Abstract
Elucidation of the evolutionary processes that constrain or facilitate adaptive divergence is a central goal in evolutionary biology, especially in non-model organisms. We tested whether changes in dynamics of gene flow (historical vs contemporary) caused population isolation and examined local adaptation in response to environmental selective forces in fragmented Rhododendron oldhamii populations. Variation in 26 expressed sequence tag-simple sequence repeat loci from 18 populations in Taiwan was investigated by examining patterns of genetic diversity, inbreeding, geographic structure, recent bottlenecks, and historical and contemporary gene flow. Selection associated with environmental variables was also examined. Bayesian clustering analysis revealed four regional population groups of north, central, south and southeast with significant genetic differentiation. Historical bottlenecks beginning 9168–13,092 years ago and ending 1584–3504 years ago were revealed by estimates using approximate Bayesian computation for all four regional samples analyzed. Recent migration within and across geographic regions was limited. However, major dispersal sources were found within geographic regions. Altitudinal clines of allelic frequencies of environmentally associated positively selected outliers were found, indicating adaptive divergence. Our results point to a transition from historical population connectivity toward contemporary population isolation and divergence on a regional scale. Spatial and temporal dispersal differences may have resulted in regional population divergence and local adaptation associated with environmental variables, which may have played roles as selective forces at a regional scale.
Similar content being viewed by others
Introduction
Climate change can have a strong influence on forest species distributions, and a 1500- to 1600-m upward migration of forests in Taiwan after the last glacial maximum has been reported (Liew and Chung, 2001). This upward shift in forest species distributions would have reduced the availability of suitable habitats, because numerous species and genera continued to grow at the same latitude, but shifted to a different elevational range. In 1920–1970, logging and cultivation of crops led to deforestation and forest degradation. Moreover, a survey of forest and land resources in Taiwan in 1995 reported that the area of high-stock forests had substantially decreased since a previous survey in 1978 because of the conversion of forest areas to agriculture, reforestation using foreign species (mainly Cryptomeria japonica) and the establishment of forest recreation areas (Taiwan Forestry Bureau, 1995). Many species are now confined to fragmented woodland remnants. Therefore, forests in Taiwan have been forced into smaller and more-isolated fragments due to climate change and anthropogenic activities that have altered natural landscapes, suggesting a transition of species’ distributions from historical connectivity to contemporary isolation.
Past demographic processes, such as historical bottlenecks and range expansions, and selection events have important roles in shaping patterns of genetic diversity in tree species (for example, Ingvarsson, 2008; Lascoux et al., 2008). Fitness and the adaptive potential of small populations are thought to be reduced due to the erosion of genetic variability (Reed and Frankham, 2003). Fragmentation and small effective population sizes caused by habitat loss can result in increased effects of genetic drift, inbreeding and restricted gene flow, which may threaten species survival. When selection is weak and there is a small amount of gene flow, homogenization of gene pools can theoretically occur (Crow and Kimura, 1970), and this often reduces the potential for populations to adapt and evolve to new environments (Bridle and Vines, 2006). Many plants in small and isolated populations produce limited local adaptation in response to changing environments, and usually harbor lower amounts of genetic variation (Leimu and Fischer, 2008). Nevertheless, local adaptation can occur even when the rate of gene flow is high (Von Wettberg et al., 2008). Gene flow may be asymmetrical with higher migration rates from central to peripheral populations (Holliday et al., 2012). Although in a species’ range, peripheral populations may harbor lower levels of genetic diversity than central populations (Eckert et al., 2008), they may also serve as sources of evolutionary novelty (Hampe and Petit, 2005; Parisod and Joost, 2010; Holliday et al., 2012). Selection, gene flow and genetic drift are all important factors that have roles in the adaptive phenotypic divergence among populations (Räsänen and Hendry, 2008). Disentangling the effects of genetic drift from selection following population discontinuity is crucial to understand the adaptation process. Selection can be empirically tested by examining the magnitude of the effect and mode of gene action at loci that underlie phenotypic evolution related to adaptations to various environments (Storz, 2005; Nosil et al., 2009).
R. oldhamii Maximowicz classified in the subgenus Tsutsusi is a subtropical broadleaf species endemic to Taiwan. R. oldhamii is presently widespread but discontinuously distributed in lowlands and up to 2500 m in humid environments in the understory of broadleaf forests. Differences in flowering times have been found between northern (January–May), central (July–October) and southern (June–August) populations of R. oldhamii in a survey of herbarium specimen records (Chang, 2006). Recent intensive field studies of populations showed that northern populations flower during the spring, whereas southern populations flower heterogeneously at different times of the year (Chi, 2009). Flowering time differences between populations have also been reported in other Rhododendron species (Kudo, 1993; Kameyama et al., 2001; Mejías et al., 2002). The occurrence of reproductive isolation or reproductive incompatibility is related to assortative mating, because immigrants that flower at different times from those of local individuals cannot reproduce, and as a consequence no gene flow occurs (Gavrilets and Vose, 2007). Therefore, geographic variations in flowering rhythms of R. oldhamii suggest population isolation caused by reduced gene flow among populations. Evidence suggests a direct relationship between natural selection and assortative mating resulting from differences in flowering times (Jiménez-Ambriz et al., 2007), and local adaptations associated with flowering time genes have also been reported (Keller et al., 2011, 2012).
The widespread distribution of R. oldhamii throughout geographically discontinuous habitats implies that some forms of historical connectivity existed between those habitats, and can be used to exemplify the fragmentation effects of postglacial climate change and recent anthropogenic influences on subtropical forest species in Taiwan. If habitat fragments and populations are isolated from one another due to landscape alterations, gene flow can be hindered and populations may become genetically differentiated over time. Moreover, isolated populations can become inbred, resulting in reduced fitness. Population genetic variability has key roles in the future evolutionary potential of an organism to a changing environment, because loss of genetic diversity in small populations renders them vulnerable to stochastic environmental conditions. However, R. oldhamii populations residing in a large range of elevations might have evolved local adaptations to a wide array of environmental conditions. To examine the influences of fragmentation and habitat loss, we genotyped 337 R. oldhamii individuals from 18 localities using 26 expressed sequence tag-simple sequence repeat (EST-SSR) loci to analyze the spatial and temporal dynamics of gene flow and selection in association with environmental variables in R. oldhamii populations. Our specific aims were to (1) examine whether population fragmentation resulting from altered natural landscapes caused reduced genetic diversity in R. oldhamii populations; (2) determine whether contemporary population isolation has occurred in relation to environmental heterogeneity; and (3) investigate whether ecologically relevant adaptive divergence has occurred in R. oldhamii populations.
