Abstract
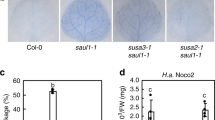
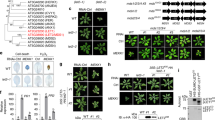
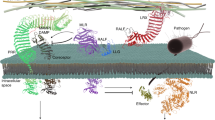
The innate immune system detects pathogen-derived molecules via specialized immune receptors to prevent infections1,2,3. Plant immune receptors include cell surface-resident pattern recognition receptors (PRRs, including receptor-like kinases (RLKs)), and intracellular nucleotide-binding domain leucine-rich repeat proteins (NLRs). It remains enigmatic how RLK- and NLR-mediated signalling are connected. Disruption of an immune-activated MEKK1–MKK1/2–MPK4 MAPK cascade activates the NLR SUMM2 via the MAPK kinase kinase MEKK2, leading to autoimmunity4,5,6,7,8,9. To gain insights into the mechanisms underlying SUMM2 activation, we used an RNA interference-based genetic screen for mekk1 autoimmune suppressors and identified an uncharacterized malectin-like RLK, named LETUM1 (LET1), as a specific regulator of mekk1–mkk1/2–mpk4 autoimmunity via complexing with both SUMM2 and MEKK2. MEKK2 scaffolds LET1 and SUMM2 for protein stability and association, and counter-regulates the F-box protein CPR1-mediated SUMM2 ubiquitination and degradation, thereby regulating SUMM2 accumulation and activation. Our study indicates that malectin-like RLK LET1 senses the perturbance of cellular homoeostasis caused by the deficiency in immune-activated signalling and activates the NLR SUMM2-mediated autoimmunity via MEKK2 scaffolding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Original data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Spoel, S. H. & Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100 (2012).
Zhang, Z. et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263 (2012).
Gao, M. et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198 (2008).
Ichimura, K., Casais, C., Peck, S. C., Shinozaki, K. & Shirasu, K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281, 36969–36976 (2006).
Suarez-Rodriguez, M. C. et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669 (2007).
Nakagami, H., Soukupova, H., Schikora, A., Zarsky, V. & Hirt, H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281, 38697–38704 (2006).
Petersen, M. et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120 (2000).
Couto, D. & Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016).
Yu, X., Feng, B., He, P. & Shan, L. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137 (2017).
Gust, A. A., Pruitt, R. & Nurnberger, T. Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791 (2017).
Cui, H., Tsuda, K. & Parker, J. E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015).
Elmore, J. M., Lin, Z. J. & Coaker, G. Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 14, 365–371 (2011).
DeYoung, B. J. & Innes, R. W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249 (2006).
Rodriguez, M. C., Petersen, M. & Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 (2010).
Meng, X. & Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266 (2013).
Tena, G., Boudsocq, M. & Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529 (2011).
Sun, T. et al. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19, e45324 (2018).
Bi, G. et al. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30, 1543–1561 (2018).
Asai, T. et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 (2002).
de Oliveira, M. V. V. et al. Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nat. Plants 2, 15218 (2016).
Yu, X. et al. The receptor kinases BAK1/SERK4 regulate Ca2+ channel-mediated cellular homeostasis for cell death containment. Curr. Biol. 29, 3778–3790 (2019).
Yang, Y. et al. RNA interference-based screen reveals concerted functions of MEKK2 and CRCK3 in plant cell death regulation. Plant Physiol. 183, 331–344 (2020).
Kong, Q. et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236 (2012).
Su, S. H. et al. Deletion of a tandem gene family in Arabidopsis: increased MEKK2 abundance triggers autoimmunity when the MEKK1-MKK1/2-MPK4 signaling cascade is disrupted. Plant Cell 25, 1895–1910 (2013).
Zhang, Z. et al. The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep. 18, 292–302 (2017).
Nissen, K. S., Willats, W. G. & Malinovsky, F. G. Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci. 21, 516–527 (2016).
Li, C., Wu, H. M. & Cheung, A. Y. FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379–2392 (2016).
Lindner, H., Muller, L. M., Boisson-Dernier, A. & Grossniklaus, U. CrRLK1L receptor-like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15, 659–669 (2012).
Franck, C. M., Westermann, J. & Boisson-Dernier, A. Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 69, 301–328 (2018).
