 transposon structure inserts into the non-coding region downstream of aphA6, which is evidenced by the 3-bp GTT target site duplication at the point of insertion as shown in pNDM-BJ0118. A 64 bp direct repeat was observed flanking the ISAba125-blaNDM-1 insertion in pNDM-44551 and each monomer was composed of the first 20 bp of the CDS of aphA6 or blaNDM-1 together with its upstream 44 bp sequence (Fig. 2a), which was consistent with the previous report that blaNDM-1 is a chimeric gene resulted from the in-frame fusion of a preexisting blaNDM-1 with aphA629.
A total of 11 single nucleotide polymorphism (SNP) sites were present in the aphA6 CDS of pNDM-44551 relative to pNDM-BJ01, resulting in three amino acid (a.a.) changes of L84F, A156T and R163K. In addition, a 67 bp deletion occurred within the 3′ flanking region of aphA6 of pNDM-44551 compared with pNDM-BJ01 (Fig. 2b). The aphA6 CDS together with its 3′ flanking region of pNDM-44551 was almost identical to the counterpart of TnaphA6 (accession number JF343537, located in the chromosome of A. baumannii from Australia) and that of pWH8144 (accession number JG241792, in A. baumannii from China), with only one SNP mismatch (T to C) at the 5′ end of aphA6. By contrast, this kind of aphA6 gene considerably differed from the counterpart of pNDM-BJ01 (Fig. 2b). aphA6 has been proposed to be an ancestral gene in Acinetobacter and possesses considerable nucleotide polymorphism30. There might be two possible explanations for the aforesaid aphA6 sequence difference: 1) the original recombination of aphA6 with blaNDM-1 may be two independent events in pNDM-44551 and pNDM-BJ01 and 2) homologous recombination may occur to induce an aphA6 swap upon the spread of the aphA6-ISAba125-blaNDM-1-containing plasmid into an aphA6-carrying Acinetobacter strain.
The above observations strongly suggest that pNDM-44551 represents a close derivate of pNDM-BJ01 although pNDM-44551 might have undergone multiple evolutionary events especially within the blaNDM-1 gene cluster. An array of the pNDM-44551 variants have been characterized in Acinetobacter from the mainland of China, such as pXBC1 in A. johnsonii31, pNDM-AB in A. baumannii32 and pAP-D499 in A. pittii33. The major genetic differences in the above plasmids were confined in the surrounding regions of blaNDM-1 (Table S1). It should be noted that the ISAba125 element upstream of blaNDM-1 is usually intact in Acinetobacter but often truncated in Enterobacteriaceae (Table S1), suggesting the probable spread of the blaNDM-1 genetic platforms from Acinetobacter to Enterobacteriaceae13,18,29,34
Genetic surroundings of silent blaOXA-58
In general, blaOXA-58 is embedded in a conserved platform ISAba3-like-blaOXA-58-ISAba3 in Acinetobacter and the upstream ISAba3-like element is often interrupted by other insertion sequence (IS) elements, which in turn provide the promoters enhancing the blaOXA-58 expression to mediate higher degrees of drug resistance compared with the parent intact ISAba3-like35,36,37,38,39. Sequence analysis revealed that the blaOXA-58 gene of pOXA58-44551 was located between a downstream ISAba3 and an upstream ISAba3-like, both of which were intact; the downstream ISAba3 was followed by a gene cluster aaC3 (aminoglycoside N3′-acetyltransferase III)-ATPase (ATPase protein) (Fig. 3), which differed from the previously reported common pattern araC1 (transcription regulator)-lysE (threonine efflux protein) in blaOXA-58 genetic contexts in Acinetobacter spp. from China40 and other countries39,41. The C-terminal 23 a.a. sequence of the transposase of the upstream ISAba3-like element in pOXA58-44551 was replaced with an unknown 26 a.a. fragment and moreover the 3′-untranslated region (3′-UTR) and the right inverted repeat (IRR) were lost (Fig. 3). This entire upstream ISAba3-like element was essentially the same as the counterpart in the blaOXA-58 genetic structure in an A. pittii isolate AP04 from China40 and some unpublished blaOXA-58-containing sequences (accession numbers JX101647, FJ195389 and FJ200187), as well as in a blaOXA-97 genetic structure36. In addition, a 372 bp DNA fragment (named 372F) as the left neighbor of the upstream ISAba3-like element in pOXA58-44551 (Fig. 3) showed 100% sequence identity to that in the genome of the insect Dendroctonus ponderosae (APGK01007886.1). In all, blaOXA-58 was mobilized into the genetic platform 372F-ISAba3-like-blaOXA-58-ISAba3 (flanked by the 28 bp direct repeat; having a GC content much lower than those of the surrounding regions), which was further inserted into the backbone of pOXA58-44551 most likely through IS-mediated transposition.
