Abstract
Heterosis, also known as hybrid vigor, is the superior performance of a heterozygous hybrid relative to its homozygous parents. Despite the scientific curiosity of this phenotypic phenomenon and its significance for food production in agriculture, its genetic basis is insufficiently understood. Studying heterosis in yeast can potentially yield insights into its genetic basis, can allow one to test the different hypotheses that have been proposed to explain the phenomenon and allows better understanding of how to take advantage of this phenomenon to enhance food production. We therefore crossed 16 parental yeast strains to form 120 yeast hybrids, and measured their growth rates under five environmental conditions. A considerable amount of dominant genetic variation was found in growth performance, and heterosis was measured in 35% of the hybrid–condition combinations. Despite previous reports of correlations between heterosis and measures of sequence divergence between parents, we detected no such relationship. We used several analyses to examine which genetic model might explain heterosis. We found that dominance complementation of recessive alleles, overdominant interactions within loci and epistatic interactions among loci each contribute to heterosis. We concluded that in yeast heterosis is a complex phenotype created by the combined contribution of different genetic interactions.
Similar content being viewed by others
Introduction
Crossing two genetically divergent parents usually results in diversity among the progeny owing to newly created genetic combinations. In some traits, the phenotypic distribution of the progeny can exceed the range of its parents under a variety of environmental conditions (McClintock, 1984; Chen, 2010). A prime example of this deviation of the progeny mean from the range of its parents’ phenotype is heterosis (that is hybrid vigor), in which the heterozygous progeny outperforms the average of its homozygous parents or even the performance of its best-performing parent. Heterosis has been traditionally attributed to new heterozygous combinations with dominant effects created by the hybridization of genetically divergent parents. As heterosis is conditioned by dominant effects, it is expected to be seen more often with fitness-related traits than with traits that respond well to selection (Semel et al., 2006). Despite the importance of this phenomenon for agriculture and research on the subject that dates back more than 100 years (Darwin, 1876; Bruce, 1910; Jones, 1917), our understanding of the genetic basis for the manifestation of heterosis is still incomplete (Lippman and Zamir, 2007; Charlesworth and Willis, 2009; Baranwal et al., 2012; Schnable and Springer, 2013).
In general, the expression of a heterotic phenotype is measured in a hybrid of two parents that are homozygous in large parts of their genomes, each for different alleles affecting the trait of interest. The hybrid of these divergent parents is heterozygous in many loci. The question is then: how does this gene heterozygosity lead to a heterotic phenotype? There are a few models, all involving dominant effects, that explain how different genetic constellations of heterozygous loci can improve the performance of a hybrid as compared with its divergent homozygous parents (Xiao et al., 1995; Lippman and Zamir, 2007). Although many studies in plants have favored one model or another (Li et al., 2001; Luo et al., 2001; Semel et al., 2006), other studies have indicated that a combination of a few models is needed to explain the genetic basis underlying heterosis (Yao et al., 2012; Guo et al., 2013; Schnable and Springer, 2013).
The ‘dominance complementation model’ proposes that since the homozygous parents are genetically divergent, in the hybrid progeny, the slightly deleterious recessive alleles of one parent are complemented by the favorable dominant alleles of the other parent and vice versa. The accumulation of reciprocal complementation over multiple loci yields a heterozygous hybrid with an overall larger number of favorable dominant alleles for more genes than in either of its parents, thereby outperforming them (Bruce, 1910). The only requirement of this model is a sufficient number of loci, each with some degree of dominance, which when additively summed can account for the heterosis.
The ‘overdominance model’ explains heterosis as an interaction between alleles within a locus that leads to an advantage for the heterozygous state in the hybrid over the homozygous state of either allele in the parents (East, 1908; Crow, 1948). Heterosis can be caused by one or more of such overdominant loci. The mechanistic explanation for overdominance might involve dosage effects, as shown, for example, for a recessive mutation affecting yield in tomatoes (Krieger et al., 2010). However, not enough overdominant loci have been identified in such studies to allow conclusions about mechanisms governing intralocus interactions and the role of overdominance. Another complication in estimating the significance of intralocus interactions is that ‘pseudo-overdominant’ loci can pose as overdominant loci. These are not genuine because they contain at least two genetically linked genes, each with a dominance complementation effect, but in repulsion phase, which can be mistakenly identified as a single overdominant locus (Lippman and Zamir, 2007; Schnable and Springer, 2013).
Epistasis refers to genetic interactions between alleles of different loci that contribute to genetic variation beyond the individual contributions of each locus. More recent studies have noted the contribution of epistatic interactions to the favorable heterotic phenotype of hybrids relative to their parents (Steinmetz et al., 2002; Sinha et al., 2006; Melchinger et al., 2007, 2008; Li et al., 2012).
The ‘dominance complementation model’ is different from the ‘overdominance’ and ‘epistasis’ models. Whereas obtaining heterosis by dominance complementation requires the sum of effects from several loci, by overdominance or epistasis, because of the synergistic interaction, the effect of even one or two loci, respectively, can lead to heterosis. The possibility that one or two loci will lead to a hybrid with a performance advantage is appealing to plant and animal breeders and motivates their efforts to identify and study overdominant and epistatic loci.
A few other models have been used to explain heterosis. These mainly involve genome-wide effects of polyploidy, gene dosage and gene balance (Schnable and Springer, 2013). Furthermore, several research approaches, such as functional genomics, transcriptomics, metabolomics and proteomics, have been applied to investigate the basis of heterosis. Such studies have tested the hypothesis that due to the genetic differences among the parental genomes, cumulative positive effects of differential gene expression in the hybrid improve the function of metabolic pathways, energy production and energetic efficiency of cellular processes in ways that lead to heterosis (Hedgecock et al., 2007; Goff, 2011; Baranwal et al., 2012; Schnable and Springer, 2013).