Materials and methods
Sampling and genotyping
Young, healthy leaves of R. oldhamii plants from 18 localities (n=337) were collected for DNA extraction (Doyle and Doyle, 1987; Table 1, Figure 1a). Twenty-six EST-SSR primer pairs were used (Supplementary Table 1). The forward primer of each primer pair was 5′-end-labeled with a 6-carboxy-fluorescein (6-FAM), hexachloro-fluorescein (HEX) or tetramethyl-6-carboxyrhodamine (TAMRA) fluorescent tag. PCR was performed in a 10-μl reaction volume containing 20 ng template DNA, 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 0.01% (v/v) Tween-20, 2.5 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphate mix, 75 nM of each primer, 0.8 μg bovine serum albumin and 0.5 U of Simpler Red Taq DNA polymerase (Thermo Scientific, Waltham, MA, USA). The PCR was performed as follows: 94 °C for 10 min, 38 cycles of 30 s at 94 °C, 30 s at 45–62 °C (Supplementary Table 1) and 30 s at 72 °C, followed by a final extension of 5 min at 72 °C. PCR products were run on a MegaBACE 1000 (Amersham Biosciences, Piscataway, NJ, USA) and sizes of alleles were scored using Genetic Profiler. The presence of null alleles, stuttering and large allele drop out was checked using MICRO-CHECKER v2.2.3 with 1000 randomizations (van Oosterhout et al., 2004).
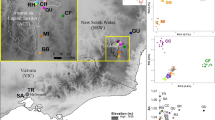
Map of sampling sites and bar plots of the STRUCTURE analysis representing assignments of genotypes to 18 Rhododendron oldhamii populations. See Table 1 for population codes. (a) R. oldhamii populations sampled in Taiwan. Populations colored black, dark gray, white and light gray, respectively, correspond to north (N), central (C), south (S) and southeast (SE) regional groups, as revealed by the Bayesian clustering results of the STRUCTURE analysis. (b) Bar plots represent assignments of genotypes for 18 R. oldhamii populations into N and S groups, the clustering of populations of the northern group into the N and C regional groups and the clustering of populations of the southern group into the S and SE regional groups. (c) Log likelihood and changes in the log likelihood for different scenarios of groupings were based on 21 microsatellite loci derived from expressed sequence tags of R. catawbiense.
Genetic diversity
Within each population at each locus, potential deviations from Hardy–Weinberg equilibrium (HWE) was tested by applying a modified Fisher’s exact test (Guo and Thompson, 1992) with 107 Markov chain iterations using GENEPOP v4.0 (Raymond and Rousset, 1995). Linkage disequilibrium between each pair of loci was tested with a likelihood-ratio statistic based on 5 × 107 Markov chain iterations implemented in GENEPOP. Bonferroni correction was applied for multiple comparisons (Rice, 1989). Standardized allelic richness (AR) was estimated using the program HP-RARE (Kalinowski, 2005). The number of alleles (AT), and observed (HO) and expected (HE) heterozygosity were analyzed using MSA v4.05 (Dieringer and Schlötterer, 2003). Inbreeding coefficient (FIS) was calculated based on neutral loci only, after removing selective outliers (see ‘Results’) using FSTAT v2.9.3.2 (Goudet; http://www2.unil.ch/popgen/softwares/fstat.htm) and deviation from HWE was estimated using a probability test (Goudet, 2001).
Environmental data
From 390 meteorological stations of the Central Weather Bureau of Taiwan, we obtained data on 11 environmental variables recorded in 1990–2011. The variables were mean temperature, maximum temperature, minimum temperature, mean wind speed, precipitation (PRE), relative humidity, cloud cover, time of sunshine, days of maximum temperature >30 °C (D30), days of minimum temperature <10 °C (D10) and wet days (RainD, that is, number of days with >0.1 mm rain per month). These environmental variables in relation to temperature, PRE and the monsoon were selected for analysis because they were reported to have major roles in vegetation type differences in Taiwan (Liew et al., 2006; Lee and Liew, 2009). A universal spherical model using the Kriging method in ArcGIS was used to interpolate values of environmental variables at an unobserved location from nearby meteorological observations (Carrera-Hernández and Gaskin, 2007). For each locality, monthly mean values of the 11 environmental variables covering a latitude/longitude grid within 1 km2 were extracted from their associated distribution maps/layers. A multivariate analysis of variance based on the Pillai–Bartlett statistic implemented in JMP v7.0 (SAS Institute, Salt Lake City, UT, USA) was used to test differences of the 11 environmental variables among all populations investigated.
Identification of selective outliers based on FST neutrality test methods
Several methods were developed to identify loci under selection (selective outliers) based on the FST neutrality test (Storz, 2005; Nosil et al., 2009). With these methods, the null distribution of FST under neutral expectations was generated using coalescent simulations. Loci with unusually high or low FST values that did not fit the neutral drift simulations were, respectively, recognized as positive or balancing selective outliers. In an effort to understand the adaptive divergence of R. oldhamii populations, we performed analyses using three different FST-based methods.
FDIST2, based on an infinite island model and robust to different demographic scenarios, was first used to identify selective outliers (Beaumont and Nichols, 1996; Beaumont and Balding, 2004). A neutral distribution of FST conditioned on heterozygosity was generated using 106 iterations of coalescent simulations based on the calculated FST for each sampled locus. Unusually high or low FST values conditioned on heterozygosity for each locus were, respectively, considered positive or balancing outliers. Significant P-values of outlier loci were evaluated at the 95% confidence level by applying the Bonferroni correction. Second, BAYESFST, which does not assume equal FST among study populations, was used. Potential selective outliers were evaluated by extracting posterior probability distributions of the locus effect (αi) based on 2000 samplings of Markov chain Monte Carlo simulations (Beaumont and Balding, 2004). Bonferroni correction was applied to determine the statistical significance of positive selection, if the 2.5% quantile locus effect was positive, and balancing selection, if the 97.5% quantile locus effect was negative. Three independent runs using different parameter values until convergence were applied to all study populations. Considering population subdivisions, the hierarchical island model test was further applied to identify selective outliers using a method similar to that of FDIST2 (Beaumont and Nichols, 1996; Excoffier et al., 2009) implemented in ARLEQUIN v3.5 (Excoffier and Lischer, 2010). Genetic differentiation calculated from the empirical data set was integrated into the hierarchical island model to detect outliers across all populations, and 95% confidence limits were extracted from 5 × 105 simulated loci.
