Abstract
Glioblastoma multiforme (GBM) is the most malignant and lethal primary brain tumor in adults accounting for about 50% of all gliomas. The only treatment available for GBM is the drug temozolomide, which unfortunately has frequent drug resistance issue. By analyzing the hub genes of GBM via weighted gene co-expression network analysis (WGCNA) of the cancer genome atlas (TCGA) dataset, and using the connectivity map (CMAP) platform for drug repurposing, we found that multiple azole compounds had potential anti-GBM activity. When their anti-GBM activity was examined, however, only three benzimidazole compounds, i.e. flubendazole, mebendazole and fenbendazole, potently and dose-dependently inhibited proliferation of U87 and U251 cells with IC50 values below 0.26 μM. Benzimidazoles (0.125−0.5 μM) dose-dependently suppressed DNA synthesis, cell migration and invasion, and regulated the expression of key epithelial-mesenchymal transition (EMT) markers in U87 and U251 cells. Benzimidazoles treatment also dose-dependently induced the GBM cell cycle arrest at the G2/M phase via the P53/P21/cyclin B1 pathway. Furthermore, the drugs triggered pyroptosis of GBM cells through the NF-κB/NLRP3/GSDMD pathway, and might also concurrently induced mitochondria-dependent apoptosis. In a nude mouse U87 cell xenograft model, administration of flubendazole (12.5, 25, and 50 mg · kg−1 · d−1, i.p, for 3 weeks) dose-dependently suppressed the tumor growth without obvious adverse effects. Taken together, our results demonstrated that benzimidazoles might be promising candidates for the treatment of GBM.
Similar content being viewed by others
Introduction
Glioblastoma multiforme (GBM) is the most aggressive, frequent and lethal subtype of malignant gliomas originating in the central nervous system [1]. The current therapy for GBM patients is surgical resection followed by adjuvant radiation therapy and chemotherapy with oral temozolomide (TMZ) [2]. Compared with radiotherapy alone, TMZ administration with radiotherapy improved the median survival time from 12.1 months to 14.6 months, and increased the two-year survival rate from 10.4% to 26.1% [3]. However, TMZ still has many side effects and tumor cells can become resistant to the drug [4]; therefore, there is an urgent clinical need for the development of new drugs as GBM therapeutics.
Given the high attrition rates, substantial costs, and slow pace of new drug discovery and development, repurposing of approved drugs is increasingly becoming an attractive proposition [5]. Drug repurposing is an economical and efficient drug development method to identify novel indication of approved or investigational drugs [6]. Various data-driven and experimental approaches have been suggested for the identification of repurposable drug candidates. The Connectivity Map (CMAP) is a platform that connects small molecules, genes, and diseases using gene-expression signatures, which has been widely used for investigation of network pharmacology and drug repurposing [7].
Weighted gene co-expression network analysis (WGCNA) is a method for the analysis and comparison of the gene expression patterns of multiple samples [8]. It can be used for identifying clusters of highly correlated genes, for summarizing such clusters using the module eigengene or an intramodular hub gene, for relating modules to one another and to external sample traits, and for determining module membership measures [9]. In our study, by identifying the GBM hub genes from the cancer genome atlas (TCGA) dataset via WGCNA and using the CMAP platform for drug repurposing, it was predicted that multiple azole compounds had potential anti-GBM activity.
Azole compounds are well known as antibacterial and antifungal agents [10] and recently several azoles were found to modulate tubulin with antitumor potential [11]. However, it is not known whether azoles have anti-GBM activity and the underlying mechanisms of action have not been systematically studied. Representative azole compounds were selected from benzimidazoles, triazoles, and imidazoles for the anti-GBM activity test. Interestingly, only benzimidazole compounds were found to inhibit proliferation of GBM cell, and three such compounds, flubendazole, fenbendazole, and mebendazole, were chosen for mechanistic analysis.
Drug repurposing was conducted using the CMAP platform with analysis of GBM hub genes by WGCNA and we found that azole compounds had anti-GBM potential. Further experiments confirmed that only benzimidazole-like azoles had anti-GBM activity, while triazoles and imidazoles were inactive. The benzimidazole compounds inhibited proliferation of GBM cell in a time- and dose-dependent manner. RNA-sequencing analyses revealed that benzimidazole might regulate cell migration, cell cycle, programmed cell death, and other biological processes of GBM cells. Benzimidazoles inhibited the migration and invasion of GBM cells, regulated the expression of key EMT markers, and dose-dependently arrested the cell cycle at the G2/M phase through the P53/P21/Cyclin B1 pathway. Moreover, these azole compounds were able to trigger pyroptosis of GBM cells via the NF-κB/NLRP3/GSDMD pathway, and at the same time, they might induce mitochondrial-dependent apoptosis. In addition, xenograft tumor transplantation experiments were conducted in nude mice to determine if flubendazole significantly suppressed tumorigenicity with no obvious adverse effects in vivo. Our results demonstrated that benzimidazoles might be promising candidates for the treatment of GBM.
Materials and methods
Data collection and preprocessing
RNA-Seq data and clinical information on GBM patients were downloaded from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). After removing repetitive samples and those without clinical information, 169 GBM tumors and 5 normal brain samples were used for WGCNA. The read counts were used to represent the gene expression level [12]. Data processing was performed using the R Studio software (Version 3.6.3, https://www.rstudio.com/) according to the policies of TCGA for data access to protect patient privacy.