The yeast Saccharomyces cerevisiae is a prime eukaryotic model organism, for which extensive genetic and molecular knowledge provides an advantage in studying the genetic basis of complex phenotypic phenomena such as heterosis (Botstein et al., 1997). Vegetative growth rate correlates with the yeast’s fitness (St Onge et al., 2007; Hillenmeyer et al., 2010), and as heterosis is more often found in fitness-related traits, growth under multiple conditions is suitable for studying heterosis. In addition, evidence from genome sequencing has suggested that in nature yeast propagates mainly by asexual reproduction, and thus yeast isolates can often be regarded as distantly related inbred populations, each adapted to its ecological niche (Schacherer et al., 2007; Liti et al., 2009; Warringer et al., 2011). Hybrids between such isolates provide a good opportunity to study heterosis and can illustrate the relative importance of the different genetic mechanisms governing this complex phenotypic phenomenon.
Materials and methods
Growth media and conditions
YEPD-rich medium (1% (w v−1) yeast extract, 2% (w v−1) peptone, 2% (w v−1) dextrose) was used, unless otherwise specified. For selection during strain construction, YEPD was supplemented with either hygromycin B (300 mg l−1) or Geneticin (G418 sulfate; 400 mg l−1) or both. Yeast growth rate was measured under five different environmental conditions: YEPD, YEPE (1% yeast extract, 2% peptone, 3% (v v−1) ethanol), YEPGal (1% yeast extract, 2% peptone, 2% (w v−1) galactose) and YEPGly (1% yeast extract, 2% peptone, 3% (v v−1) glycerol), all at 30 °C. Growth under YEPD was also measured at 37 °C. Solid media were prepared by the addition of 2% (w v−1) agar to the liquid media.
Sporulation and dissection of diploids
To obtain haploids, diploid cells were grown overnight in patches on YEPD plates and scraped into sporulation solution (0.75% (w v−1) potassium acetate) for 2–3 days. To prepare for dissection, a 15-μl aliquot of the sporulation culture was incubated with 15 μl zymolyase (2 mg ml−1) for 30 min at 30 °C to digest the ascus wall. Treated tetrads were dissected to individual haploid spores using a micromanipulator microscope (Singer Instruments, Somerset, UK) on YEPD plates.
PCR procedures
DNA templates, in the form of either 5 μl of water-suspended cells or 50 ng of purified DNA, were used in a total reaction volume of 20 μl containing 1.66 μM of each forward and reverse primer, 0.25 mM MgCl2, 0.66 mM dNTP mix, 2 μl buffer, 0.5 μl Taq DNA polymerase and polymerase chain reaction (PCR)-grade water to complete the volume. The PCR amplification profile was 94 °C for 10 min, followed by touchdown cycles of 94 °C for 30 s, annealing from the higher to lower temperature for 1 min with a decrease of 0.5 °C per each of 14 cycles and extension at 72 °C for 2 min. The touchdown cycles were followed by 23 cycles at the lower annealing temperature and a final elongation step at 72 °C for 10 min. PCR products were verified by electrophoresis on a 1.5% TBE agarose gel containing ethidium bromide.
Transformation of genes
To construct the haploid parental strains, MX4 drug-resistant cassettes (HygB or Kan) were inserted into the HO locus of the wild strains following their self-diploidization. Gene replacement with a drug cassette was performed following the standard LiAc method (Gietz and Woods, 2002). Yeast transformants were selected on drug-containing YEPD plates and successful integration was confirmed by PCR using position-specific primers. The primers used in the transformation procedure are detailed in Supplementary Table S1.
Hybrid-collection construction
Sixteen wild strains were selected for this study (Supplementary Table S2) from the larger collection of fully sequenced strains previously described by Liti et al. (2009). The stages of strain-collection construction are detailed in Supplementary Figure S1. Briefly, the diploid wild strains were sporulated and single haploid spores were dissected. As these wild strains were homothallic, the spores contained an active HO gene and could self-diploidize to form homozygous diploids. The diploid of each strain was transformed with the KanMX4 or HygBMX4 drug-resistant cassette replacing the HO gene, and drug-resistant diploids were sporulated. Dissection of tetrads enabled selection of eight haploids per strain, four of each of the opposite mating types. Mating type and haploid/diploid state were determined by PCR using MAT locus primers (Supplementary Table S1). To verify the genetic background of the haploid parental strains before crossing and establishment of the hybrid collection, a set of 10 hypervariable microsatellite markers (Supplementary Table S1) was used as detailed by Richards et al. (2009). Verified haploid isolates of each strain with opposite mating types and different drug resistances were mated to yield eight replicates of parental homozygous diploids per strain. Crosses between all pairwise combinations of 16 parental haploids yielded 120 hybrid diploid strains, each in eight replicates (Supplementary Table S2). Thus, altogether 1088 strains were included in the measurements of growth rate. The cross-scheme included reciprocal crosses to eliminate possible effects of drug resistance and mating type (Supplementary Figure S1).
Construction of reciprocal backcross populations
Two populations of reciprocal backcrosses were constructed for each of the two genetically divergent pairs of parental lines. One pair of parental strains was SK1 and S1001 and the other was YJM145 and S1001 (Supplementary Table S2). YJM145 and S1001 are diploids (McCusker et al., 1994; Dean et al., 2008), isogenic to the fully sequenced YJM789 and S288c strains, respectively. The F1 hybrid of each parental pair was sporulated and about 180 haploid spores (segregants) were dissected from each cross. Each of the segregants was crossed two times, once to each haploid parent, to generate two reciprocal backcross populations of diploids per cross. Growth of reciprocal SK1 × S1001 backcrosses was measured at 30 °C and 37 °C (Supplementary Table S4, data available from the Dryad Digital Repository: doi:10.5061/dryad.4j07v), whereas that of the YJM145 × S1001 backcrosses was measured at 37 °C and 40 °C (Supplementary Table S5, data available from the Dryad Digital Repository: doi:10.5061/dryad.4j07v). We measured the different crosses at different temperatures to obtain more pronounced growth differences: YJM145 and the hybrid YJM145 × S1001 grow well at high temperature, whereas SK1 and the hybrid SK1 × S1001 are not as proficient at high temperature.
Doubling-time measurements
Strain doubling times (DTs) were measured in 96-well plates containing liquid medium by incubation with shaking and repeated measurements of optical density (OD595) using the Infinite 200 multimode reader (Tecan, Männedorf, Switzerland). Measurement cycles lasted 463 s, including shaking, incubation and the OD reading. A specific script was used to extract the DT of the culture in each well. The script extracted the OD values in the logarithmic growth phase (OD595 0.15–0.5) of the culture and calculated the DT using the formula: DT (h)=(log 2 × number of cycles × 463 s/(log(second OD)−log(first OD)))/3600. DT values were log transformed (Log2 DT) before statistical analyses.