Schematic alignment of blaOXA-58 genetic surroundings.
Shown are the blaOXA-58 gene cluster from pOXA58-44551 (upper panel) and that of the most common pattern (lower panel) characterized previously35,36,37,38,39,40. Genes and their transcriptional orientations are represented by horizontal arrows. The primers used for PCR/sequencing are marked with thin black arrows as well as corresponding primer names.
The evidences described below from different aspects denoted that blaOXA-58 was expressed either very weakly or not at all in 44551. First, the addition of EDTA (the inhibitor of MBLs including NDM1) in the agar plates for bacterial cultivation made 44551 lose almost the entire resistance to the tested β-lactams, but the addition of excess NaCl (the inhibitor of OXA-58) had no effect on the resistance profile of 44551 (Table 1), indicating that the production of NDM-1 enzyme contributed the majority to β-lactams resistance while blaOXA-58 contributed little to the resistance. Second, the ISAba3-like element upstream of blaOXA-58 in pOXA58-44551 was intact, suggesting a lack of the blaOXA-58-driven promoter which was usually provided by other inserted IS elements in the typical OXA-58-encoding genetic structures35,36,37,38,39 (Fig. 3). This postulation was further supported by a recent surveillance study, which showed that as many as 25 of the total of 32 blaOXA-58-positive Acinetobacter isolates recovered from 23 Chinese provinces remained susceptible to carbapenems and all of them contained an intact ISAba3-like element upstream of blaOXA-58, while the remaining non-susceptible isolates showed an insertion of additional IS element into the upstream ISAba3-like element42. Third, repeated attempts to transform the plasmid pBBRIMCS3-OXA-58 (containing the ISAba3-like-blaOXA-58 fragment cloned from pOXA58-44551) into E. coli DH5α failed to generate an ampicilin-resistant, blaOXA-58-haboring clone. Finally, northern blot with a probe specific to blaOXA-58 showed very weak or almost invisible signals in A. pittii 44551, but a very strong band could be detected in the BL21-OXA58 strain, which was an E. coli BL21 strain carrying the plasmid pET-28a-blaOXA58 (Fig. 4a and 4b). Consistent with the northern blot results, RT-qPCR assay revealed that the relative mRNA abundance of blaOXA-58 in strain 44551 was about 30-fold lower than that in BL21-OXA58, with the 16S rRNA genes being the internal control (Fig. 4c).
Detection of blaNDM-1 and blaOXA-58 transcripts.