Identification of differentially expressed genes (DEGs)
The R package, edgeR, was used to identify the DEGs between tumor and normal samples [13]. The empirical probability distribution of the fold-changes correlated with significant DEGs was used to define a |log2 Fold Change| with a threshold >1.5 and false discovery rate (FDR) < 0.05 [14].
Construction of gene co-expression network and module detection with WGCNA
The gene co-expression networks based on DEGs were established using the WGCNA package [8]. After sorting the TCGA expression matrix data, an appropriate soft-threshold power β was selected with the “pick soft threshold” function in the WGCNA, and a scale-free topology plot was generated based on the appropriate soft-threshold power. Lastly, a cluster dendrogram plot and clustering tree of co-expressed gene modules of DEGs were created with the following major parameters: cutHeight = 0.6, minSize = 10. The module eigengene (ME) was obtained using WGCNA to indicate the expression profiles of module genes.
Relationship between modules and clinical traits
WGCNA can be used to determine the relevancy between modules and clinical parameters. Since the ME can summarize the module gene expression, we calculated the correlation between MEs and the input clinical parameters. This process is called module clinical trait relationship (MCTR) analysis. To investigate the MCTR of gene modules, we firstly sorted the corresponding module genes according to the modules constructed by WGCNA, then, the ME for each module was calculated and correlated with clinical parameters, with significance at P < 0.05.
Construction of module genes network and hub genes identification
The edge file, which included the source node and target node genes, was generated by WGCNA for protein–protein interaction (PPI) analysis in Cytoscape (Version 3.8.2, https://cytoscape.org/). To identify high-level genes that play a key role in the PPI network, the Cytohubba package based on Cytoscape was used to perform the hub gene analysis, and the top 50 genes in all modules were considered as hub genes. The PPI network was further analyzed in the STRING database (https://www.string-db.org/).
Anti-GBM drug repositioning using the CMAP platform
By identifying the DEGs in GBM and performing WGCNA on node genes, we were able to upload the upregulated and down-regulated differential node genes of GBM to the CMAP database (https://portals.broadinstitute.org/cmap/). This allowed the identification of potential therapeutic drugs, which can reverse the genes of key GBM nodes.
The tested azole compounds and pan-caspase inhibitor
The following azole compounds used in the anti-GBM activity test were purchased from Topscience (Shanghai, China): econazole, T0444, 27220-47-9; miconazole, T0344, 22916-47-8; ketoconazole, T0679, 65277-42-1; fluconazole, T1388, 86386-73-4; voriconazole, T0120, 137234-62-9; posaconazole, T6211, 171228-49-2; flubendazole, T0009, 31430-15-6; mebendazole, T1155, 31431-39-7; and fenbendazole, T1141, 43210-67-9. The pan-caspase inhibitor, Z-VAD-FMK, was purchased from MedChemExpress (NJ, USA), HY-16658B, 161401-82-7.
Cell culture
Human GBM cell lines, U87-MG and U251-MG were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, CA, USA) with 10% fetal bovine serum (FBS, Gibco, New Zealand). U87-MG and U251-MG cells were cultured in a 5% CO2 incubator at 37 °C.
Cell proliferation assay
A CCK-8 kit (C0038, Beyotime, Shanghai, China) was used to measure cell proliferation according to manufacturer’s instructions. Cells were cultured in 96-well plates (3 × 103 cells/well) at 37 °C with 0–10 μmol/L azole compounds for 12, 24 or 48 h, followed by 10 μL of CCK-8 reagent and incubated for 1 h. A450 was measured and IC50 values were calculated using GraphPad Prism 7 software (GraphPad Software Inc., San Diego, CA, USA).
U87 and U251 cells were pretreated with 50 μmol/L Z-VAD-FMK for 30 min to block caspase activity. Then U87 and U251 cells were treated with 0, 0.125, 0.25, and 0.5 μmol/L of flubendazole for 24 h, followed by 10 μL of CCK-8 reagent and incubated for 1 h. Then we followed the above method to measure cell proliferation.
DNA synthesis assay
The EdU Apollo 567 in vitro imaging kit (RiboBio, Guangzhou, China) was used in accordance with the manufacture’s protocol. U87 and U251 cells were treated with 0, 0.125, 0.25, and 0.5 μmol/L of benzimidazoles for 24 h, then EdU was added to each well and cells were incubated for 2 h. Cells were fixed with 4% paraformaldehyde for 30 min and permeablized with 0.5% Triton-X 100 for 10 min. Cells were stained with Apollo 567 and Hoechst 33342 for 30 min and scanned with a high content imaging system (Cellomics ArrayScan VTI, Thermo Fisher Scientific, Carlsbad, CA, USA).
Lactate dehydrogenase (LDH) assay
The level of lactate dehydrogenase released from cells was measured using a cytotoxicity LDH assay kit (Dojindo, Japan). U87 and U251 cells were seeded into 96 well plates at 1 × 105/well and treated with 0, 0.125, 0.25, and 0.5 μmol/L of benzimidazoles for 24 and 48 h. After treatment with azole, the medium was removed and replaced with 100 μL of tetrazolium working solution, the cells were incubated at room temperature for 30 min, then the reaction was stopped by addition of 50 μL stop solution to each well and the absorbance was measured at 490 nm.