Test for associations of EST-SSR alleles with environmental variables
Environmental variables that might have played roles as selective forces were uncovered by the spatial analysis method (Joost et al., 2008). In practice, spatial analysis method uses univariate logistic regressions to test associations of allelic frequencies at marker loci with values of environmental variables. All possible pairwise combinations of alleles vs environmental variables were considered. P-values of both the likelihood ratio and Wald tests with Bonferroni correction were used to evaluate the significance of the associations.
Genetic structure
Genetic structure and demographic analyses were performed using 21 neutral loci (excluding selective outliers, see ‘Results’). Pairwise FST was estimated and the significance was tested by Fisher’s exact tests after 5 × 104 permutations using ARLEQUIN v3.5 (Excoffier and Lischer, 2010). Moreover, the hierarchical genetic structure of the data was partitioned into among-group, within-group and within-population components of the total analysis of molecular variance with 5 × 104 permutations using ARLEQUIN. Bayesian clustering was used for the purpose of inferring the population structure using STRUCTURE v2.2 (Pritchard et al., 2000). For each level of K (=1–19), 10 complete analyses each with 106 iterations and 105 burn-in steps, containing population information, were performed under a no-admixture model. Different clustering scenarios were evaluated using the mean log probability (L(K); Pritchard et al., 2000) and the change in log probability, ΔL(K) (Evanno et al., 2005).
Migration and drift
To test the fit of migration and drift to the evolutionary dynamics of contemporary R. oldhamii populations, relative roles of immigration vs drift were assessed using the 2MOD program (Ciofi et al., 1999). The 2MOD program evaluates: (1) the gene flow model, which assumes drift-immigration equilibrium of gene frequencies within populations, and (2) the drift model, which assumes that drift is the sole reason for population divergence since some time in the past.
To understand long-term migration rates among regional populations and among populations within a given region, we used MIGRATE-N v3.1.3 (http://popgen.sc.fsu.edu/Migrate/Migrate-n.html). Long-term migration rates, M, 4Ne generations in the past based on a coalescent approach, were estimated using the maximum-likelihood mode. MIGRATE-N tests all possible combinations of symmetrical or no migration between each regional population group and between populations within a region with the likelihood ratio test. MIGRATE-N estimates the long-term effective population size (Ne) of each population group and populations within a region as parameter θ (that is, 4Neμ, where μ is the mutation rate per site). MIGRATE-N also estimates the bi-directional M among population groups and among populations within a region (M=m/μ, where m is the immigration rate per generation). In practice, we sampled 1 of every 200 reconstructed genealogies for each locus in the maximum-likelihood runs for 10 short and 3 long chains. For short and long chains, 1000 and 104 genealogies were, respectively, recorded, with the first 200 and 2000 genealogies discarded as burn-in.
To characterize recent migration among R. oldhamii populations and among population groups, rates and directions of dispersal that occurred in the last few generations were estimated using a Bayesian Markov chain Monte Carlo approach in BAYESASS v1.3 (Wilson and Rannala, 2003). This software assesses proportions of individuals assigned to their hypothesized population (population group) and to other populations (other population group). The Markov chain Monte Carlo chain consisted of 3 × 107 iterations with a sampling frequency of 2 × 104, and the first 107 steps were discarded as burn-in.
Changes in population sizes
To understand whether R. oldhamii populations experienced bottlenecks in the distant past, we estimated the times of the beginning and end of bottlenecks separately for the four sets of data according to STRUCTURE groupings (see ‘Results’) using the approximate Bayesian computation (ABC) implemented in the software DIYABC v1.04.33beta (Cornuet et al., 2008, 2010). Two scenarios including one assuming constant population size and one allowing a population size change at t generations in the past were defined. A generalized stepwise mutation model of microsatellite mutation (Estoup et al., 2002) with a mean rate uniformly distributed between 10−4 and 10−3 substitutions/generation were used. To avoid any influence from limiting prior assumptions on posterior distributions, we simulated the two models using a broad prior for each parameter. In the bottleneck scenario, the prior on Ne-ancestral was bounded between 1 and 105. The prior on Ne-bottleneck was bounded between 1 and 1000. The Ne-contemporary was bounded between 1 and 105. To estimate the time of bottleneck events (in the number of generations) for the four regional groups of R. oldhamii, we set the time when the bottlenecks began and ended, respectively, up to 24 000 and 12 000 years ago in the Pleistocene based on a generation time of 12 years for Rhododendron plants (Cross, 1975). The time associated with the beginning of the bottleneck defined as Tbottleneck-start was bounded between 1 and 2000 generations, and the time associated with the end of the bottleneck defined as Tbottleneck-end was bounded between 1 and 1000 generations. In the scenario of a constant population size, Ne-constant was bounded between 1 and 105 through time (1–2000 generations). All distributions of parameters mentioned above were uniform. For each model, 106 genetic data sets were simulated with the parameters described above. We used four summary statistics for the observed and simulated data sets: the mean number of alleles, the mean heterozygosity, the mean allelic size variance and the mean M-ratio index (Garza and Williamson, 2001). We kept the 104 data sets with the smallest normalized Euclidean distances under the local linear regression method of Beaumont et al. (2002) to build posterior parameter distributions using Locfit 2.0 (http://cran.r-project.org/web/packages/locfit/) in the R environment (R Development Core Team, 2010). The 95% confidence intervals (CIs) for the posterior probability of each scenario were estimated using a logistic regression approach (Fagundes et al., 2007; Cornuet et al., 2008). Type I and II error rates and statistical measures of performance, including the mean relative bias and the root of the relative mean square error, were calculated for model checking (Excoffier et al., 2005).