Total RNAs were extracted from strains 44551, BL21-NDM and BL21-OXA58. RNA samples were analyzed on 1.2% agarose gel followed by EtBr staining and then subjected to Northern blot hybridization with the DIG-labeled probe specific to blaNDM-1 (a) and blaOXA-58 (b). IPTG was added in the cell cultures to induce the expression of blaNDM-1 or blaOXA-58. Lane 1: 44551; 2: BL21-NDM in the absence of IPTG; 3: BL21-NDM in the presence of IPTG; 4: BL21-OXA58 in the absence of IPTG; 5: BL21-OXA58 in the presence of IPTG. The calculated size of blaNDM-1 or blaOXA-58 transcript is about 1 kb. The EtBr staining of the 23S and 16S rRNA genes (2.9 kb and 1.5 kb, respectively) was used as loading control (lower panels). The relative mRNA abundances of blaNDM-1 or blaOXA-58 in 44551, BL21-NDM and BL21-OXA58 induced with IPTG, respectively, were detected by RT-qPCR (c). The 16S rRNA genes of 44551 and BL21 were employed as the internal control. The normalized mRNA abundance of blaNDM-1 in 44551 was set as 1. Results are expressed as mean ± SD.
Concluding remarks
In the present study, blaNDM-1 and blaOXA-58 were found to be harbored on two different plasmids in a single clinical A. pittii isolate named 44551, with the former gene expressing well and the latter one silent. Each of these two resistance genes was embedded in a genetic structure differing partly from its previously characterized variants. The blaNDM-1-carrying plasmid in 44551, conferring a high level of carbapenem resistance, showed a strong ability of horizontal transfer into Acinetobacter and Enterobacteriaceae. The silent blaOXA-58 has the potential to evolve into the active form due to additional IS element insertion driven by environmental pressures such as the presence of carbapenems. The fact that A. pittii 44551 colonized in the lower respiratory tract of the indicated hospitalized patient increased the possibility of the strain 44551 dissemination into hospital settings. Since the Acinetobacter strains co-harboring blaNDM-1 (active) and blaOXA-58 (either active or silent) have the potential to widely spread in China42, increased surveillance of these kinds of bacteria in hospital and community settings is urgently needed.
Methods
Clinical Acinetobacter isolates
Each clinical sample was inoculated onto MacConkey agar plates and the dominant strain was recovered and identified using Vitek II (BioMérieux, Durham). Discrimination of Acinetobacter was performed by one-tube multiplex PCR specific for A. baumannii identification43 and by 16S-23S rRNA intergenic spacer sequencing for other types of Acinetobacter44. All the primers used in this study are listed in Table S2.
Detection of carbapenemase genes and their genetic contexts
The MBL genes blaIMP, blaVIM, blaSIM and blaNDM were screened by multiplex PCR45. The OXA genes blaOXA-23, blaOXA-24 and blaOXA-58 were detected individually by PCR. The plasmid sample was prepared from 44551 using a BAC/PAC DNA Isolation Maxi Kit (Omega Bio-Tek) and the flanking regions were sequenced by primer walking from both ends of blaNDM-1 or blaOXA-58. All PCR amplicons were subjected to DNA sequencing with an ABI 3700 sequencer.
Plasmid construction and electrotransformation
The blaNDM-1 or blaOXA-58 coding region together with its immediately upstream IS element was amplified from 44551 and then cloned into the cloning vector pBBRIMCS346, generating the recombinant plasmid pBBRIMCS3-NDM-1 or pBBRIMCS3-OXA-58, respectively. The entire coding region of blaOXA-58 was cloned into the expression vector pET-28a, generating pET-28a-blaOXA-58. pBBRIMCS3-NDM-1 or pBBRIMCS3-OXA-58 was transformed into E. coli DH5α through electrotransformation, while pET-28a-blaOXA-58 was transformed into E. coli BL21, with an attempt to obtain the E. coli clone expressing the corresponding carbapenemase enzyme.