Migration and invasion assays
The effect of benzimidazoles on migration and invasion of U87 and U251 cells was checked using a transwell (#3422, Corning Costar, Carlsbad, CA, USA) assay. Cells were detached, centrifuged and resuspended in DMEM at 5 × 104 cells/mL and 200 μL of cell suspension was used for the migration and invasion assays. For the invasion assay, the bottom of the transwell insert was coated with Matrigel (1:7 dilution, Corning Biocoat, USA) and the cells were then seeded into the upper chamber. The subsequent steps were the same as the migration assay. After incubation for 24 h, the cells that passed through the transwell membrane were fixed with 4% paraformaldehyde for 15 min, and then stained with 1% crystal violet for 20 min. Cells were imaged under a microscope (Nikon, Japan) and representative fields were counted.
Mitochondrial membrane potential assay
U251 and U87 cells cultured in 60-mm dishes were treated with 0, 0.125, 0.25, and 0.5 μmol/L of benzimidazoles for 24 h. Cells were harvested, washed, and stained with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1) at 37 °C for 30 min. Mitochondrial membrane potential of the cells was measured by flow cytometry (BD Accuri C6, San Diego, CA, USA).
PI/Annexin V FITC assay
The rate of apoptosis and pyroptosis in U87 and U251 was determined using a Propidium iodide (PI)/annexin V-FITC kit (TransGen Biotech, Beijing, China). Cells cultured in 60-mm dishes were treated with 0, 0.125, 0.25 and 0.5 μmol/L of benzimidazoles for 24 h. GBM cells were pretreated with the caspase inhibitor, Z-VAD-FMK (50 μmol/L), for 30 min and exposed to 0.25 μmol/L flubendazole for 24 h. Then, cells were harvested, washed with phosphate-buffered saline (PBS) and stained with PI and annexin V-FITC for 15 min.
Cell cycle assay
Cells cultured in 60-mm dishes were treated with 0, 0.125, 0.25, and 0.5 μmol/L of benzimidazoles for 24 h, after which the cells were harvested, washed with PBS and fixed in 70% ethanol overnight at 4 °C. After centrifugation, the supernatants were discarded and cells were resuspended and incubated with PI at 37 °C for 30 min. Cell cycle was determined by flow cytometry (BD Accuri C6, San Diego, CA, USA). Data were analyzed by FlowJo V10 software (Tristar, CA, USA).
RNA sequencing and differential gene enrichment
U251 and U87 cells incubated with 0.25 μmol/L flubendazole for 24 h were lysed in TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and prepared for next generation sequencing by Novogene Corporation using an Illumina DNA sequencer (Beijing, China). Total RNA was qualified by NanoDrop spectrophotometer and Agilent 2100 bioanalyzer to ensure purity and integrity. NEB libraries were built and qualified, then pooled and sequenced using an Illumina sequencer. The annotation, GO and KEGG pathway analyses were performed using the database for annotation, visualization and integrated discovery (DAVID, Version 6.8, https://david.ncifcrf.gov/). Node genes were determined by maximal clique centrality (MCC) using CytoHubba in Cytoscape 3.8.2.
Western blotting
Cells were lysed in RIPA buffer (Beyotime, Shanghai, China) with protease inhibitor cocktail (CWBIO, Beijing, China) on ice for 30 min. Cell lysates were centrifuged at 12,000 r/min for 15 min at 4 °C and supernatants were collected. Protein concentration was determined, and equal amounts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride (PVDF) membranes, and blocked in 5% skim milk for two hours. The PVDF membranes were then incubated overnight with appropriate dilutions of primary antibodies at 4 °C. After washing five times with TBST, the blots were incubated with HRP-conjugated secondary antibodies (7074, anti-rabbit IgG; 7076, anti-mouse IgG, Cell Signaling Technology, Danvers, USA). Protein bands were visualized with SuperECL (Applygen, Beijing, China) and image density was quantitated using a Tanon 4600 imaging system (Tanon, Beijing, China).
Primary antibodies used in this study were: E-cadherin (3195), N-cadherin (13116), β-Catenin (8480), MMP2 (40994), Slug (9585), GAPDH (5174), P53 (48818), P21 (2947), cdc25C (4688), cdc2 (9116), p-cdc2 (9114), Cyclin B1 (12231), MYD88 (50010), NF-κB (8242), NLRP3 (15101), Caspase 1 (3866), GSDMD (97558), Lamin B1 (13435), β-Actin (3700), Bcl-2 (15071), Bcl-xL (2764), Bad (9268), Bim (2933), PARP (9532), Cleaved PARP (5625), Caspase 3 (14220), Cleaved-Caspase 3 (9661), Caspase 7 (12827), Cleaved-Caspase 7 (8438), Cleaved-Caspase 9 (20750) were purchased from Cell Signaling Technology (Boston, MA, USA). IL-1β (ab254360) was from Abcam (Cambridge, UK).
Xenograft model of nude mice
All animal experiments were conducted in accordance with the principles of the NIH Guide for the Care and Use of Laboratory Animals and approved protocols of Institute of Materia Medica, CAMS & PUMC. Female BALB/c-nu nude mice (16–18 g) were bought from Vital River Laboratory Animal Technology (Beijing, China) and housed at the Institute of Materia Medica animal barrier facility. U87 cells (1 × 108/mL) were subcutaneously implanted in the right flanks of the mice. After 2 weeks, when the average volume of the tumors was about 100 mm3, the mice were randomly divided into five groups (n = 6): no treatment, treated with 25 mg/kg, i.p. temozolomide (TMZ), or treated with flubendazole (FBZ) once a day for 3 weeks at 12.5, 25 and 50 mg/kg, i.p. Body weight and tumor volume were recorded every 2 days. The tumor volume V, in mm3, was calculated by the equation, V = 0.5 × a × b2 (where a = length and b = width in mm). At the end of the experiment, blood was collected from the canthus with a capillary tube and all mice were euthanized. Tests for 17 blood biochemistry parameters were performed using an automated hematology analyzer (MEK-7222K, NIHON KOHDEN, Japan). The weights of tumors and the heart, liver, spleen, kidney, lung and brain were recorded.