Heterosis measurements
To determine modes of inheritance and heterosis levels, a few statistics were estimated:
-
Mid-parent value (m)—mean of the homozygote parents’ phenotype: (P1+P2)/2.
-
Additive genetic deviation (a)—the difference between each of the homozygous parents and the mid-parent value: (P1 or P2)−(m).
-
Dominant genetic deviation (d)—the deviation of the heterozygous hybrid (Hyb) from the mid-parent value: Hyb−(m).
The degree of dominance (d/a) defined the inheritance mode as follows: (d/a=0)—codominance; (0<d/a<1) or (−1<d/a<0)—partial dominance of the faster or slower parent, respectively; (d/a=1 or −1)—full dominance of the faster or slower parent, respectively and (d/a>1) or (d/a<−1)—over- or underdominance of the hybrid, respectively.
Determination of mode of inheritance
On the basis of the DT of the hybrid and its parents, each of the 120 hybrids under each of the five conditions was assigned to one of seven mode-of-inheritance categories. Confidence interval (CI) values (P=0.05) around the mean Log2 DT were estimated for every strain based on the growth of the eight replicates. If the slower CI limit of the hybrid was faster than the faster CI limit of the faster parent, then the hybrid was categorized as overdominant. If the faster CI limit of the hybrid was slower than the slower CI limit of the slower parent, the hybrid was categorized as underdominant. If the hybrid CI overlapped the faster parent CI, the hybrid was categorized as dominant. If the hybrid CI overlapped the slower parent CI, the hybrid was categorized as recessive. If the hybrid CI was between the slower CI limit of the faster parent and the mid-parent value, the hybrid was categorized as partially dominant. If the hybrid CI was between the faster CI limit of the slower parent and the mid-parent value, the hybrid was categorized as partially recessive. If the mid-parent value was in the CI range of the hybrid, the hybrid was categorized as codominant. It should be noted that categorizing hybrids based on overlaps in CI is a conservative method that might underestimate how many hybrids are heterotic in each condition.
Analysis of variance components
To estimate the variance components from the diallel structure of our collection, we used the restricted maximum-likelihood method as implemented in the statistical software JMP 8 (SAS Institute, Cary, NC, USA). The Log2 DT values of the parental strains were analyzed in a full bifactorial design, separately for each condition. The two parental factors and their interaction were defined as random effects. To estimate the variance components of the five conditions together, the randomized parental factors and their interaction were analyzed as random factors nested within the fixed-conditions factor. The following variance components were extracted from the variance estimates of the analyses: additive variance (VA) was the variance among homozygote parents, dominant variance (VD) was the variance of the interaction among parents (hybrids), genetic variance was the sum of both (VG=VA+VD), residual variance (VResidual) was the variance due to random effects and phenotypic variance (VTotal) was the total variance, the sum of all components together.
Analyses of strain phylogeny, heterotic combination network and sharing of heterotic mates
The genetic distances between strains were based on pairwise single-nucleotide polymorphism differences across their genome sequence, adopted from Liti et al. (2009) genome-sequencing study. A phylogenetic tree based on pairwise distances was generated for our subset of 16 parental strains using the neighbor-joining method as implemented in MEGA 5 evolutionary analysis software (Tamura et al., 2011). The network of heterotic combinations was constructed using the Cytoscape program (Saito et al., 2012).
The three clusters of genetically similar strains corresponded to the Malaysian, North American and European/North American clusters in the phylogenetic tree. Sharing was calculated between pairs within each cluster, and between pairs from different clusters as controls, using the EstimateS program (Chao and Lin, 2012).
Results and discussion
Growth variability and mode of inheritance in the strain collection
Our constructed collection of strains included 16 homozygous diploid parents and their 120 diploid hybrids, each in eight biological replicates. The growth rate of each strain, calculated as DT, was measured at least two times under each of five environmental conditions (YEPD 30 °C, YEPD 37 °C, YEPE, YEPGal and YEPGly). The phenotypic data set is given in Supplementary Table S3 (data available from the Dryad Digital Repository: doi:10.5061/dryad.4j07v).
The mean Log2 DT values of the 16 parental strains and the 120 hybrids varied considerably under each of the five conditions (Figures 1a and b and Supplementary Figure S2), reflecting the genetic variability in growth-related genes among these different genetic backgrounds and their combinations. Furthermore, the variability in growth rates of the parents reflected the different adaptations evolved in each strain to the conditions in its original habitat. Notably, the growth-rate distributions of both parents and hybrids (Figures 1a and b) were bimodal with a small overlap between the faster growth in YEPD and YEPGal and the slower growth in YEPE and YEPGly, the latter two conditions containing nonfermentable carbon sources. Thus, comparing strain performance across conditions mandated normalizing the data.
Growth and heterosis in the strain collection. (a) Frequency distribution of DTs of the 16 parental strains across the five conditions. (b) Frequency distribution of DTs of the 120 hybrid strains across all five conditions. In (a and b), the larger values from YEPGly and YEPE measurements are darkened. (c) Medians and means of the degree of dominance (d/a) for the seven mode-of-inheritance categories. The categories were defined as: overdominant (OD), fully dominant (Dom), partially dominant (PDom), codominant (CoD), partially recessive (PRec), fully recessive (Rec) and underdominant (UD). (d) Frequency distribution of the degree of dominance across all conditions showing a median of 0.93 and a mean of 1.40. Note: a value of 1 denotes full dominance.
To study the prevalence of dominant inheritance across conditions, we defined the performance of each hybrid relative to its parents in two separate ways. First, we calculated a continuous statistic for the level of dominance (d/a). Second, we assigned each hybrid in each condition to one of seven discrete mode-of-inheritance categories. To test the concordance between the two measures of hybrid performance, we calculated the mean and median degree of dominance across conditions in each mode-of-inheritance category. The mean and median decreased with a shift from the overdominant to underdominant categories (Figure 1c), indicating agreement between these two ways of defining hybrid performance. This agreement validated the standardization of growth rates across conditions, allowing us to use both the categorical assignment and the continuous value of each hybrid for further analyses across all conditions.