Conjugal transfer
A. pittii 44551 harboring blaNDM-1 and blaOXA-58 was used as the donor and E. coli J53 (NaN3 resistant), EC600 (rifampin resistant) and A. baumannii MZPB were used as the recipients. MZPB is a homemade polymyxin-resistant derivate of a clinical isolate A. baumannii MZ and remains susceptible to β-lactams. Membrane mating experiments were performed on Mueller-Hinton (MH) agar47. After 18 h of incubation, the mixed cultures were suspended in MH broth and plated onto MH agar containing ampicillin (50 μg/ml) and NaN3 (200 μg/ml) for J53, ampicillin (50 μg/ml) and rifampin (500 μg/ml) for EC600 and ampicillin (50 μg/ml) and polymyxin (20 μg/ml) for MZPB. Conjugants were picked randomly from the original selective plates and inoculated into the selective LB broth, these initial cultures were used for PCR-based screening for the presence of blaNDM-1 and blaOXA-58. To exclude the donor strain contamination, the initial culture of positive conjugants were spread onto the Amp+/PB+ plate, then the second-passage colonies were randomly picked for each strain and subjected for PCR detection of blaNDM-1 and blaOXA-58. The strain species were differentiated by Random Amplified Polymorphic DNA with two short primers M13 and AP248.
Antimicrobial susceptibility testing
The MIC values for each indicated strain cultured on the MH plates were measured using Etest (AB bioMérieux, Solna, Sweden). The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines and the British Society for Antimicrobial Chemotherapy (SAC) breakpoints49,50.
S1-PFGE and Southern blot
Bacterial genomic DNA was prepared in agarose plugs and digested with S1 nuclease (Takara). The linearized plasmids and partially digested genomic DNA were separated through the CHEF-Mapper XA PFGE system (Bio-Rad). The DNA fragments were stained with ethidium bromide (EtBr), transferred to a Hybond N+ membrane (GE Amersham Biosciences) and hybridized with the DIG-labeled probe specific to blaNDM-1 or blaOXA-58. Probe labeling and signal detection were carried out with DIG high primer DNA labeling and detection starter kit II according to the manufacturer's instructions (Roche Diagnostics).
RNA extraction and Northern blot
By using Trizol Reagent (Life Technologies), total RNA was isolated from the overnight culture of each indicated strain with or without addition of 1 mM IPTG. The RNA samples were analyzed on a formaldehyde-containing 1.2% agarose gel. Subsequent EtBr staining, membrane transfer, probe labeling and hybridization and signal detection were carried out as above.
RT-qPCR
5 μg of RNA treated with DNase I (Promega) was subjected to reverse transcription by using random hexamers and PrimeScript RT reagents (Takara). Simultaneously, a control reverse transcription reaction without reverse transcriptase was performed to rule out genomic DNA contamination. The cDNA reactions were diluted 1:5 in water as the template for PCR detection of blaNDM-1 or blaOXA-58 and diluted 1:500 for amplification of the 16S rRNA genes. Each PCR reaction contained 2 μl of cDNA, 8 μl of forward and reverse primers (each at 0.75 μM) and 10 μl of SYBR green PCR Supermix (Bio-Rad). Three independent bacterial cultures (total RNA samples) were employed as biological replicates and each RNA sample was analyzed in triplicate in PCR. The PCR parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s, using Bio-Rad CFX96 thermocycler. The detecting mRNA levels of blaNDM-1 or blaOXA-58 were normalized to those of the 16S rRNA genes.
Plasmid replicon typing
Plasmid pOXA58-44551 was recovered from the S1-PFGE gel and used as the template for PCR detection of replicase genes using the A. baumannii PCR-based replicon typing scheme27. The variants belonging to the same group of replicases were recognized by further PCR amplicon sequencing.
Nucleotide sequence accession numbers
The blaNDM-1 or blaOXA-58 gene cluster reported herein was deposited in GenBank with the accession number KF208467 or KF208466, respectively.
References
Nemec, A. et al. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162, 393–404 (2011).
Abbott, I., Cerqueira, G. M., Bhuiyan, S. & Peleg, A. Y. Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev. Anti. Infect. Ther. 11, 395–409 (2013).
Poirel, L. & Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836 (2006).
Mendes, R. E., Bell, J. M., Turnidge, J. D., Castanheira, M. & Jones, R. N. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63, 55–59 (2009).