Statistical analysis
The results are represented as mean ± SD. Statistically significant differences between groups were determined by one-way ANOVA using GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA), and P < 0.05 was considered statistically significant.
Results
Azoles as potential anti-GBM drugs
By processing and analyzing the GBM RNA-seq data in TCGA, and filtering under conditions of log (FC) > 2, P < 0.01, 2720 down-regulated genes and 3474 upregulated genes were obtained (Fig. 1a). Differential gene enrichment analysis showed that the abnormally expressed genes of GBM were mainly enriched in IL-23-mediated signaling events, peptide ligand-binding receptors, and rhodopsin-like receptors. The cellular components of the DEGs were mainly enriched in the areas of the plasma membrane, the extracellular milieu, and exosomes (Fig. 1b). Using the systems biology method of WGCNA, the key node genes were screened by constructing a gene co-expression network. The weighted gene co-expression network was constructed using 6194 DEGs and 169 GBM samples. Nine co-expressed gene modules were identified by WGCNA, and each module was color-coded (Fig. 1c). WGCNA can also be used to determine the correlations between modules and clinical parameters. As shown in Fig. 1d, ME-yellow was negatively correlated with the age of GBM patients (r = −0.18, P = 0.03) and ME-purple was negatively correlated with the living status of GBM patients (r = −0.19, P = 0.02). The Cytoscape software was used to obtain a network in the edge and node gene list files from DEGs based on WGCNA method, and the top 50 hub genes were defined via the CytoHubba package (Fig. 1e). The protein-protein interaction network was further analyzed using the STRING database (Fig. 1f). Lastly, drug repurposing was conducted based on top 50 GBM node genes and the CMAP platform and it was determined that multiple azoles could be potential anti-GBM drugs (Table 1).
a Volcano map of differentially expressed genes from analysis of GBM transcriptome data in TCGA. b Biological pathways and cellular component enrichment of the differential genes. c Cluster module of the WGCNA of GBM transcriptome data. d Relationship between the WGCNA module and clinical characteristics of GBM patients. e Hub genes from the module genes of WGCNA identified with the CytoHubba package. f Protein–protein interactions between the hub genes
Benzimidazoles inhibit proliferation of GBM cells
Three imidazoles (econazole, miconazole, and ketoconazole), three triazoles (fluconazole, voriconazole, and posaconazole) and three benzimidazoles (flubendazole, mebendazole, fenbendazole) were selected to test for anti-GBM activity (Fig. 2a, Supplementary Fig. S1). Surprisingly, only the three benzimidazole compounds showed good activity in inhibiting the proliferation of GBM cells, while the half inhibition rate of the remaining compounds was >10 μmol/L (Supplementary Table S6). The three benzimidazoles dose-dependently changed the morphology of the U87 (Fig. 2b) and U251 cells (Fig. 2c). The cells shrank and rounded up, presenting a typical apoptotic morphology; however, in the early stages of cell death, balloon-like membrane protrusions were observed that were distinct from classic apoptotic blebbing. The CCK-8 assay showed that the benzimidazoles inhibited the proliferation of U87 (Fig. 2d) and U251 cells (Fig. 2e) in a dose- and time-dependent manner. The IC50 values of flubendazole, mebendazole, and fenbendazole for the U87 cells at 24 h were 0.1701, 0.2066, and 0.1819 μmol/L, respectively and those for U251 cells were 0.2243, 0.2532, and 0.2259 μmol/L. The EdU-DNA synthesis assay showed that benzimidazoles inhibited DNA synthesis in U87 (Fig. 2f, Supplementary Fig. 4a) and U251 cells (Fig. 2g, Supplementary Fig. 4b) in a dose-dependent manner.
a Structures of flubendazole, mebendazole and fenbendazole. Benzimidazoles’ effects on morphology of U87 cells after 24 h (b), and on morphology of U251 cells at 24 h (c). IC50 values (CCK-8 assay) of benzimidazoles on U87 cells (d), and on U251 cells (e). Benzimidazoles inhibited DNA synthesis (EdU assay) in U87 cells (f) and in U251 cells (g) at 24 h. Scale bar = 100 μm
Differential gene expression and enrichment analysis of U87 and U251 cells treated with flubendazole
The three benzimidazole compounds have similar structures, but flubendazole has a slightly stronger effect than the other two compounds; therefore, RNA sequencing was performed on U87 and U251 cells treated with 0.25 μmol/L flubendazole for 24 h. Compared with gene expression in the control cells, there were 139 upregulated genes and 235 downregulated genes found in U87 cells (Supplementary Fig. 2a). In the U251 cells, there were 196 upregulated genes and 593 downregulated genes (Supplementary Fig. 2b). Gene ontology analysis of the DEGs of U87 cells revealed that flubendazole mainly affected biological processes, such as programmed cell death, RNA polymerase II promoters and NF-κB transcription factor activity. The cellular components of DEGs of U87 cells were mostly found in the plasma membrane and extracellular region, whose molecular functions were principally enriched in calcium ion binding, signal transducer activity, and GTPase activity (Supplementary Fig. 2c). The biological processes of the DEGs in U251 cells were mainly concentrated in DNA-templated transcription, and the regulation of cell proliferation and programmed cell death. The cellular components of DEGs of U251 cells were mostly present in the extracellular region, the plasma membrane and the extracellular space, wherein the molecular functions were principally enriched in sequence-specific DNA binding, and transcription factor activity (Supplementary Fig. 2d). The common DEGs between U87 cells and U251 cells after flubendazole treatment were mainly enriched in cell proliferation, cell cycle, cell migration, NF-κB and P53 signaling pathways, which indicates potential directions for further mechanistic studies of benzimidazoles (Supplementary Fig. 2e).