Dominance and heterosis in hybrid growth
First, we quantified the amount of heterosis observed in this strain collection. A degree-of-dominance value > 1 reflected heterotic performance. The distribution of the degree-of-dominance values for all hybrids and conditions had a median of 0.93 and a mean of 1.4, reflecting a skew in the distribution toward dominance (Figure 1d). In fact, more than 60% of the hybrid–condition combinations included a dominant effect, as their degree-of-dominance value was significantly larger than zero. Further support for the abundance of dominant effects came from the high fraction of dominant genetic variance found in the variance component analysis (Table 1). Out of the total phenotypic variance (VTotal), the genetic variance (VG) component constituted from 46% in YEPGly to 88% in YEPD 37 °C. Growth in different environments involves functions of general growth genes and condition-specific ones. Both the number of condition-specific genes and the strength of the evolutionary constraints on the genes can vary among conditions. The combination of these two factors results in a larger or smaller fraction of genetic variance among conditions.
Furthermore, the fraction of the dominant genetic variance component (VD) out of VG ranged between 53% in YEPGly and 75% in YEPE. For all conditions together, VD/VG was 63%, in agreement with the abundance of dominant effects found in the distribution of hybrid performance.
The proportion of overdominant (best-parent heterosis) hybrids was 35% on average and ranged from 23% (28/120 hybrids) in YEPGal to 47% in YEPD 30 °C (Supplementary Figure S3). Grouping together all of the dominance categories (over-, fully and partially dominant) placed the proportion of dominant inheritance between 72% of the hybrids in YEPD 37 °C and 38% in YEPGly. The lower fraction of overdominant hybrids in YEPGal vs YEPD 30 °C (Supplementary Figure S3) is interesting as the VD component in galactose is larger than in YEPD 30 °C (Table 1). Given that both the residual variance and the fraction of all dominance categories combined were similar under these two conditions, we hypothesize that interactions such as overdominance and epistasis contributed more to heterosis in YEPD 30 °C than in YEPGal.
In contrast to the prevalence of overdominance and dominance in general, the proportion of underdominant hybrids was only 5% and together with fully and partially recessive hybrids constituted 18% of the overall hybrid phenotypes.
The number of hybrids in each mode-of-inheritance category varied considerably among parental strains (Supplementary Figure S4). Many parental strains formed hybrids that were heterotic under multiple conditions. We found 3, 9 and 23 parental combinations that were heterotic under five, four and three environmental conditions, respectively. Among parents of the 12 hybrids that were heterotic under four or five conditions, we found enrichment of slow-growing parental strains (discussed further on). In addition to these multicondition heterotic hybrids, 24 and 41 hybrids were heterotic under two and a single condition, respectively. Overall, 83% of the hybrids were heterotic under at least one of the five conditions.
In contrast to the average 35% overdominant hybrids found here, <5% of best-parent heterosis was reported in hybrids of parental strains sampled from the same yeast collection (Zorgo et al., 2012). Six parental strains and two conditions were common to both studies. In the overlapping part taken from our data, we found overdominance in 37% of the hybrid–condition combinations. There are likely several reasons for the discrepancy in the prevalence of heterosis between our and Zorgo et al. (2012) study. First, the set of strains only partially overlapped. Second, the method of heterosis calculation made a difference; calculating best-parent heterosis as in Zorgo et al. (2012) on the overlap taken from our data yielded a significant but not perfect correlation coefficient of 0.75 with our calculation of d/a. Third, the setup and nature of the growth conditions were different. Only 18 out of 56 conditions in the other study were complex conditions, the rest being simple in the sense that they either depleted a nutrient from or added a toxin to the medium. Fitness and growth are complex traits, and heterosis is expected to occur more frequently in fitness-related traits. Growth under complex environmental conditions reflects cellular fitness better than growth in the presence or absence of a specific molecule, because in the latter case a polymorphism in a single gene might account for the whole difference in phenotype (Hillenmeyer et al., 2010).
Taken together, the high fraction of the dominant variance component, the proportion of overdominant hybrids and the multicondition heterotic hybrids demonstrated the prevalence of dominant effects and heterosis in this hybrid collection and indicated its suitability for studying the genetic models underlying heterosis in yeast.
Genetic distance between parents and heterosis in their hybrids
Our collection included isolates from various natural niches and geographical locations as shown by the phylogenetic tree (Figure 2a). The phylogeny of our strain subset matches that of a larger reported and discussed collection (Liti et al., 2009). In our strain phylogeny, there were three clusters, North American, Malaysian and European/North American, each containing genetically similar strains that probably did not hybridize in nature to strains from other clusters and therefore have pure genomes. Other strains in our collection had mosaic genomes composed of regions from genetically divergent strains as determined by Liti et al. (2009).
Genetic distances and heterosis. (a) Phylogenetic tree of the 16 parental strains. Denoted next to each edge tip are the strain code and name, its geographical origin and genome constitution (pure or mosaic). Clusters of strains with very similar genomes are boxed. The phylogeny is based on the single-nucleotide polymorphism (SNP) differences among strains as in Liti et al. (2009). (b) Distribution of genetic distances between the 120 pairwise combinations of the 16 parental strains. (c) Mean genetic distance and CI between parents of hybrids included in each of the inheritance modes as defined in Figure 1c. The overdominant (OD) group was significantly different from the codominant (CoD) group (Tukey–Kramer honest significance difference test, P<0.05), and the rest were not different from each other. The numbers above the whiskers denote the number of hybrids constituting that group.
A wide range in genetic distances between pairs of parental strains is represented in this collection (Figure 2b). We tested whether genetic distances between parents correlate with heterosis in their hybrids. The Pearson’s linear correlation coefficient between the degree-of-dominance values of hybrids and the genetic distances between their parents was low under all conditions, but significantly different from zero in YEPD 30 °C and 37 °C (r=0.18, P=0.047 and r=0.26, P=0.005, respectively). Similarly, the correlation between the degree-of-dominance values of hybrids and the square of the genetic distances between their parents was insignificant, except for in YEPD 30 °C. Consistent with the low correlation, all mode-of-inheritance groups presented in Figure 1c had similar mean genetic distances (Figure 2c). Therefore, neither the amount of dominance nor the tendency to obtain heterosis was strongly correlated with genetic distances between parents.