Results
Genetic diversity
In total, 232 alleles were found across 18 R. oldhamii populations over the 26 analyzed loci. In a total of 468 population-locus pairwise comparisons, 4 were found to significantly depart from HWE and 6 showed significant linkage disequilibrium after Bonferroni correction. No systematic departure from HWE or linkage disequilibrium was found. The number of alleles per locus ranged 2–28. The average number of alleles per population was lowest at Renluen (RL; 2.154) and highest at Chiayang (CY, 3.962; Table 1). The mean value of AR for each population ranged 1.958 (RL) to 2.691 (Huoyanshan, HYS). HE ranged 0.240 (RL) to 0.336 (Wuling, WL). HO ranged 0.217 (Wushe, WS) to 0.337 (WL). FIS ranged 0.082 (CY) to 0.178 (WS) with significant departures from HWE toward homozygote excess observed in the Lushan (LS) and WS populations. No correlations of altitude with AR, HE, HO or FIS were found (Spearman’s r=0.215, 0.046, 0.434 and 0.072; P=0.984, 0.388, 0.841 and 0.764; respectively). Moreover, these population genetic parameters also had no correlations with longitude or latitude (longitude: Spearman’s r=0.216, −0.060, 0.066 and −0.112; P=0.368, 0.810, 0.771 and 0.652; latitude: Spearman’s r=0.184, 0.027, −0.058 and 0.124; P=0.441, 0.904, 0.810 and 0.603; respectively).
Environmental variables and selective outliers
The multivariate analysis of variance showed significant differences in all environmental variables except PRE among R. oldhamii populations based on the Pillai–Bartlett statistic (V=4.1892, F187, 2178=7.1639, P<2.2e-16) at P<0.05 (Supplementary Figure 1, Supplementary Table 2). Congruent evidence for positive selection at two loci (5643 and 5685) and for balancing selection at three loci (1655, 1915 and 6455) was found using different detection methods (Table 2). Positively selected loci showed strong associations with a number of environmental variables such as temperature, wind speed, PRE, cloud cover and the number of wet days at a significance threshold of 2.993E-10 corresponding to a 99.999% CI. Moreover, protein-coding gene sequences of the two positively selected loci 5643 and 5685 were functionally annotated to the receptor protein kinase-like protein and cystatin-related domain protein with respective E-values of 2E-04 and 7E-14, using the BlastX tool of NCBI (Supplementary Table 1).
Genetic structure
Based on 21 neutral loci, a global analysis of pooled data from all populations showed that individuals of the 18 R. oldhamii populations, although with some evidence for admixture, could be assigned to two separate groups (Figure 1b), that is, northern and southern, according to changes in the log probability (ΔL(K); Figure 1c). However, as Evanno’s ΔL(K) method only finds the uppermost level of structure in a given data set, we then separately reanalyzed the northern and southern groups. Subdivision into two separate clusters each, with some admixing, for the northern and southern groups was revealed (Figure 1b). One major cluster in the northern group, which was comprised of most of the northern populations, was classified as the north regional group, and included populations Ergirshan (EGS), Baling (BL), Tsanguanliao, Tsaigongkeng (TGK), Shihtoushan, Wuliaojian and HYS. Population WL formed an independent cluster from populations assigned to the northern group and was classified as a central regional group. The third major cluster constituted most populations of the southern group classified as the south regional group, which contained populations CY, Chingjing (CJ), LS, WS, Chungheng, Hueisun, RL and Leleku. The southeast (SE) regional group comprised populations Yeinping (YP) and Wuru (WR) of the southern group. Variable levels of genetic diversity estimates for the four regional groups were found (Supplementary Table 3). Populations of the north and south regional groups had similar levels of heterozygosity (Wilcoxon two-sample test, P=0.355), but south regional populations had significantly lower levels of allelic richness (Wilcoxon two-sample test, P=0.049).
The majority of genetic variation was partitioned within populations, based on either two or four regional groups (84.70% or 84.63%, respectively; Supplementary Table 4). Genetic variation partitioned among populations was significant for both two- and four-regional groups (ΦST=0.15305, P<0.001; ΦST=0.15370, P<0.001, respectively). Moreover, most pairwise FST estimates were significant after applying Bonferroni correction, except a few comparisons between populations within the same regional group (Supplementary Table 5). Estimates of the pairwise FST were also significant for four regional samples, for which the SE regional group most greatly diverged from the other regional groups (Supplementary Table 6). Furthermore, the grouping of four geographic regional clusters was further supported by the significant analysis of molecular variance (ΦCT=0.0988, P<0.001; Supplementary Table 4).
Gene flow and bottlenecks
The test of the population history using 2MOD strongly supported a long-term gene flow model against a pure drift model (P (gene flow)=1.00, P (drift)=0.00), indicating that R. oldhamii populations have been under drift-immigration equilibrium for a long period of time. Historical migration rates (M) calculated using MIGRATE-N revealed significant asymmetrical migration among the four regional groups (Figure 2) and among populations within the north, south and SE regional groups according to non-overlapping 95% CIs (Supplementary Figure 2). Estimating historical migration rates for total populations requires large computing times, so we calculated pairwise comparisons within the north, south and SE regional samples separately (because only one population was in the central region, no calculation was performed).
Graphical representation of results of migrations estimated using MIGRATE-N among the four regional population groups. Maximum-likelihood estimates and 95% confidence intervals (in parentheses) of the long-term migration rate (M) and mutation-scaled effective population size (θ) using the MIGRATE-N program are shown. Bold values indicate that migration was asymmetrical as determined from non-overlapping 95% confidence intervals; arrows represent the direction of gene flow among regions or among sites. C, central; N, north; S, south; SE, southeast.
Estimates of M among regional samples ranged 0.77–5.14 and averaged 2.30. Lower values (0.77) of M were found for dispersal out of the central regional group; however, M values of >1 were found for other estimates of among-region migrations (Figure 2). Long-term θ values for the four regional groups were comparable and ranged 0.92–1.04. Nm (the product of θ and M divided by 4) estimated with MIGRATE-N was low among the four regional groups and ranged 0.18–1.30. When separately analyzed using data of the north, south and SE regional groups, respective average long-term migration rates among populations within regional groups were 1.246 (0.51–2.25), 1.067 (0.25–2.35) and 1.054 (0.91–1.20) (Supplementary Figure 2a–c, respectively). Respective average long-term θ values for the north, south and SE regional groups were 0.876 (0.77–0.99), 0.804 (0.62–0.90) and 0.847 (0.79–0.91). Moreover, respective average long-term Nm values for the north, south and SE regional groups were 0.276 (0.098–0.557), 0.218 (0.039–0.529) and 0.238 (0.180–0.273).
Frequent among-region long-term migration rates were reflected in the high proportions of common alleles found among regional groups (Figure 3). However, regional group-specific alleles were not uncommon either and reached >20% in the north and south regional groups. Furthermore, alleles of positively selected loci 5643 and 5685 were also found to be population and region specific (Figure 4). Allele 262 of locus 5643 and alleles 159 and 165 of locus 5685 showed significant positive correlations of altitude with allelic frequency (Spearman’s r=0.513, 0.472 and 0.552; P=0.029, 0.040 and 0.018, respectively), whereas allele 163 of locus 5685 showed a significant negative correlation of altitude with allelic frequency (Spearman’s r=−0.541, P=0.025).