Benzimidazoles inhibited migration and invasion of GBM cells
The migration and invasion ability of cancer cells in vitro is an important indicator of their degree of malignancy and serves as a target for testing anticancer drug activity. Using transwell assays, we confirmed that the benzimidazoles dose-dependently inhibited the migration and invasion of U87 and U251 cells (Fig. 3a–d) at 24 h. Results from Western blots showed that flubendazole dose-dependently reduced the expression of the EMT markers, N-cadherin, β-catenin, the Slug transcription factor and MMP2, and increased the level of E-cadherin in U87 and U251 cells (Fig. 3e, f). These results indicated that benzimidazoles could inhibit the metastatic characteristics of GBM cells by downregulating the expression of key EMT markers.
Benzimidazoles inhibited the migration of U87 cells (a) and the invasion of U87 cells (b) at 24 h. Benzimidazoles inhibited the migration of U251 cells (c) and the invasion of U251 cells (d) at 24 h. Expression of EMT-related markers after flubendazole treatment in U87 cells (e) and in U251 cells (f) at 24 h
Benzimidazoles arrested the GBM cell cycle at the G2/M phase
Flow cytometry was used to explore the effects of benzimidazoles on the cell cycle of GBM cells. The three compounds were able to arrest the cell cycle at the G2/M phase of U87 and U251 cells (Fig. 4a, b) in a dose-dependent manner, but their effects were slightly greater in U251 cells. To explore the molecular mechanism of the compounds’ G2/M phase arrest in GBM cells, samples of U87 and U251 cells treated with 0, 0.125, 0.25, and 0.5 μmol/L flubendazole for 12 and 24 h were collected and analyzed. The results showed that flubendazole upregulated the expression of tumor suppressor protein P53/P21 and inhibited the expression of cyclin B1 in both U87 and U251 cells (Fig. 4c, d) in a dose- and time-dependent manner. Flubendazole also downregulated the expression and phosphorylation of cdc2 in U251 cells, while this effect was not significant in U87 cells. Thus, benzimidazoles appear to arrest the GBM cell cycle at G2/M phase via their effects on the P53/P21/cyclin B1 pathway.
Benzimidazoles triggered pyroptosis of GBM cells
Pyroptosis is a form of programmed cell death, and the activation of pyroptosis is closely related to specific inflammatory processes [15]. The features of pyroptosis include gasdermin family-mediated pore formation in the plasma membrane, cell swelling, and plasma membrane disruption, together with release of the proinflammatory cytokines, IL-1β and IL-18 [16]. Consistent with characteristic pyroptotic cell morphology [17], balloon-like membrane protrusions were seen in U87 and U251 cells (Fig. 5a, b) after treatment of benzimidazoles, which were distinct from classic apoptotic blebbing. Moreover, LDH was released by the swollen U87 and U251 cells (Fig. 5c, d) in a time- and dose-dependent manner, indicating plasma membrane rupture and leakage. When caspase 1 was activated, it cleaved gasdermin D (GSDMD-FL) after Asp276 for mouse or Asp275 in humans [18], to generate an N-terminal cleavage product (GSDMD-N) that triggered pyroptosis by forming membrane pores and releasing inflammatory cytokines such as interleukin-1β (IL-1β) [19, 20]. Activation of inflammasomes, especially the NLR family pyrin domain-containing 3 (NLRP3) [21], plays an important role in pyroptosis. NLRP3 promotes caspase-1-mediated IL-1β and IL-18 release and cleavage of GSDMD. Nuclear factor-κB (NF-κB) mainly promotes the production of proinflammatory factors such as pro-IL-1β, pro-IL-18, NLRP3, and caspase-1 [22]. Our results showed that flubendazole promoted the expression of NLRP3 and caspase 1 in U87 and U251 cells in a dose-dependent and time-dependent manner and significantly upregulated the expression of GSDMD-N. After flubendazole treatment, the expression of Myd88, NF-κB and IL-1β, was also found to be significantly increased (Fig. 5e, f). We further checked the expression of NF-κB in the cytoplasm and nucleus after treatment of flubendazole. It was shown in Fig. 5g that flubendazole significantly promoted the nuclear translocation of NF-κB, suggesting that benzimidazoles could trigger pyroptosis of GBM cells via the NF-κB/NLRP3/GSDMD pathway.
