Abstract
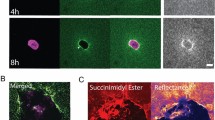
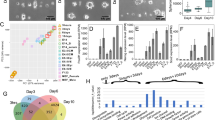
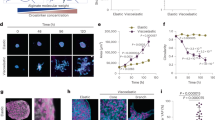
The isotropic or anisotropic organization of biological extracellular matrices has important consequences for tissue function. We study emergent anisotropy using fibroblasts that generate varying degrees of matrix alignment from uniform starting conditions. This reveals that the early migratory paths of fibroblasts are correlated with subsequent matrix organization. Combined experimentation and adaptation of Vicsek modelling demonstrates that the reorientation of cells relative to each other following collision plays a role in generating matrix anisotropy. We term this behaviour ‘cell collision guidance’. The transcription factor TFAP2C regulates cell collision guidance in part by controlling the expression of RND3. RND3 localizes to cell–cell collision zones where it downregulates actomyosin activity. Cell collision guidance fails without this mechanism in place, leading to isotropic matrix generation. The cross-referencing of alignment and TFAP2C gene expression signatures against existing datasets enables the identification and validation of several classes of pharmacological agents that disrupt matrix anisotropy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Primary accession data files are deposited at the NCBI Gene Expression Omnibus under GSE121536. The data that support the findings of this study are available from the corresponding author on reasonable request.
Code availability
The code for the computational model continues to be developed and is available via the following link: https://github.com/wershofe/FibroblastMatrixModel. The version pertaining to this manuscript is called cellCellModel.
References
Mouw, J. K., Ou, G. & Weaver, V. M. Extracellular matrix assembly: a multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15, 771–785 (2014).
Conklin, M. W. et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011).
Conklin, M. W. et al. Collagen alignment as a predictor of recurrence after ductal carcinoma in situ. Cancer Epidem. Biomar. 27, 138–145 (2018).
Provenzano, P. P. et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 (2006).
Drifka, C. R. et al. Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget 7, 76197–76213 (2016).
Goetz, J. G. et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell 146, 148–163 (2011).
Acerbi, I. et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 7, 1120–1134 (2015).
Riching, K. M. et al. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 107, 2546–2558 (2014).
Han, W. et al. Oriented collagen fibers direct tumor cell intravasation. Proc. Natl Acad. Sci. USA 113, 11208–11213 (2016).
Provenzano, P. P. et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6, 11 (2008).
Abercrombie, M. & Heaysman, J. E. Observations on the social behaviour of cells in tissue culture: I. speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp. Cell Res. 5, 111–131 (1953).
Stramer, B. & Mayor, R. Mechanisms and in vivo functions of contact inhibition of locomotion. Nat. Rev. Mol. Cell Biol. 18, 43–55 (2016).
Davis, J. R. et al. Inter-cellular forces orchestrate contact inhibition of locomotion. Cell 161, 361–373 (2015).
Carmona-Fontaine, C. et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957–961 (2008).
Theveneau, E. et al. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 15, 763–772 (2013).
Xu, Q., Mellitzer, G., Robinson, V. & Wilkinson, D. G. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature 399, 267–271 (1999).
Elsdale, T. & Bard, J. Cellular interactions in mass cultures of human diploid fibroblasts. Nature 236, 152–155 (1972).
Elsdale, T. R. Parallel orientation of fibroblasts in vitro. Exp. Cell Res. 51, 439–450 (1968).
Bertin, E., Baskaran, A., Chate, H. & Marchetti, M. C. Comparison between Smoluchowski and Boltzmann approaches for self-propelled rods. Phys. Rev. E 92, 042141 (2015).
Wegner, S. et al. Effects of grain shape on packing and dilatancy of sheared granular materials. Soft Matter 10, 5157–5167 (2014).
Prost, J. The Physics of Liquid Crystals (Oxford Univ. Press, 1995).
Ginelli, F., Peruani, F., Bar, M. & Chate, H. Large-scale collective properties of self-propelled rods. Phys. Rev. Lett. 104, 184502 (2010).
Peruani, F., Deutsch, A. & Bar, M. Nonequilibrium clustering of self-propelled rods. Phys. Rev. E 74, 030904 (2006).