Previous studies have reported that the more genetically distant the parental lines are, the higher the chances for heterosis in their hybrids (Cheres et al., 2000; Zhang et al., 2010). The expectation of finding more heterosis in crosses between distantly related parents relies on the assumption that dominance complementation is more likely to occur between distantly related parents. As this expectation was not met in our collection, we inferred that dominance complementation was not a major mechanism contributing to heterosis in yeast, and thus overdominance and epistasis might have a larger role to the extent that a substantial correlation between heterosis in hybrids and parental genetic distances was not observed.
Heterotic profiles of genetically similar strains
To further explore the relationship between genetic similarity and heterosis, we determined whether genetically similar strains form similar heterotic combinations. On the background of the phylogenetic tree, we depicted the network of heterotic combinations for the strains with the largest (Figure 3a) and smallest (Figure 3b) number of heterotic combinations. The difference in density of the two networks is evident from the number of edges as well as from their thickness, which represents the number of conditions under which a particular heterotic combination was measured.
Heterosis profiles of genetically similar or different parental strains. (a–c) Edges connect pairs of strains that formed a heterotic hybrid. The heterotic combination network is overlaid on the background of the phylogenetic tree. The number of conditions under which a specific hybrid was heterotic is denoted by the different line types (see legend). (a) The network of heterotic combinations of the three parents with the largest number of heterotic combinations (27, 19 and 37; circled). (b) The heterotic combination network of the three parents with the fewest heterotic combinations (8, 4, and 18; circled). (c) A comparison between the networks of 18 and 19, which are highly similar genetically, but are among the strains with the fewest and largest number of heterotic combinations, respectively. The three clusters of genetically similar strains are boxed. (d) The fraction of the shared heterotic combinations between pairs of strains within each of the three clusters of genetically similar strains. As controls, the sharing was calculated for pairs from different clusters (genetically different strains). In gray circles are pairwise sharing values, whereas the black squares denote the mean of the pairwise comparisons.
The three strains that yielded the largest number of heterotic hybrids (strains 27, 19 and 37) were genetically divergent, as were the three strains with the fewest heterotic combinations (8, 4 and 18). One might have expected that divergent parental strains will form different heterosis networks. However, closely related strains did not form similar networks either. The most striking example was the genetically similar strains 18 and 19 that both belong to the Malaysian cluster yet formed very different heterosis networks. Whereas strain 18 formed 14 heterotic combinations and belonged to the group with the fewest heterotic combinations, strain 19 formed 38 heterotic combinations and belonged to the group with the largest number of heterotic combinations (Figure 3c).
To obtain a quantitative estimate of the relationship between genetic similarity of strains and similarity in their heterotic partners, we calculated how many times two strains shared a heterotic combination with the same third strain. We performed these comparisons between pairs from the same genetic cluster and as controls, we compared the same set of strains between pairs from different clusters. We found that the overlap in shared heterotic combinations between pairs of similar strains was quite variable, ranging between 3 and 49%. Furthermore, the similarity in profiles was not different between genetically similar and genetically distant pairs of parents (Figure 3d). These findings held true for each of the genetic clusters alone and for all three clusters together. Another quantitative measure was to correlate degree-of-dominance vectors between genetically similar strains and as a control between genetically distant ones (Supplementary Figure S5a and b). The results of this analysis corroborated those of the heterotic-profile sharing.
Thus, from our data it is clear that genetically similar strains do not have a strong tendency to form heterosis with the same mates. As genetically similar strains share alleles that affect their growth performance, forming heterotic hybrids with different mates implies that only a few polymorphic genes can cause heterosis. As overdominance in even one gene can yield heterosis, our results suggest that in hybrids of genetically similar parents, overdominance can explain heterosis better. However, it would be difficult to explain the large amount of heterotic combinations and the variability in these combinations among similar strains such as 18 and 19 with the effect of a few genes. Thus, we propose that in such cases, synergistic epistatic interactions between a few genes also contribute to heterosis.
Ecology of parental strains and heterosis in their hybrids
We sought to further characterize variation in forming heterotic hybrids among the 16 parental strains. To enable comparisons despite the differences in DTs among conditions, we ranked the heterotic hybrids under each condition such that 1 was assigned to the fastest growing, 2 to the second fastest and so on. Figure 4a shows two measures for each parental strain per condition: the number of heterotic hybrids and their mean rank. This analysis showed that the variation in heterotic combinations among parental strains could be attributed to the number of heterotic hybrids per parental strain under each condition, as well as to the number of conditions under which each hybrid was heterotic.
The number of heterotic hybrids and the mean of their growth rank under each condition by parental strains. (a) All heterotic hybrids under each condition were ranked according to their growth rate (fastest ranked 1, second-fastest ranked 2, etc.). The mean of this rank is shown on the y axis for each parental line (x axis). The size of the bubble corresponds to the number of heterotic hybrids that each parent created under each condition. (b) Correlations between the growth rate of the parental strains and either the number or the growth rate of their heterotic hybrids. The growth of the parental lines was ranked under each condition (fastest ranked 1, second-fastest ranked 2, etc.) and summed over conditions (x axis). The overall number of heterotic hybrids (y axis, blue full circles) was positively correlated with the growth rate of the parent (r=0.78, P=0.0003). The mean rank of the heterotic hybrids of each parent (from a) was averaged across the five conditions (y axis, red empty circles). This mean of heterotic hybrid rank was positively correlated (r=0.65, P=0.006) with the growth rate of the parent. Arrows next to axes explain the trend of the changes.