Huck, N., Moore, J. M., Federer, M. & Grossniklaus, U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159 (2003).
Boisson-Dernier, A. et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288 (2009).
Miyazaki, S. et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19, 1327–1331 (2009).
Guo, H. et al. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 106, 7648–7653 (2009).
Ge, Z. et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600 (2017).
Stegmann, M. et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289 (2017).
Mang, H. et al. Differential regulation of two-tiered plant immunity and sexual reproduction by ANXUR receptor-like kinases. Plant Cell 29, 3140–3156 (2017).
Guo, H. et al. FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 28, 3316–3324 (2018).
Kessler, S. A. et al. Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971 (2010).
Nitta, Y. et al. MEKK2 inhibits activation of MAP kinases in Arabidopsis. Plant J. 103, 705–714 (2020).
Cheng, Y. T. et al. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad. Sci. USA 108, 14694–14699 (2011).
Gou, M. et al. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69, 411–420 (2012).
Lu, D. et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442 (2011).
Zhang, J. et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185 (2007).
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B. & Sussman, M. R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411 (2014).
Xiao, Y. et al. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274 (2019).
Li, F. et al. Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16, 748–758 (2014).
Bajar, B. T., Wang, E. S., Zhang, S., Lin, M. Z. & Chu, J. A guide to fluorescent protein FRET pairs. Sensors 16, 1488 (2016).
He, P., Shan, L. & Sheen, J. in Plant–Pathogen Interactions (ed. Ronald, P.C.) 1–9 (Springer, 2007).
Shu, C. et al. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21, 1137–1148 (2013).
Bucherl, C., Aker, J., de Vries, S. & Borst, J. W. Probing protein–protein Interactions with FRET–FLIM. Methods Mol. Biol. 655, 389–399 (2010).
Halter, T. et al. The leucine-rich repeat receptor kinase BIR2 Is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143 (2014).
Acknowledgements
We thank the ABRC for Arabidopsis T-DNA insertion library and various mutant seeds. We thank P. Krysan (University of Wisconsin, United States), Y. Zhang (University of British Columbia, Canada) and J. Hua (Cornell University, United States) for Arabidopsis seeds. We thank C. Franck and C. Zipfel for the critical reading of the manuscript and members of the laboratories of L.S. and P.H. for discussions and comments on the experiments. The work was supported by National Institutes of Health (NIH) grant no. R01GM092893 and National Science Foundation (NSF) grant no. MCB-1906060 to P.H. and NIH grant no. R01GM097247 and the Robert A. Welch Foundation grant no. A-1795 to L.S. Y.H. and D.G. were partially supported by China Scholarship Council (CSC) and G.C.M was partially supported by INCT/CNPq Fellowship, Brazil.
Author information
Authors and Affiliations
Contributions
Y.H., J.L., L.S. and P.H. conceived the project, designed experiments and analysed data. J.L., Y.H., L.K., X.Y., B.F., D.L., B.Z., G.C.M., P.Y. and D.G. performed experiments and analysed data. W.M.W, E.P.B.F. and P.L. analysed data and provided critical feedback. L.S. and P.H. wrote the manuscript with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Table 1.
Supplementary Data 1
Statistical source data.
Supplementary Data 2
Statistical source data.
Supplementary Data 3
Statistical source data.
Supplementary Data 4
Unprocessed western blots.
Supplementary Data 5
Statistical source data.
Supplementary Data 6
Unprocessed western blots.
Supplementary Data 7
Unprocessed western blots.
Supplementary Data 8
Unprocessed western blots.
Supplementary Data 9
Unprocessed western blots.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Liu, J., Huang, Y., Kong, L. et al. The malectin-like receptor-like kinase LETUM1 modulates NLR protein SUMM2 activation via MEKK2 scaffolding. Nat. Plants 6, 1106–1115 (2020). https://doi.org/10.1038/s41477-020-0748-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-020-0748-6
This article is cited by
-
MAPK Cascades in Plant Microbiota Structure and Functioning
Journal of Microbiology (2024)
-
Phytocytokine signalling reopens stomata in plant immunity and water loss
Nature (2022)
-
Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in Nicotiana benthamiana
BMC Plant Biology (2021)
-
The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes
Nature Communications (2021)
-
Malectin-like receptor kinases as protector deities in plant immunity
Nature Plants (2021)