Grossmann, R., Peruani, F. & Bar, M. Mesoscale pattern formation of self-propelled rods with velocity reversal. Phys. Rev. E 94, 050602 (2016).
Grossman, D., Aranson, I. S. & Ben Jacob, E. Emergence of agent swarm migration and vortex formation through inelastic collisions. New J. Phys. 10, 023036 (2008).
Li, X. et al. On the mechanism of long-range orientational order of fibroblasts. Proc. Natl Acad. Sci. USA 114, 8974–8979 (2017).
Duclos, G., Garcia, S., Yevick, H. G. & Silberzan, P. Perfect nematic order in confined monolayers of spindle-shaped cells. Soft Matter 10, 2346–2353 (2014).
Drifka, C. R. et al. Human pancreatic stellate cells modulate 3D collagen alignment to promote the migration of pancreatic ductal adenocarcinoma cells. Biomed. Microdev. 18, 105 (2016).
Pankov, R. et al. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170, 793–802 (2005).
Vicsek, T., Czirok, A., Ben-Jacob, E., Cohen, I. I. & Shochet, O. Novel type of phase transition in a system of self-driven particles. Phys. Rev. Lett. 75, 1226–1229 (1995).
Ohlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Costa, A. et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 33, 463–479 (2018). e410.
Su, S. et al. CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 172, 841–856 (2018). e816.
Provenzano, P. P., Inman, D. R., Eliceiri, K. W., Trier, S. M. & Keely, P. J. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 95, 5374–5384 (2008).
Teixeira, A. I., Abrams, G. A., Bertics, P. J., Murphy, C. J. & Nealey, P. F. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 116, 1881–1892 (2003).
Attieh, Y. et al. Cancer-associated fibroblasts lead tumor invasion through integrin-beta3-dependent fibronectin assembly. J. Cell Biol. 216, 3509–3520 (2017).
Riento, K., Guasch, R. M., Garg, R., Jin, B. & Ridley, A. J. RhoE binds to ROCK I and inhibits downstream signaling. Mol. Cell Biol. 23, 4219–4229 (2003).
Hodge, R. G. & Ridley, A. J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17, 496–510 (2016).
Butler, M. T. & Wallingford, J. B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375–388 (2017).
Yang, N. & Friedl, A. Syndecan-1-induced ECM fiber alignment requires integrin alphavbeta3 and Syndecan-1 ectodomain and heparan sulfate chains. PLoS ONE 11, e0150132 (2016).
Peruani, F. et al. Collective motion and nonequilibrium cluster formation in colonies of gliding bacteria. Phys. Rev. Lett. 108, 098102 (2012).
Borzsonyi, T. et al. Orientational order and alignment of elongated particles induced by shear. Phys. Rev. Lett. 108, 228302 (2012).
Levay, S. et al. Frustrated packing in a granular system under geometrical confinement. Soft Matter 14, 396–404 (2018).
Yang, N., Mosher, R., Seo, S., Beebe, D. & Friedl, A. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 178, 325–335 (2011).
Gilkes, D. M., Bajpai, S., Chaturvedi, P., Wirtz, D. & Semenza, G. L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 288, 10819–10829 (2013).
Hidalgo-Carcedo, C. et al. Collective cell migration requires suppression of actomyosin at cell–cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat. Cell Biol. 13, 49–58 (2011).
Fortier, M. et al. RhoE controls myoblast alignment prior fusion through RhoA and ROCK. Cell Death Differ. 15, 1221–1231 (2008).
Roycroft, A. & Mayor, R. Molecular basis of contact inhibition of locomotion. Cell Mol. Life Sci. 73, 1119–1130 (2016).
Mayor, R. & Carmona-Fontaine, C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 20, 319–328 (2010).
Wartiovaara, J., Leivo, I. & Vaheri, A. Expression of the cell surface-associated glycoprotein, fibronectin, in the early mouse embryo. Dev. Biol. 69, 247–257 (1979).
Shi, Q. et al. Rapid disorganization of mechanically interacting systems of mammary acini. Proc. Natl Acad. Sci. USA 111, 658–663 (2014).
Ahmadzadeh, H. et al. Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proc. Natl Acad. Sci. USA 114, E1617–E1626 (2017).
Checa, S., Rausch, M. K., Petersen, A., Kuhl, E. & Duda, G. N. The emergence of extracellular matrix mechanics and cell traction forces as important regulators of cellular self-organization. Biomech. Model. Mechan. 14, 1–13 (2015).