Moreover, estimates of recent migration among populations mostly overlapped 0 at the 95% CI, indicating little or no contemporary migration except for significant recent migration detected within regional groups from specific sources using BAYESASS (Supplementary Table 7). Population TGK served as a migration source within the north regional group, and population CY served as a migration source within the south regional group. Recent migration was also estimated using data from the four regional-groups. No recent among-region migration was observed except from the north to central regional group with a mean of 0.293 (95% CI: 0.24–0.33), which was significant based on the 95% CI for a significant migration rate (1.55E-4, 0.218).
In order to test whether populations of R. oldhamii regional groups had experienced bottlenecks in the distant past, we used an ABC framework to test two scenarios: (1) R. oldhamii populations experienced bottlenecks in the distant past; and (2) R. oldhamii populations have maintained constant population size. Historical bottlenecks evaluated using the ABC had posterior probabilities of 0.998–1, whereas the model of constant population sizes had posterior probabilities that ranged 0–0.0002. Type I and II error rates for selection of historical bottlenecks were 0.06 and 0.024 for north regional samples, 0.054 and 0.044 for central regional samples, 0.048 and 0.028 for south regional samples and 0.022 and 0.042 for SE regional samples, respectively. The posterior median number of generations when bottlenecks began for the north regional group was estimated to be 905 (95% confidence limits: 151, 1883; Supplementary Table 8). For the central regional group, the posterior median number of generations when bottlenecks began was estimated to be 988 (95% confidence limits: 217, 1908). For the south regional group, the posterior median number of generations when bottlenecks began was estimated to be 764 (95% confidence limits: 80, 1798). The time when population size decline began for the SE regional group was estimated to be 1091 (95% confidence limits: 287, 1949) generations. The posterior median number of generations when bottlenecks ended for the north regional group was estimated to be 180 (95% confidence limits: 2, 601). For the central regional group, the posterior median number of generations when bottlenecks ended was estimated to be 292 (95% confidence limits: 2, 740). For the south regional group, the posterior median number of generations when bottlenecks ended was estimated to be 226 (95% confidence limits: 3, 660). The posterior median number of generations when bottlenecks ended for the SE regional group was estimated to be 132 (95% confidence limits: 1, 495).
Discussion
Genetic diversity, bottlenecks and inbreeding
Maintenance of genetic variation in a population is important for long-term persistence, especially in the face of novel selective pressures (Amos and Balmford, 2001; Hedrick, 2004). In plants, genomic SSRs revealed much-higher genetic diversity than that revealed by EST-SSRs (for example, Chabane et al., 2005; Peleg et al., 2008). In this study, relatively lower polymorphism was detected using 26 pairs of EST-SSR primers (HE=0.284, ranged 0.240–0.336) in R. oldhamii populations compared with that utilizing genomic-derived SSR markers in R. brachycarpum (HE=0.815, ranged 0.676–0.913; Hirao, 2010) and R. ripense (HE=0.800, ranged 0.794–0.809; Kondo et al., 2009) of Japan, and R. simsii of China (HE=0.754, ranged 0.278–0.944; Tan et al., 2009). The level of R. oldhamii’s genetic diversity based on EST-SSRs is lower than averages observed for plants in general (0.61), endemic (0.42), widespread (0.62), outcrossing (0.65) and long-lived perennials (0.68) based on genomic-derived microsatellite data (Nybom, 2004). Moreover, R. oldhamii showed a relatively lower level of population genetic diversity detected by EST-SSRs compared with that of cold-temperate congeners of the R. pseudochrysanthum complex (average HE=0.394) and R. formosanum (average HE=0.395) that occur in Taiwan examined with a similar set of EST-SSRs (Liang et al., unpublished data).
Among 81 studies of plant species, 64% of those studies reported that central populations harbored higher levels of genetic diversity compared with peripheral populations (Eckert et al., 2008). Our results showed no general pattern consistent with the abundant-center hypothesis. It is likely that in Taiwan, species may have persisted in the same areas with smaller population sizes during the last glacial maximum because only certain mountain peaks in Taiwan were covered by ice sheets (Siame et al., 2007), and a general pattern of genetic diversity consistent with the abundant-center hypothesis attributed to postglacial recolonization was not revealed. Nevertheless, EST-SSR variations may have been targets of selection and represent functional divergence in response to environmental heterogeneity (Hoffmann and Sgrò, 2011).
ABC estimates of the time of population size declines suggest a long-term decline in Ne, which began after the end of the Pleistocene with a posterior median ranging 9168–13 092 years ago and ending in the late Holocene with a posterior median ranging 1584–3504 years ago for all four regional group samples analyzed. Postglacial bottlenecks were also reported in other long-lived plant species (for example, Tsuda and Ide, 2005; Holliday et al., 2010). However, estimates of when the time bottlenecks began and ended might not be correct due to the simplistic models implemented and analyses based on a few summary statistics in the ABC (for example, Ingvarsson, 2008).
Nevertheless, the generally low diversity observed in R. oldhamii reflects fragmentation (loss and isolation) caused by reductions in population sizes from previously larger contiguous populations in the distant past. Although current R. oldhamii populations as a whole may still be large, fragmentation at the range-wide scale is probable because no correlations of genetic diversity measures (AR, HE, HO and FIS) with altitude, longitude or latitude were found. Range-wide fragmentation is most likely related to postglacial range shifts caused by climate change. Moreover, substantial loss of allelic richness of south regional populations compared with north regional populations might not be related to differences in sampling sizes (Wilcoxon two-sample test, P=0.9315). It is probable that south regional populations suffered greater isolation and smaller genetic neighborhoods due to climate change and recent anthropogenic disturbances.
Given that Rhododendron species predominantly outcross via insect pollination (Ono et al., 2008; Hirao, 2010), negative FIS values that significantly deviate from HWE toward a heterozygote excess for R. oldhamii populations are expected. However, no significant negative FIS values were found for any R. oldhamii population. Positive FIS values were found for populations EGS, BL, CY, CJ, LS, WS, YP and WR, among which only populations LS and WS significantly departed from HWE toward a homozygote excess. Self-fertilization of Rhododendron plants has been reported to be inhibited due to seed abortion in the post-zygotic stage (Hirao, 2010). It is likely that selection against inbreeding may have played a role in mitigating the deleterious effects of small population sizes and resulted in our detecting significant excesses of homozygotes in only 2 of the 18 populations examined. Nevertheless, our results indicate that mating was restricted within populations, which might have been caused by an increasing selfing rate along ranges of plant species in response to climate change (Levin, 2012).