We then looked into why such variability in heterotic combinations exists among parental strains and tested the relationships among three factors: ecological origin, growth phenotype and degree of dominance. Assuming that similar ecological contexts placed similar evolutionary constraints on fitness-related genes, while relaxing the constraint on other genes, one would expect to find similar performance of strains from similar niches, and similarity in the mates with which they form heterotic combinations. This expectation was not met. We created matrices of phenotypic values for pairs of strains, first based on DT and second on the degree of dominance, separately for each condition. Clustering analyses based on either degree-of-dominance or DT matrices gave different groupings of the parental strains under each condition, rather than consistent groupings according to ecological origins (data not shown). Furthermore, the clustering results based on the degree of dominance differed from those based on DT under each condition separately. Thus, we found here that at least growth performance of the strains (parental and hybrids) and number of heterotic hybrids per parental strain cannot be well explained by their ecological origin. Strains 18 and 19 were isolated in Malaysia; both were associated with Bertram palm nectar and are closely related; nevertheless, each formed a very different heterotic network (Figure 3c). Significant similarity in growth phenotype between genetically similar strains and between strains of similar ecological origin was also lacking in a study using a smaller set of wild strains (Ziv et al., 2013). Therefore, ecological origin was not a strong determinant of the recessive mutations represented in this set of parental strains, and complementation of recessive mutations did not make a major contribution to heterosis.
Performance of parental strains and heterosis in their hybrids
We further compared the growth rates between parental and hybrid strains (Table 2). On average, hybrids grew faster than parents did and this advantage was significant under all conditions tested except YEPGly. Faster average growth of hybrids compared with their parental strains in ethanol concentrations higher than the 3% used here has been reported previously (Timberlake et al., 2011). Nevertheless, looking at the distribution of growth values, we found that while in YEPGly and YEPD 30 °C, the fastest-growing strains were heterotic hybrids, in YEPD 37 °C, YEPE and YEPGal, a homozygous parental strain had the fastest growth rate.
With the growth performance of hybrids and parental strains in mind, when considering utilization of heterosis in crop plants and livestock crossbreeding, two important questions come to mind. First, what set of parental strains will yield more heterotic hybrids and second, what will the quality of those heterotic hybrids be? We assigned each of the 16 parental strains a rank under each condition and summed the ranks for all conditions such that fast-growing parents had a lower total rank, whereas slower-growing parents had a higher one. We found a positive and significant Pearson’s correlation (r=0.78, P=0.0003) between the rank of the parents and the total number of heterotic hybrids they produced across all conditions (Figure 4b). Therefore, slower-growing parents produced a larger number of heterotic hybrids.
However, faster-growing parents also formed heterotic hybrids. For each parent, we averaged the performance rank of its heterotic hybrids across all conditions. A positive and significant correlation (r=0.65, P=0.006) was found between the total rank of the parents and the mean rank of their heterotic hybrids (Figure 4b). Such positive correlations were also found for each condition separately and these condition-specific correlations were significant for three of the five conditions. This correlation indicated that faster-growing parents formed faster-growing heterotic hybrids.
Slower-growing parents might contain more recessive growth alleles; as such, the finding that slower-growing parents form a larger number of heterotic hybrids is consistent with the dominance complementation model, as suggested previously for yeast (Zorgo et al., 2012). The larger the number of recessive mutations one strain has, the more counterpart strains will have the opportunity to complement them with dominant alleles.
However, faster-growing parents that contain fewer recessive alleles formed faster-growing heterotic hybrids. Explaining such cases of heterosis by dominance complementation alone is less feasible. Whereas heterosis due to dominance complementation requires the sum of the effects of several heterozygous genes, overdominance could explain heterosis with a large effect of even a single gene. Faster-growing parents already have favorable alleles in many genes that slower-growing parents do not have. Our findings therefore support the notion that heterosis in hybrids of faster-growing strains is due to fewer loci; this notion is better suited to the synergistic effects of overdominance and epistasis. Heterosis due to a large effect of a single overdominant locus has been reported in plants, such as tomato, wheat, maize and rice (Lippman and Zamir, 2007; Charlesworth and Willis, 2009; Krieger et al., 2010; Li et al., 2013). Epistasis can also explain heterosis in hybrids of faster-growing strains as was found for growth at high temperature in yeast (Steinmetz et al., 2002; Sinha et al., 2006).
The phenotype of reciprocal backcrosses
In a backcross population relative to the fully heterozygous hybrid, on average, only half of the alleles are retained in a heterozygous state. If dominance complementation were the major mechanism contributing to heterosis, retaining only half of the complemented loci would lead to an expected backcross average that is midway between the hybrid and the backcross parent. Improvement of the hybrid compared with the respective parent was set at 100%, and we tested whether the improvement of the BC1 population relative to the respective parent was around 50%. We used two sets of reciprocal backcross populations, each under two conditions. The phenotypic data sets of these populations are given in Supplementary Tables S4 and S5 data available from the Dryad Digital Repository: doi:10.5061/dryad.4j07v.
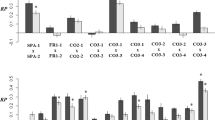
The SK1 × S1001 hybrid (7 × 37 in Supplementary Table S2) was heterotic in YEPD 30 °C and YEPD 37 °C. We compared the mean growth of each BC1 population to that of the hybrid and the respective backcross parent (Figure 5a). For example, the segregants were backcrossed to parent 37 (BC1-37) and the mean DT of this backcross population was 26.8% (1.268 times) faster than that of the 37 × 37 parent in YEPD 37 °C (denoted by y in Figure 5a). The 37 × 7 hybrid grew 56.2% faster than the 37 × 37 parent (denoted by x in Figure 5a). The ratio between the relative differences (0.268/0.562=0.477) was about 50%, indicating a regression of BC1-37 to the midway. Values around 0.5 were also obtained for BC1-7 under YEPD 37 °C and BC1-37 under YEPD 30 °C (Table 3). BC1-7 under YEPD 30 °C regressed toward the 7 × 7 parent more than a halfway (0.345).
Growth analysis of reciprocal backcross populations. The mean Log2 DT (y axis) of the hybrid, diploid parental strains and reciprocal backcross populations (x axis) at different temperatures. The horizontal lines are drawn at the level of the best parent (BP) in each condition. The values of the backcrosses (BC1) are means of about 180 diploid strains per population. Asterisk above a BC1 bar denotes that the mean of the BC1 population was significantly different (P<0.05) from the value of the hybrid and the respective backcross parent. Denoted by curly brackets and small letters y and x is an example of the differences in growth rates, the ratio of which was tested to support or disprove the dominance complementation model. The analyses were carried out in two genetic backgrounds in which the hybrid was heterotic. The background of (a) 7 × 37 and (b) of 38 × 37.