Sawhney, R. K. & Howard, J. Slow local movements of collagen fibers by fibroblasts drive the rapid global self-organization of collagen gels. J. Cell Biol. 157, 1083–1091 (2002).
Wang, H., Abhilash, A. S., Chen, C. S., Wells, R. G. & Shenoy, V. B. Long-range force transmission in fibrous matrices enabled by tension-driven alignment of fibers. Biophys. J. 107, 2592–2603 (2014).
Calvo, F. et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 (2013).
Hirata, E. et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer Cell 27, 574–588 (2015).
Lo, C. M., Wang, H. B., Dembo, M. & Wang, Y. L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144–152 (2000).
Lachowski, D. et al. Substrate rigidity controls activation and durotaxis in pancreatic stellate cells. Sci. Rep. 7, 2506 (2017).
Wong, S., Guo, W. H. & Wang, Y. L. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc. Natl Acad. Sci. USA 111, 17176–17181 (2014).
Doyle, A. D., Wang, F. W., Matsumoto, K. & Yamada, K. M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 184, 481–490 (2009).
Wu, C. et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987 (2012).
Gaggioli, C. et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9, 1392–1400 (2007).
Franco-Barraza, J., Beacham, D. A., Amatangelo, M. D. & Cukierman, E. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr. Protoc. Cell Biol. 71, 10.9.1–10.9.34 (2016).
Tambe, D. T. et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469–475 (2011).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We are indebted to Bioinformatics and Biostatistics, Light Microscopy (in particular D. Barry), Advanced Sequencing Facilities, Cell Services and the Biological Research Facility at the Francis Crick Institute for scientific and technical support throughout the project. We thank C. Mein (Barts and the London School of Medicine and Dentistry) for support and advice with RNA sequencing. E.S., D.P., P.A.B. and E.W. were funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001144, FC001003), the UK Medical Research Council (FC001003, FC001144) and the Wellcome Trust (FC001003, FC001144). E.S. and D.P. also received funding from Breast Cancer Now (2013NovPR182). X.T. received funding from by the Spanish Ministry of Science and Innovation (Severo Ochoa Award), the Generalitat de Catalunya (Cerca Program), the European Research Council (CoG-616480), the European Commission (FET Proactive 731957) and Obra Social ‘La Caixa’. A.L. received financial support through the Junior Leader Postdoctoral Fellowship Programme from ‘La Caixa’ Banking Foundation.
Author information
Authors and Affiliations
Contributions
D.P., E.W. and E.S. conceived and developed the project. D.P. performed the experiments and analysed the data. E.W. and P.A.B. developed the computational model and analysed the data. S.B. performed the bioinformatic analysis of RNASeq and LINCS data. A.L. performed the traction force microscopy and analysis in Figs. 1f and 4e. R.P.J. wrote the code for analysis of fibre alignment. S.G. performed the tracking of some cell collision data. D.P. and E.S. wrote the manuscript with assistance from E.W., A.L., R.P.J., X.T. and P.A.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1 and 2.
Supplementary Video 1
Alignment emerges prior to confluence and early migration paths predict final organization.
Supplementary Video 2
Actomyosin is supressed at cell contacts during collision guidance.
Supplementary Video 3
Actomyosin is concentrated at cell contacts during CIL.
Supplementary Video 4
Myosin is supressed at cell contacts during nematic gliding.
Rights and permissions
About this article
Cite this article
Park, D., Wershof, E., Boeing, S. et al. Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions. Nat. Mater. 19, 227–238 (2020). https://doi.org/10.1038/s41563-019-0504-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-019-0504-3
This article is cited by
-
How multiscale curvature couples forces to cellular functions
Nature Reviews Physics (2024)
-
Some Effects of Fiber Dispersion on the Mechanical Response of Incompressible Soft Solids
Journal of Elasticity (2022)
-
Silk Fibroin Nacre
Advanced Fiber Materials (2022)
-
The matrix in cancer
Nature Reviews Cancer (2021)
-
Dual anisotropicity comprising 3D printed structures and magnetic nanoparticle assemblies: towards the promotion of mesenchymal stem cell osteogenic differentiation
NPG Asia Materials (2021)