Historical connectivity and contemporary isolation
Genetic signatures of current range-wide patterns of genetic diversity often display influences of past vegetational changes in many plants (Hu et al., 2009). Multiple postglacial migration sources are common in many plant species in Taiwan, because plant species retreated into refugia in different geographic areas (Wu et al., 2006; Kuo et al., 2010). Our population estimates of θ from MIGRATE-N ranged 0.62–0.99 (Supplementary Figure 2a–c), which are comparable to those of long-lived Eucalyptus (Field et al., 2011) and annual Helianthus species (Kane et al., 2009). The long-term mutation-scaled Ne ranged 1550–2475 individuals per population, suggesting large long-term effective population sizes in R. oldhamii, when a mutation rate of 10−4 per allele per generation commonly found in plant microsatellites was used in the calculations (Marriage et al., 2009). Contemporary populations of R. oldhamii in Taiwan are possibly descendants of remnant populations surviving colder periods of the Pleistocene because population samples from different regional groups shared high proportions of common alleles. Similar sets of ancestral alleles might have persisted throughout glacial/interglacial periods, resulting in overall long-term inter-regional and intra-regional migration rates of >1. However, higher long-term inter-regional than intra-regional migration rates estimated by MIGRATE-N might have been caused by the high frequencies of common alleles that occurred among regions. Moreover, low levels of migrants and asymmetrical migrations especially among populations within geographic regions may have resulted from fragmentation due to an upward shift in range distributions inflicted by postglacial climate change and recent human interference. In addition, higher levels of emigration from south to north and from south to SE were found, which may be interpreted as consistent with more migrants from central to peripheral populations than the reverse (Holliday et al., 2012). Although asymmetrical migration is commonly found in natural systems, central populations functioning as main sources of gene flow should not be applied to the regional systems, because our results showed no general pattern of migrational direction among populations within regional groups.
Population pairwise FST levels within the north and south regional groups were relatively lower (average pairwise FST=0.049 and 0.088, respectively) compared with FST=0.124 averaged over all pairwise comparisons, suggesting a pattern of a relatively stable genetic constitution of populations over time within geographic regions. Moreover, significantly lower levels of average pairwise FST values (t-test, P=0.004) and a higher average long-term migration rate estimated by MIGRATE-N (Wilcoxon two-sample test, P=0.0448) among north regional populations than among south regional populations indicate higher historical migration rates among north regional populations. However, no significant difference in the θ estimated by MIGRATE-N was found between the north and south regional groups (Wilcoxon two-sample test, P=0.554). The higher level of historical migration might have been related to the more homogeneous and overlapping flowering times among north regional populations (Chi, 2009).
Forest fragmentation inflicted by postglacial climate change and recent anthropogenic activities may have been severe and have influenced patterns and magnitudes of gene flow among populations and among geographic regions of a species. Results of the 2MOD analysis strongly favored the gene flow model, suggesting that R. oldhamii populations are overall not genetically isolated, and that there is a sufficient amount of gene flow occurring between populations. However, genetic isolation was revealed especially among regional samples by significant levels of genetic differentiation among populations of regional groups (ΦCT=0.0988, average pairwise FST=0.1115). Contemporary isolation was further supported by, generally, no recent inter- or intra-regional migration detected by BAYESASS, which is the expected pattern of habitat destruction impeding gene exchange between and within geographic regions. The presence of regional- and population-specific alleles may have resulted from an ongoing process of isolation, or have been caused by the loss of those alleles in other populations or regions due to habitat destruction. Nevertheless, these results suggest declining gene flow within and among geographic regions across deforested areas attributable to both geographic topology (for example, roads, rivers, and mountains) and ecological factors (for example, unsuitable habitats), resulting in reduced population connectivity and increased population divergence. Therefore, historical population connectivity and contemporary isolation dictated the present genetic structure of R. oldhamii.
Ecologically relevant adaptive divergence
Loci detected as selective outliers by different methods should typically be true, whereas those identified by only one method could be false positives generated by an individual method (Foll and Gaggiotti, 2008). In this study, congruence was found and lends confidence to the selective outliers identified with different FST-based neutrality test methods. In addition, positively selected outliers were found to be strongly associated with a number of environmental variables, based on an analysis using the logistic regression approach using spatial analysis method. A correlation of population genetic variability with environmental variables is an indication of local adaptation and selection (Linhart and Grant, 1996). Our results suggest that ecologically relevant selective forces were involved in population adaptive divergence of R. oldhamii. Spatial heterogeneity in environmental variables, revealed by the multivariate analysis of variance, is expected to cause genetic divergence among populations, which may create sufficient variations in fitness-related traits and the consequent formation of local adaptations (for example, Eckert et al., 2010).
Population divergence might have been caused by random genetic drift as observed in allele 265 of locus 5643, which was nearly fixed in all populations of the north, central and SE regional groups. However, only intermediate frequencies of this allele were present in south regional populations, thus precluding drift as a factor causing divergence. On the other hand, intermediate frequencies of allele 262 of this locus were found in south regional populations, in contrast to only minor frequencies found in populations of other regional groups, suggesting positive selection of this allele among south regional populations. Similar situations were also found for alleles of locus 5685, which displayed differential levels of allelic frequencies in populations of different regional groups, suggestive of regional-specific selection at these alleles. Moreover, altitudinal clines of gene frequency may originate in response to an ecological gradient or as a result of long-range dispersal among populations with differing genetic compositions (Felsenstein, 1976). In R. oldhamii, altitudinal clines of allelic frequencies of positively selected loci were strongly associated with environmental variables, indicating rapid evolutionary adaptations counteracting climate change and fragmentation of natural landscapes (Hoffmann and Sgrò, 2011).
Moreover, offspring fitness of dispersed seeds to neighboring regions (populations) may have been low due to competition with local seedlings. A pattern of restricted dispersal of R. oldhamii could have been caused by small effective population sizes or possibly by the short distance Rhododendron seeds can disperse (Hirao, 2010) and ineffective pollen flow (Marshall et al., 2010), resulting in the beginning of reproductive isolation, and this may have facilitated within-population mating because of fragmentation and population divergence (Jiménez-Ambriz et al., 2007). Environmental variables might have played roles as selective forces, enforcing population adaptive divergence, in rapidly evolving small populations of R. oldhamii even when selection was weak. Populations that are demographically isolated and are responding to local selection may promote genetic divergence among populations of R. oldhamii.