The YJM145 × S1001 (38 × 37) hybrid was heterotic in YEPD 37 °C and 40°C (Figure 5b). The mean of the BC1-37 population regressed 0.575 of the difference between the hybrid and the respective 37 × 37 parent in YEPD 37 °C and 0.312 at 40 °C (Table 3). The mean of BC1-38 population regressed to 0.369 of the difference and all the way (−0.066) to the 38 × 38 parent at 37 °C and 40 °C, respectively. In the latter case, the advantage of the hybrid was completely eliminated in the BC1 population.
Our results indicated that dominance complementation is probably more important in the 37 × 7 hybrid (three of four cases regressed about halfway), whereas overdominance and epistasis were more significant in the 37 × 38 hybrid (one of four cases regressed halfway). The differences between the two hybrids demonstrate the complex genetics underlying heterosis and reveal the combined contribution of multiple genetic mechanisms to heterosis, even within a single hybrid.
Conclusions
We constructed a collection of 120 S. cerevisiae hybrids by crossing 16 parental strains representing the wide geographical, ecological and genetic distributions of this yeast species (Liti et al., 2009). We used this hybrid collection to study the extent of heterosis in yeast and found it to be considerable. We then analyzed the data to explore which of the genetic models explaining heterosis (Lippman and Zamir, 2007; Baranwal et al., 2012; Schnable and Springer, 2013) fits the observed patterns of hybrid performance in this collection. We found evidence of all three main genetic models—dominance complementation, overdominance and epistasis—contributing and often acting together to yield heterosis in yeast.
In support of the contribution of dominance complementation were a larger number of heterotic hybrids for slow-growing parental strains and cases in which the mean BC1 growth regressed halfway between the hybrid and the backcross parent.
Overdominance refers to an intralocus interaction between alleles, whereas epistasis refers to the interaction between alleles of different loci. Findings supporting the contribution of these interactions to heterosis were lack of correlation between genetic distances and heterosis levels and differences in heterotic partners among genetically similar strains, as well as among strains originated from similar ecological niches, regression of more than halfway for the means of BC1 populations and heterosis in hybrids of well-performing parental strains.
Previous studies involving yeast hybrids have observed heterosis (Timberlake et al., 2011; Zorgo et al., 2012), but not to the extent reported here. The presence of a single loss-of-function mutation per hybrid is consistent with finding many partially and fully dominant hybrids, as suggested by Zorgo et al. (2012), but not with the amount of overdominant hybrids found here. Thus, our study indicates that more than major loss-of-function mutations contribute to genetic variation in yeast strains. Loss-of-function is an extreme form of deleterious variation. A computational analysis reported that 12% of the coding and 20% of the noncoding variations in yeast are deleterious (Doniger et al., 2008). The abundance of deleterious variations and the prevalence of dominance in hybrids are two sides of the same coin (Charlesworth and Willis, 2009) contributing to the detected extent of heterosis. Deleterious mutations can contribute to heterosis by dominance complementation and also by other mechanisms. A good example of this comes from heterosis in tomatoes, where heterozygosity with a loss-of-function allele in a single locus yields an overdominant phenotype (Krieger et al., 2010).
Our results therefore support a more complex view of the yeast genetic makeup that better explains the prevalence of heterosis. This complex view is evident from several of our findings. As the strains used in this study diverged considerably in sequence and originated from a variety of ecological niches (Liti et al., 2009), many new allelic combinations, which often led to hybrid advantage, were formed in their crosses. Considerable differences in allele content were even found between strains from similar ecological niches as indicated here by their different heterotic profiles and previously by their differences in growth rate (Ziv et al., 2013). Furthermore, the abundance of deleterious mutations supported the prevalence of heterosis, but in a complex way, rather than in accordance with the simple expectation that larger divergence is more likely to yield heterosis. Finally, we provided evidence for all three major genetic models—dominance complementation, overdominance and epistasis—taking part in yeast heterosis. Therefore, viewing heterosis as a trait conditioned by a complex genetic basis and dominated by nonadditive effects is consistent with our findings.
Our work establishes a basis for further understanding aspects of heterosis with significance beyond yeast. From this collection, specific hybrids can be chosen to identify specific genes that contribute to heterosis by any of the discussed models. Yeast, for which more advanced genetic tools and methodologies exist compared with crop plants and farm animals, can then be used to study the molecular basis of heterosis and especially of overdominant and epistatic interactions. Studying heterosis in yeast is expected to enhance our mechanistic understanding of heterosis genetics and to decipher the ways in which it can be applied to increase food production.
Data archiving
Three Supplementary Tables containing the DT data of three experiments, namely, strains under five conditions (Supplementary Table S3), 7 × 37 BC1 population (Supplementary Table S4) and 38 × 37 BC1 population (Supplementary Table S5) are deposited and available from the Dryad Digital Repository: doi:10.5061/dryad.4j07v.
References
Baranwal VK, Mikkilineni V, Zehr UB, Tyagi AK, Kapoor S . (2012). Heterosis: emerging ideas about hybrid vigour. Expl Bot 63: 6309–6314.
Botstein D, Chervitz SA, Cherry JM . (1997). Yeast as a model organism. Science (New York, NY) 277: 1259–1260.
Bruce AB . (1910). The mendelian theory of heredity and the augmentation of vigor. Science (New York, NY) 32: 627–628.
Chao A, Lin CW . (2012). Nonparametric lower bounds for species richness and shared species richness under sampling without replacement. Biometrics 68: 912–921.
Charlesworth D, Willis JH . (2009). The genetics of inbreeding depression. Nat Rev Genet 10: 783–796.
Chen ZJ . (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71.
Cheres MT, Miller JF, Crane JR, Knapp SJ . (2000). Genetic distance as a predictor of heterosis and hybrid performance within and between heterotic groups in sunflower. Theoret Appl Genet 100: 889–894.
Crow JF . (1948). Alternative hypotheses of hybrid vigor. Genetics 33: 477–487.
Darwin CR . (1876) The Effect of Cross and Self-Fertilization in the Vegetable Kingdom. John Murray: London.
Dean EJ, Davis JC, Davis RW, Petrov DA . (2008). Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet 4: e1000113.
Doniger SW, Kim HS, Swain D, Corcuera D, Williams M, Yang S-P et al. (2008). A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet 4: e1000183.