U87 cells (a) and U251 cells (b) were treated with benzimidazoles and imaged. Arrows indicate ballooned cell membranes characteristic of pyroptotic cells (scale bar = 50 μm). (I) control, (II) flubendazole (0.5 μmol/L, 24 h), (III) mebendazole (0.5 μmol/L, 24 h), and (IV) fenbendazole (0.5 μmol/L, 24 h). Benzimidazoles induced release of LDH from U87 cells in a dose- and time-dependent manner (c) and from U251 cells (d). e, f Benzimidazoles triggered pyroptosis in GBM cells via the NF-κB/NLRP3/GSDMD pathway. The percentage of annexin V-PI double-positive cells (pyroptotic) is labeled in red. g Flubendazole significantly promoted the nuclear translocation of NF-κB. *P < 0.05, **P < 0.01, ***P < 0.001 vs control
Benzimidazoles induced mitochondria-dependent apoptosis of GBM cells
JC-1 staining detected by flow cytometry showed that benzimidazoles dose-dependently reduced the mitochondrial membrane potential (MMP, ΔΨm) in U87 and U251 cells (Fig. 6a, b). Early apoptosis is usually accompanied by a decrease in mitochondrial membrane potential [23]. The Bcl-2 family proteins have been mainly classified into the pro-apoptotic Bad and anti-apoptotic Bcl-2 proteins that control ΔΨm and the activation of the caspase cascade [24]. Results from Western blot showed that the expression of Bcl-2 and Bcl-xL was significantly decreased after flubendazole treatment, and the expression of Bad and Bim was upregulated compared with the control (Fig. 6c, d). Flubendazole time-dependently and dose-dependently reduced the expression of PARP, caspase 3 and caspase 7, and induced the expression of cleaved PARP, cleaved caspase 3 and cleaved caspase 7 (Fig. 6e, f). These data suggest that benzimidazoles, besides triggering pyroptosis in GBM cells, also lead to apoptosis.
Flow cytometry using JC-1 staining showed that benzimidazoles reduced mitochondrial membrane potential in U87 cells (a) and U251 cells (b). Effect of flubendazole on expression of Bcl-2 family proteins in U87 cells (c) and U251 cells (d). Effect of flubendazole on the expression of caspase family proteins in U87 cells (e) and in U251 cells (f). **P < 0.01, ***P < 0.001 vs Control
The effect of benzimidazoles was blocked by caspase inhibitor Z-VAD-FMK
Annexin V- PI double staining was used to distinguish early from late apoptotic and necrotic cells. The annexin V single-positive staining might indicate cells at an early stage of apoptosis, while annexin V-PI double-positive staining might represent cells at a later stage of apoptosis or pyroptosis (Fig. 7a, b). Z-VAD-FMK is a pan-caspase inhibitor, which could be used to block apoptosis and pyroptosis [25, 26]. Z-VAD-FMK could prevent cell death induced by flubendazole. The CCK-8 assay showed that Z-VAD-FMK dose-dependently reversed the cytotoxic effects and the changes of cell morphology triggered by flubendazole on U87 and U251 cells (Fig. 7c–e). Annexin V-PI double staining indicated that Z-VAD-FMK prevented the apoptotic and pyroptotic effects exerted by flubendazole on U87 and U251 cells (Fig. 7f). These results demonstrated that benzimidazoles could inhibit proliferation of glioblastoma cells by inducing apoptotic and pyroptotic cell death.
Flow cytometry analysis of benzimidazole-treated U87 cells stained with annexin V-FITC and PI (a) and U251 cells (b). The CCK-8 assay showed that Z-VAD-FMK prevented flubendazole cytotoxicity in U87 cells (c) and in U251 cells (d). e Z-VAD-FMK reversed flubendazole-mediated changes of cell morphology in U87 and U251 cells. f Z-VAD-FMK blocked flubendazole-induced apoptosis and pyroptosis in U87 and U251 cells. *P < 0.05, **P < 0.01 vs -Z-VAD-FMK
Flubendazole inhibited the growth of glioblastoma in vivo
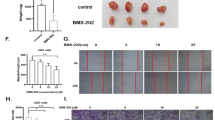
To explore the anti-GBM effect of benzimidazoles in vivo, flubendazole was selected for testing in a nude mouse xenograft model (Fig. 8a). Compared with mice injected with vehicle alone, those treated with flubendazole showed dose-dependent growth inhibition of the tumors. The weight and volume of tumors were significantly decreased with administration of flubendazole. In addition, the beneficial effect of flubendazole was almost equivalent to that of the positive control drug, TMZ, and the tumor had almost disappeared after 24 days of administration of 50 mg/kg flubendazole (Fig. 8b–d). There was no significant change in body weight (Fig. 8e) or in the relative weight, appearance, or morphology of organs including heart, liver, kidney, lung, and spleen (Fig. 8f) during the flubendazole treatment regimen. Physiological blood tests showed no obvious systemic toxicity. Flubendazole also reversed the decrease in lymphocyte ratio caused by transplanted tumors in nude mice, which was not affected by TMZ (Supplementary Fig. 3). Taken together, these results indicated that flubendazole could inhibit growth of GBM tumor with no observable side effects in vivo.
a Schematic of xenograft tumor experiment for the antitumor effects of flubendazole in vivo. b Images of xenograft tumors. c Changes in tumor volumes during the flubendazole administration period. d Tumor weights at the end of the experiment. e Changes in body weight during the flubendazole administration period. f Relative organ weights at the end of the experiment. The data were presented as mean ± SD with **P < 0.01, ***P < 0.001 vs. vehicle group
Discussion
GBM is the brain tumor with the highest degree of malignancy, the worst prognosis, and is highly prone to recurrence and metastasis [27, 28]. TMZ is the only therapeutic drug approved for GBM treatment, and drug resistance often occurs in clinical use, leading to treatment failure [29, 30]. Drug repurposing is an efficient strategy for testing approved or investigational drugs for new uses. It offers distinct advantages over developing an entirely new drug by lowering overall development costs and reducing development times [6]. In this study, by analyzing the hub genes of GBM via WGCNA analysis of the TCGA dataset and using the Connectivity Map platform for drug repurposing, it was predicted that multiple azole compounds had potential anti-GBM activity.