Protein gene-coding sequences of the two positively selected loci had functional annotations corresponding to a receptor protein kinase-like protein and cystatin-related domain protein. The receptor protein kinase constitutes a diverse group of proteins that have roles in signal transduction pathways responding to extracellular environments essential for a variety of plant processes, including development (Becraft, 1998; Li, 2010) and abiotic stresses (Marshall et al., 2012). Receptor-like kinases are known to regulate cell wall function related to plant defense mechanism (Postel et al., 2010; Steinwand and Kieber, 2010). A local adaptation of leucine-rich repeat receptor-like kinases was implied (Diévart et al., 2011). The cystatin-related domain protein belongs to the cystatin superfamily, the members of which function as cysteine protease inhibitors that protect cells from unwanted proteolysis in response to invasion by plant pathogens (Turk et al., 2001; Turk et al., 2002). Moreover, site-directed mutagenesis at positively selected amino-acid sites of plant cystatin was found to enhance defense potency against herbivore attacks (Goulet et al., 2008). Both positively selected loci identified are involved in plant defenses against biotic stresses, and local adaptations involving plant resistance genes were also documented in R. formosanum, which is also endemic to Taiwan (Liao et al., 2012).
Conclusions
Long-term effects of postglacial warming might have caused low levels of population genetic diversity and significant genetic structuring in subtropical R. oldhamii populations. The general lack of recent detectable among-population migration in R. oldhamii within and among regions, reflecting contemporary population isolation, is probably due to habitat heterogeneities as evidenced by significant differences in many environmental variables. Our results suggest that R. oldhamii populations are experiencing a transition from historical population connectivity toward contemporary population isolation and divergence on a regional scale. Furthermore, differential spatial and temporal dispersals might have evoked genetic differences in populations of R. oldhamii in different geographic regions, resulting in local adaptations associated with environmental variables.
Data Archiving
Genotype data were deposited at Dryad: http://dx.doi.org/10.5061/dryad.nc221.
References
Amos W, Balmford A . (2001). When does conservation genetics matter? Heredity 87: 257–265.
Beaumont MA, Balding DJ . (2004). Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13: 969–980.
Beaumont MA, Nichols RA . (1996). Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263: 1619–1626.
Beaumont MA, Zhang W, Balding DJ . (2002). Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035.
Becraft PW . (1998). Receptor kinases in plant development. Trends Plant Sci 3: 384–388.
Bridle JR, Vines TH . (2006). Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol Evol 22: 140–147.
Carrera-Hernández JJ, Gaskin SJ . (2007). Spatio temporal analysis of daily precipitation and temperature in the basin of Mexico. J Hydrol 336: 231–249.
Chabane K, Ablett GA, Cordeiro GM, Valkoun J, Henry RJ . (2005). EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Resour Crop Evol 52: 903–909.
Chang Y-M . (2006) The investigation of the flowering pattern of Rhododendron oldhamii Maxim., MSc thesis, Providence University, Taichung, Taiwan.
Chi WT . (2009) The analysis of flowering rhythm and its relationship with the distribution of populations of Rhododendron oldhamii Maxim. in western Taiwan, MSc thesis, National Taiwan University, Taipei, Taiwan.
Ciofi C, Beaumont MA, Swingland IR, Bruford MW . (1999). Genetic divergence and units for conservation in the Komodo dragon Varanus komodoensis. Proc R Soc B 266: 2269–2274.
Cornuet J-M, Santos F, Beaumont MA, Robert CP, Marin J-M, Balding DJ et al. (2008). Inferring population history with DIYABC: a user-friendly approach to Approximate Bayesian Computations. Bioinformatics 24: 2713–2719.
Cornuet J-M, Ravigné V, Estoup A . (2010). Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinformatics 11: 401.
Cross JR . (1975). Biological flora of the British Isles: Rhododendron ponticum L. J Ecol 63: 345–364.
Crow JF, Kimura M . (1970) An Introduction to Population Genetics Theory. Harper and Row: New York.
Dieringer D, Schlötterer C . (2003). MICROSATELLITE ANALYSER: a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3: 167–169.
Diévart A, Gilbert N, Droc G, Attard A, Gourgues M, Guiderdoni E et al. (2011). Leucine-rich repeat receptor kinases are sporadically distributed in eukaryotic genomes. BMC Evol Biol 11: 367.
Doyle JJ, Doyle JL . (1987). A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull 19: 11–15.
Eckert AJ, van Heerwaarden J, Wegrzyn JK, Nelson CD, Ross-Ibarra J, González-Martínez SC et al. (2010). Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185: 969–982.
Eckert CG, Samis KE, Lougheed SC . (2008). Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17: 1170–1188.
Estoup A, Jarne P, Cornuet J-M . (2002). Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol 11: 1591–1604.
Evanno G, Regnaut S, Goudet J . (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Estoup A, Cornuet JM . (2005). Bayesian analysis of an admixture model with mutations and arbitraily linked markers. Genetics 169: 1727–1738.
Excoffier L, Hofer T, Foll M . (2009). Detecting loci under selection in a hierarchically structured population. Heredity 103: 285–298.
Excoffier L, Lischer HEL . (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567.
Fagundes NJR, Ray N, Beaumont MA . (2007). Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA 104: 17614–17619.
Felsenstein J . (1976). The theoretical population genetics of variable selection and migration. Ann Rev Genet 10: 253–280.
Field DL, Ayre DJ, Whelan RJ, Young AG . (2011). Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida. Heredity 106: 841–853.
Foll M, Gaggiotti O . (2008). A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993.
Garza JC, Williamson EG . (2001). Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10: 305–318.
Gavrilets S, Vose A . (2007). Case studies and mathematical models of ecological speciation. 2. Palms on an oceanic island. Mol Ecol 16: 2910–2921.
Goudet J . (2001) FSTAT, a program to estimate and test gene diversities and fixation indices. Version 2.9.3. URL http://www2.unil.ch/popgen/softwares/fstat.htm.
Goulet M-C, Dallaire C, Vaillancourt L-P, Khalf M, Badri AM, Preradov A et al. (2008). Tailoring the specificity of a plant cystatin toward herbivorous insect digestive cysteine proteases by single mutations at positively selected amino acid sites. Plant Physiol 146: 1010–1019.