East EM . (1908). Inbreeding in corn. In: Reports of the Connecticut Agricultural Experiment Station for Years 1907–1908 pp 419–428.
Gietz RD, Woods RA . (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In: Guthrie C, Fink GR (eds). Guide to Yeast Genetics and Molecular and Cell Biology, Part B Vol. 350, Elsevier Science: San Diego, CA, USA. pp 87–96.
Goff SA . (2011). A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. N Phytol 189: 923–937.
Guo X, Guo Y, Ma J, Wang F, Sun M, Gui L et al. (2013). Mapping heterotic loci for yield and agronomic traits using chromosome segment introgression lines in cotton. J Integr Plant Biol 55: 759–774.
Hedgecock D, Lin JZ, DeCola S, Haudenschild CD, Meyer E, Manahan DT et al. (2007). Transcriptomic analysis of growth heterosis in larval Pacific oysters (Crassostrea gigas). Proc Natl Acad Sci USA 104: 2313–2318.
Hillenmeyer ME, Ericson E, Davis RW, Nislow C, Koller D, Giaever G . (2010). Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol 11: R30.
Jones DF . (1917). Dominance of linked factors as a means of accounting for heterosis. Genetics 2: 466–479.
Krieger U, Lippman ZB, Zamir D . (2010). The flowering gene single flower truss drives heterosis for yield in tomato. Nat Genet 42: 459–463.
Li W, Zhu H, Challa GS, Zhang Z . (2013). A non-additive interaction in a single locus causes a very short root phenotype in wheat. Theoret Appl Genet 126: 1189–1200.
Li Y, Zhang X, Ma C, Shen J, Chen Q, Wang T et al. (2012). QTL and epistatic analyses of heterosis for seed yield and three yield component traits using molecular markers in rapeseed (Brassica napus L.). Genetika 48: 1171–1178.
Li ZK, Luo LJ, Mei HW, Wang DL, Shu QY, Tabien R et al. (2001). Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158: 1737–1753.
Lippman ZB, Zamir D . (2007). Heterosis: revisiting the magic. Trends Genet 23: 60–66.
Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA et al. (2009). Population genomics of domestic and wild yeasts. Nature 458: 337–341.
Luo LJ, Li ZK, Mei HW, Shu QY, Tabien R, Zhong DB et al. (2001). Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158: 1755–1771.
McClintock B . (1984). The significance of responses of the genome to challenge. Science (New York, NY) 226: 792–801.
McCusker JH, Clemons KV, Stevens DA, Davis RW . (1994). Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136: 1261–1269.
Melchinger AE, Utz HF, Piepho HP, Zeng ZB, Schon CC . (2007). The role of epistasis in the manifestation of heterosis: a systems-oriented approach. Genetics 177: 1815–1825.
Melchinger AE, Utz HF, Schon CC . (2008). Genetic expectations of quantitative trait loci main and interaction effects obtained with the triple testcross design and their relevance for the analysis of heterosis. Genetics 178: 2265–2274.
Richards KD, Goddard MR, Gardner RC . (2009). A database of microsatellite genotypes for Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 96: 355–359.
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S et al. (2012). A travel guide to Cytoscape plugins. Nat Methods 9: 1069–1076.
Schacherer J, Ruderfer DM, Gresham D, Dolinski K, Botstein D, Kruglyak L . (2007). Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS One 2: e322.
Schnable PS, Springer NM . (2013). Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64: 71–88.
Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N et al. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103: 12981–12986.
Sinha H, Nicholson BP, Steinmetz LM, McCusker JH . (2006). Complex genetic interactions in a quantitative trait locus. PLoS Genet 2: e13.
St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW et al. (2007). Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39: 199–206.
Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH et al. (2002). Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Timberlake WE, Frizzell MA, Richards KD, Gardner RC . (2011). A new yeast genetic resource for analysis and breeding. Yeast 28: 63–80.
Warringer J, Zorgo E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT et al. (2011). Trait variation in yeast is defined by population history. PLoS Genet 7: e1002111.
Xiao J, Li J, Yuan L, Tanksley SD . (1995). Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754.
Yao H, Dogra Gray A, Auger DL, Birchler JA . (2012). Genomic dosage effects on heterosis in triploid maize. Proc Natl Acad Sci USA 110: 2665–2669.
Zhang T, Ni X-l, Jiang K-f, Deng H-f, He Q, Yang Q-h et al. (2010). Relationship between heterosis and parental genetic distance based on molecular markers for functional genes related to yield traits in rice. Rice Sci 17: 288–295.
Ziv N, Siegal ML, Gresham D . (2013). Genetic and nongenetic determinants of cell growth variation assessed by high-throughput microscopy. Mol Biol Evol 30: 2568–2578.
Zorgo E, Gjuvsland A, Cubillos FA, Louis EJ, Liti G, Blomberg A et al. (2012). Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol Biol Evol 29: 1781–1789.
Acknowledgements
We thank Dr Gianni Liti for generously providing the sequenced wild-type yeast strains, Prof Avigdor Cahaner for his advice on the variance components analysis of the diallel structure, Prof Joseph Hillel for his critical discussion on the manuscript and the three anonymous reviewers for their constructive comments who improved this paper. This research was supported by Israeli Science Foundation Grant Nos. 928/07 and 95/08 (Bikura program) to LD. SS was supported by an intramural grant from the Lejwa Fund of the Hebrew University of Jerusalem.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Shapira, R., Levy, T., Shaked, S. et al. Extensive heterosis in growth of yeast hybrids is explained by a combination of genetic models. Heredity 113, 316–326 (2014). https://doi.org/10.1038/hdy.2014.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2014.33
This article is cited by
-
Simulation of heterosis in a genome-scale metabolic network provides mechanistic explanations for increased biomass production rates in hybrid plants
npj Systems Biology and Applications (2019)
-
Hybridization is a recurrent evolutionary stimulus in wild yeast speciation
Nature Communications (2019)
-
Multiple interacting QTLs affect disease challenge survival in common carp (Cyprinus carpio)
Heredity (2019)
-
Heterosis as a consequence of regulatory incompatibility
BMC Biology (2017)
-
Novel brewing yeast hybrids: creation and application
Applied Microbiology and Biotechnology (2017)