Interestingly, we found that only benzimidazole compounds had anti-GBM activity, while imidazoles and triazoles did not. Flubendazole, fenbendazole and mebendazole all demonstrated good time- and dose-dependent anti-GBM activity. Mebendazole had been reported to potentiate radiation therapy in triple-negative breast cancer [31]. Mebendazole inhibited tumor growth and prevented lung metastasis in models of advanced thyroid cancer [32] and resulted in increasing survival in two preclinical models of GBM [33]. Fenbendazole was identified as a microtubule-destabilizing agent and caused human NSCLC cell death [34]. Flubendazole elicited anti-cancer effects via targeting EVA1A-modulated autophagy and apoptosis in triple-negative breast cancer [35, 36]. It also exerted potent antitumor activity in colorectal cancer cells by inhibiting STAT3 and activating autophagy [37].
Here we found that all benzimidazole compounds had the ability to inhibit the proliferation and metastasis of GBM cells. To determine the molecular mechanism of their effects, RNA-sequencing analyses were performed. The data revealed that benzimidazole regulated cell migration, cell cycle, programmed cell death and other biological processes of GBM cells. Benzimidazoles inhibited the migration and invasion of GBM cells and regulated the expression of key EMT markers, which indicated that benzimidazoles could inhibit the metastasis of GBM. They also dose-dependently arrested the cell cycle at G2/M phase of GBM cells through the P53/P21/cyclin B1 pathway. In addition, a nude mouse xenotransplantation tumor experiment was conducted to assess whether flubendazole significantly suppressed tumorigenicity with no obvious adverse effect in vivo. Taken together, our results demonstrated that benzimidazoles might be promising chemotherapy drugs for the treatment of GBM. The future holds the promise of the development of benzimidazole compounds for use as an alternative drug for treating TMZ-resistant GBM patients or in combination with TMZ to treat patients who are less responsive to TMZ.
Programmed cell death (PCD) is a regulated process for inducing cell death that involves specific genes encoding signals or activities [38]. PCD usually includes apoptosis, autophagy, pyroptosis, ferroptosis, etc., which involves tightly structured signaling cascades and molecularly defined effector mechanisms [39]. RNA sequencing data revealed that the benzimidazole compounds can regulate the PCD process in GBM, and we showed that they induced mitochondrial-dependent apoptosis and pyroptosis in a dose-dependent manner. These benzimidazoles appeared to act via the NF-κB/NLRP3/GSDMD pathway to trigger pyroptosis in GBM cells and concurrently, they might induce mitochondria-dependent apoptosis of GBM cells involving the Bcl-2, PARP and caspase-related pathway. Recent studies have established that pyroptosis can be induced by proteolytically processed GSDMD upon activation of caspase-1 during apoptosis [15], and we found that GSDMD was cleaved to generate N-terminal fragments following pharmacologic inhibition, with kinetics comparable to those of caspase-1 or PARP cleavage [18]. Targeting specific cell death pathways has emerged as a promising strategy for the treatment of cancer [40, 41], but so far, there have been only a limited number of drugs available that target necroptosis, pyroptosis or autophagy. Additional compounds are being tested and are likely to lead to future therapies for a variety of diseases [42]. Benzimidazoles, especially flubendazole, are likely to be potent anti-GBM candidates with a novel mechanism of action as they can induce concurrent apoptotic and pyroptotic cell death (Fig. 9).
In conclusion, benzimidazoles are promising candidates for the treatment of GBM via arresting cell cycle and inducing concurrent apoptotic and pyroptotic cell death. It is of great importance to develop benzimidazoles for combination therapy or as alternative drugs for GBM patients who are resistant or less responsive to TMZ.
Change history
15 August 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41401-022-00971-x
References
Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–48.
Perry JR, Laperriere N, O’Callaghan CJ, Brandes AA, Menten J, Phillips C, et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376:1027–37.
Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009-2013. Neuro Oncol. 2016;18:v1–v75.
Karachi A, Dastmalchi F, Mitchell DA, Rahman M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro Oncol. 2018;20:1566–72.
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58.
Langedijk J, Mantel-Teeuwisse AK, Slijkerman DS, Schutjens MH. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today. 2015;20:1027–34.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52. e17
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559.
Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, et al. Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by Weighted Gene Co-Expression Network Analysis (WGCNA). Cancers. 2019;12:37.
Peng XM, Cai GX, Zhou CH. Recent developments in azole compounds as antibacterial and antifungal agents. Curr Top Med Chem. 2013;13:1963–2010.
Torres FC, Garcia-Rubino ME, Lozano-Lopez C, Kawano DF, Eifler-Lima VL, von Poser GL, et al. Imidazoles and benzimidazoles as tubulin-modulators for anti-cancer therapy. Curr Med Chem. 2015;22:1312–23.
Guo Y, Li CI, Ye F, Shyr Y. Evaluation of read count based RNAseq analysis methods. BMC Genom. 2013;14:S2.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Vavoulis DV, Francescatto M, Heutink P, Gough J. DGEclust: differential expression analysis of clustered count data. Genome Biol. 2015;16:39.
Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42:245–54.
Vande Walle L, Lamkanfi M. Pyroptosis. Curr Biol. 2016;26:R568–R72.
Rathkey JK, Benson BL, Chirieleison SM, Yang J, Xiao TS, Dubyak GR, et al. Live-cell visualization of gasdermin D-driven pyroptotic cell death. J Biol Chem. 2017;292:14649–58.
Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, et al. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180:941–55. e20
Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–8.
Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5.
Moossavi M, Parsamanesh N, Bahrami A, Atkin SL, Sahebkar A. Role of the NLRP3 inflammasome in cancer. Mol Cancer. 2018;17:158.
Liu Z, Gan L, Xu Y, Luo D, Ren Q, Wu S, et al. Melatonin alleviates inflammasome-induced pyroptosis through inhibiting NF-kappaB/GSDMD signal in mice adipose tissue. J Pineal Res. 2017;63:e12414.
Charlot JF, Pretet JL, Haughey C, Mougin C. Mitochondrial translocation of p53 and mitochondrial membrane potential (Delta Psi m) dissipation are early events in staurosporine-induced apoptosis of wild type and mutated p53 epithelial cells. Apoptosis. 2004;9:333–43.
Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63.
Pero ME, Zullo G, Esposito L, Iannuzzi A, Lombardi P, De Canditiis C, et al. Inhibition of apoptosis by caspase inhibitor Z-VAD-FMK improves cryotolerance of in vitro derived bovine embryos. Theriogenology. 2018;108:127–35.
Schneider KS, Gross CJ, Dreier RF, Saller BS, Mishra R, Gorka O, et al. The inflammasome drives GSDMD-independent secondary pyroptosis and IL-1 release in the absence of caspase-1 protease activity. Cell Rep. 2017;21:3846–59.
Lee E, Yong RL, Paddison P, Zhu J. Comparison of glioblastoma (GBM) molecular classification methods. Semin Cancer Biol. 2018;53:201–11.
Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35:2402–9.
Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3:198–210.
Wang S, Yao F, Lu X, Li Q, Su Z, Lee JH, et al. Temozolomide promotes immune escape of GBM cells via upregulating PD-L1. Am J Cancer Res. 2019;9:1161–71.
Zhang L, Bochkur Dratver M, Yazal T, Dong K, Nguyen A, Yu G, et al. Mebendazole potentiates radiation therapy in triple-negative breast cancer. Int J Radiat Oncol Biol Phys. 2019;103:195–207.
Williamson T, Mendes TB, Joe N, Cerutti JM, Riggins GJ. Mebendazole inhibits tumor growth and prevents lung metastasis in models of advanced thyroid cancer. Endocr Relat Cancer. 2020;27:123–36.
Bai RY, Staedtke V, Aprhys CM, Gallia GL, Riggins GJ. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncol. 2011;13:974–82.
Dogra N, Kumar A, Mukhopadhyay T. Fenbendazole acts as a moderate microtubule destabilizing agent and causes cancer cell death by modulating multiple cellular pathways. Sci Rep. 2018;8:11926.
Zhen Y, Zhao R, Wang M, Jiang X, Gao F, Fu L, et al. Flubendazole elicits anti-cancer effects via targeting EVA1A-modulated autophagy and apoptosis in triple-negative breast cancer. Theranostics. 2020;10:8080–97.
Oh E, Kim YJ, An H, Sung D, Cho TM, Farrand L, et al. Flubendazole elicits anti-metastatic effects in triple-negative breast cancer via STAT3 inhibition. Int J Cancer. 2018;143:1978–93.
Lin S, Yang L, Yao Y, Xu L, Xiang Y, Zhao H, et al. Flubendazole demonstrates valid antitumor effects by inhibiting STAT3 and activating autophagy. J Exp Clin Cancer Res. 2019;38:293.
D’Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43:582–92.
Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–64.
Zhang X, Zhang P, An L, Sun N, Peng L, Tang W, et al. Miltirone induces cell death in hepatocellular carcinoma cell through GSDME-dependent pyroptosis. Acta Pharm Sin B. 2020;10:1397–413.
Lu H, Zhang S, Wu J, Chen M, Cai MC, Fu Y, et al. Molecular targeted therapies elicit concurrent apoptotic and gsdme-dependent pyroptotic tumor cell death. Clin Cancer Res. 2018;24:6066–77.
Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell. 2020;78:1045–54.
Acknowledgements
This work was supported by Beijing Natural Science Foundation (7212157), CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007), National Natural Science Foundation of China (81803584, 81703536), Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025, 2018ZX09711001-012).
Author information
Authors and Affiliations
Contributions
GHD, JHW designed research; LWR, WL, XJZ, JYL, YHY performed research; LWR, SL, SZ, WQF and BX analyzed data; LWR wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, Lw., Li, W., Zheng, Xj. et al. Benzimidazoles induce concurrent apoptosis and pyroptosis of human glioblastoma cells via arresting cell cycle. Acta Pharmacol Sin 43, 194–208 (2022). https://doi.org/10.1038/s41401-021-00752-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00752-y
Keywords
This article is cited by
-
PCSK5 downregulation promotes the inhibitory effect of andrographolide on glioblastoma through regulating STAT3
Molecular and Cellular Biochemistry (2024)
-
Pyroptosis, ferroptosis, and autophagy cross-talk in glioblastoma opens up new avenues for glioblastoma treatment
Cell Communication and Signaling (2023)
-
Flubendazole exhibits anti-glioblastoma effect by inhibiting STAT3 and promoting cell cycle arrest
Scientific Reports (2023)
-
Rosmarinic acid, the active component of Rubi Fructus, induces apoptosis of SGC-7901 and HepG2 cells through mitochondrial pathway and exerts anti-tumor effect
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Pyroptosis-related prognosis model, immunocyte infiltration characterization, and competing endogenous RNA network of glioblastoma
BMC Cancer (2022)