Moro, M. et al. An outbreak caused by multidrug-resistant OXA-58-positive Acinetobacter baumannii in an intensive care unit in Italy. J. Hosp. Infect. 68, 97–99 (2008).
Castanheira, M., Wanger, A., Kruzel, M., Deshpande, L. M. & Jones, R. N. Emergence and clonal dissemination of OXA-24- and OXA-58-producing Acinetobacter baumannii strains in Houston, Texas: report from the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 46, 3179–3180 (2008).
Poirel, L. et al. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 202–208 (2005).
Cornaglia, G., Giamarellou, H. & Rossolini, G. M. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect. Dis. 11, 381–393 (2011).
Gupta, V. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin. Investig. Drugs 17, 131–143 (2008).
Bushnell, G., Mitrani-Gold, F. & Mundy, L. M. Emergence of New Delhi metallo-beta-lactamase type 1-producing enterobacteriaceae and non-enterobacteriaceae: global case detection and bacterial surveillance. Int. J. Infect. Dis. 17, e325–333, 10.1016/j.ijid.2012.11.025 (2013).
Yong, D. et al. Characterization of a new metallo-beta-lactamase gene, blaNDM-1 and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054 (2009).
Villa, L., Poirel, L., Nordmann, P., Carta, C. & Carattoli, A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67, 1645–1650 (2012).
Poirel, L., Dortet, L., Bernabeu, S. & Nordmann, P. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407 (2011).
Wailan, A. M. & Paterson, D. L. The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev. Anti. Infect. Ther. 12, 91–115 (2014).
Dortet, L., Poirel, L. & Nordmann, P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014, 249856, 10.1155/2014/249856 (2014).
Pfeifer, Y. et al. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66, 1998–2001 (2011).
Poirel, L. et al. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089 (2012).
Hu, H. et al. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56, 1698–1702 (2012).
Karthikeyan, K., Thirunarayan, M. A. & Krishnan, P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65, 2253–2254 (2010).
Krizova, L., Bonnin, R. A., Nordmann, P., Nemec, A. & Poirel, L. Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. J. Antimicrob. Chemother. 67, 1550–1552 (2012).
Chen, Z. et al. Coexistence of blaNDM-1 with the prevalent blaOXA23 and blaIMP in pan-drug resistant Acinetobacter baumannii isolates in China. Clin. Infect. Dis. 52, 692–693 (2011).
Feizabadi, M. M. et al. Distribution of blaTEM, blaSHV, blaCTX-M genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb. Drug Resist. 16, 49–53 (2010).
Perez-Perez, F. J. & Hanson, N. D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162 (2002).
Poirel, L., Cabanne, L., Vahaboglu, H. & Nordmann, P. Genetic environment and expression of the extended-spectrum beta-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49, 1708–1713 (2005).
Adams-Haduch, J. M. et al. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52, 3837–3843 (2008).
Hu, Q., Hu, Z., Li, J., Tian, B. & Xu, H. Detection of OXA-type carbapenemases and integrons among carbapenem-resistant Acinetobactor baumannii in a teaching hospital in China. J. Basic Microbiol. 51, 467–472 (2011).
Bertini, A. et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4168–4177 (2010).
Poirel, L., Naas, T. & Nordmann, P. Diversity, epidemiology and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54, 24–38 (2010).
Toleman, M. A., Spencer, J., Jones, L. & Walsh, T. R. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother 56, 2773–2776 (2012).
Yoon, E. J. et al. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3')-VI. MBio 5, e01972–01914, 10.1128/mBio.01972-14 (2014).
Zong, Z. & Zhang, X. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 68, 1007–1010 (2013).
Zhang, W. J. et al. Complete sequence of the blaNDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J. Antimicrob. Chemother. 68, 1681–1682 (2013).
Yang, J. et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–513 (2012).
Bogaerts, P., Huang, T. D., Rezende de Castro, R., Bouchahrouf, W. & Glupczynski, Y. Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J. Antimicrob. Chemother. 68, 2414–2415 (2013).
Chen, T. L. et al. Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in acinetobacter genomic species 13TU. Antimicrob. Agents Chemother. 54, 3107–3112 (2010).
Poirel, L., Mansour, W., Bouallegue, O. & Nordmann, P. Carbapenem-resistant Acinetobacter baumannii isolates from Tunisia producing the OXA-58-like carbapenem-hydrolyzing oxacillinase OXA-97. Antimicrob. Agents Chemother. 52, 1613–1617 (2008).
Marti, S. et al. Characterization of the carbapenem-hydrolyzing oxacillinase OXA-58 in an Acinetobacter genospecies 3 clinical isolate. Antimicrob. Agents Chemother. 52, 2955–2958 (2008).
Chen, T. L., Wu, R. C., Shaio, M. F., Fung, C. P. & Cho, W. L. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 2573–2580 (2008).
Poirel, L. & Nordmann, P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 1442–1448 (2006).
Fu, Y. et al. Characterization of a novel plasmid type and various genetic contexts of blaOXA-58 in Acinetobacter spp. from multiple cities in China. PLoS One. 9, e84680, 10.1371/journal.pone.0084680 (2014).
Evans, B. A., Hamouda, A., Towner, K. J. & Amyes, S. G. Novel genetic context of multiple blaOXA-58 genes in Acinetobacter genospecies 3. J. Antimicrob. Chemother. 65, 1586–1588 (2010).
Ji, S. et al. Prevalence of carbapenem-hydrolyzing class D beta-lactamase genes in Acinetobacter spp. isolates in China. Eur. J. Clin. Microbiol. Infect. Dis. 33, 989–997 (2013).
Chen, T. L. et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13, 801–806 (2007).
Chang, H. C. et al. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Antimicrob. Chemother. 59, 321–322 (2007).
Ellington, M. J., Kistler, J., Livermore, D. M. & Woodford, N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59, 321–322 (2007).
Kovach, M. E. et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995).
Haase, J., Kalkum, M. & Lanka, E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncP alpha plasmid RP4. J. Bacteriol. 178, 6720–6729 (1996)
Chansiripornchai, N., Ramasoota, P., Bangtrakulnonth, A., Sasipreeyajan, J. & Svenson, S. B. Application of randomly amplified polymorphic DNA (RAPD) analysis for typing avian Salmonella enterica subsp .enterica. FEMS Immunol. Med. Microbiol. 29, 221–225 (2000).
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI document M100-S22, Vol. 32 (CLSI, 2012).
Andrews, J. M. & Howe, R. A. BSAC standardized disc susceptibility testing method (version 10). J. Antimicrob. Chemother. 66, 2726–2757 (2011).
Acknowledgements
This work was supported by Guangdong Innovative Research Team Program grant 2009010058 and Meizhou Science and Technology Program grant number 2013B121 to X.M. We thank Dr. Guangxia Gao for critical reading of the manuscript. The English writing of this manuscript was polished by EnPapers.
Author information
Authors and Affiliations
Contributions
X.G., D.Z. and X.C. designed the experiments. S.Z., X.C., X.M., G.Z. and J.W. performed the experiments. X.G., X.C. and D.Z. analyzed the data. X.G., X.M., G.Z. and D.Z. contributed reagents and materials. X.G., D.Z. and X.C. wrote this manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary data
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhou, S., Chen, X., Meng, X. et al. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep 5, 8976 (2015). https://doi.org/10.1038/srep08976
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08976
This article is cited by
-
Co-Existence of Carbapenemase-Encoding Genes in Acinetobacter baumannii from Cancer Patients
Infectious Diseases and Therapy (2021)
-
Co-occurrence of carbapenemase encoding genes in Acinetobacter baumannii, a dream or reality?
BMC Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.