Guo SW, Thompson EA . (1992). Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361–372.
Hampe A, Petit RJ . (2005). Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8: 461–467.
Hedrick PW . (2004). Recent developments in conservation genetics. For Ecol Manag 197: 3–19.
Hirao AS . (2010). Kinship between parents reduces offspring fitness in a natural population of Rhododendron brachycarpum. Ann Bot 105: 637–646.
Hoffmann AA, Sgrò CM . (2011). Climate change and evolutionary adaptation. Nature 470: 479–485.
Holliday JA, Suren H, Aitken SN . (2012). Divergent selection and heterogeneous migration rates across the range of Sitka spruce (Picea sitchensis). Proc R Soc B 279: 1675–1683.
Holliday JA, Yuen M, Ritland K, Aitken SN . (2010). Postglacial history of a widespread conifer produces inverse clines in selective neutrality tests. Mol Ecol 19: 3857–3864.
Hu FS, Hampe A, Petit RJ . (2009). Paleoecology meets genetics: deciphering past vegetational dynamics. Front Ecol Environ 7: 371–379.
Ingvarsson PK . (2008). Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180: 329–340.
Jiménez-Ambriz G, Petit C, Bourrié I, Dubois S, Olivieri I, Ronce O . (2007). Life history variation in the heavy metal tolerant plant Thlaspi caerulescens growing in a network of contaminated and noncontaminated sites in southern France: role of gene flow, selection and phenotypic plasticity. New Phytol 173: 199–215.
Joost S, Kalbermatten M, Bonin A . (2008). Spatial analysis method (SAM): a software tool combining molecular and environmental data to identify candidate loci for selection. Mol Ecol Res 8: 957–960.
Kalinowski ST . (2005). HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5: 187–189.
Kameyama Y, Isagi Y, Nakagoshi N . (2001). Patterns and levels of gene flow in Rhododendron metternichii var. Hondoense revealed by microsatellite analysis. Mol Ecol 10: 205–216.
Kane NC, King MG, Barker MS, Raduski A, Karrenber S, Yatabe Y et al. (2009). Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow below between divergent Helianthus species. Evolution 63: 2061–2075.
Keller SR, Levsen N, Ingvarsson PK, Olson MS, Tiffin P . (2011). Local selection across a latitudinal gradient shapes nucleotide diversity in balsam Poplar, Populus balsamifera L. Genetics 188: 941–952.
Keller SR, Levsen N, Olson MS, Tiffin P . (2012). Local adaptation in the flowering time gene network of balsam poplar, Populus balsamifera L. Mol Biol Evol 29: 3143–3152.
Kondo T, Nakagoshi N, Isagi Y . (2009). Shaping of genetic strucuture along Pleistocene and modern river systems in the hydrochorous riparian azalea, Rhododendron ripense (Ericaceae). Am J Bot 96: 1532–1543.
Kudo G . (1993). Relationship between flowering time and fruit set of the entomophilous alpine shrub, Rhododendron aureum (Ericaceae), inhabiting snow patches. Am J Bot 80: 1300–1304.
Kuo D-C, Lin C-C, Ho K-C, Cheng Y-P, Hwang S-Y, Lin T-P . (2010). Two genetic divergence centers revealed by chloroplastic DNA variation in populations of Cinnamomum kanehirae Hay. Conser Genet 11: 803–881.
Lascoux M, Pyhäjärvi T, Källman T, Savolainen O . (2008). Past demography in forest trees: what can we learn from nuclear DNA sequences that we do not already know? Plant Ecol Divers 1: 209–215.
Lee C-Y, Liew P-M . (2009). Late Quaternary vegetation and climate changes inferred from a pollen record of Dongyuan lake in southern Taiwan. Palaeogeo Palaeoclimat Palaeoecol 287: 58–66.
Leimu R, Fischer M . (2008). A meta-analysis of local adaptation in plants. PLoS ONE 3: e4010.
Levin DA . (2012). Mating system shifts on the trailing edge. Ann Bot 109: 613–620.
Li J . (2010). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol 13: 509–514.
Liao PC, Chung JD, Chen CL, Hwang CJ, Sung YH, Chang YT et al. (2012). Natural variation, functional divergence, and local adaptation of nucleotide binding site sequences in Rhododendron (Ericaceae). Tree Genet Gen 8: 879–893.
Liew P-M, Chung N-J . (2001). Vertical migration of forests during the last glacial period in subtropical Taiwan. West Pac Earth Sci 1: 405–414.
Liew P-M, Huang S-Y, Kuo C-M . (2006). Pollen stratigraphy, vegetation and environment of the last glacial and Holocene-A record from Toushe Basin, central Taiwan. Quarter Internal 147: 16–33.
Linhart YB, Grant MC . (1996). Evolutionary significance of local genetic differentiation in plants. Ann Rev Ecol System 27: 237–277.
Marriage TN, Hudman S, Mort ME, Orive ME, Shaw RG, Kelly JK . (2009). Direct estimation of the mutation rate at dinucleotide microsatellite loci in Arabidopsis thaliana (Brassicaceae). Heredity 103: 310–317.
Marshall A, Aalen RB, Audenaert D, Beeckman T, Broadley MR, Butenko MA et al. (2012). Tackling drought stress: RECEPTOR-LIKE KINASES present new approaches. Plant Cell 24: 2262–2278.
Marshall DL, Avritt JJ, Maliakal-Witt S, Medeiros JS, Shaner MGM . (2010). The impact of plant and flower age on mating patterns. Ann Bot 105: 7–22.
Mejías JA, Arroyo J, Ojeda F . (2002). Reproductive ecology of Rhododendron ponticum (Ericaceae) in relict Mediterranean populations. Bot J Linn Soc 140: 297–311.
Nosil P, Funk DJ, Ortiz-Barrientos D . (2009). Divergent selection and heterogeneous genomic divergence. Mol Ecol 18: 375–402.
Nybom H . (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13: 1143–1155.
Ono A, Dohzono I, Sugawara T . (2008). Bumblebee pollination and reproductive biology of Rhododendron semibarbatum (Ericaceae). J Plant Res 121: 319–327.
Parisod C, Joost S . (2010). Divergent selection in trailing-versus leading-edge populations of Biscutella laevigata. Ann Bot 105: 655–660.
Peleg Z, Fahima T, Abbo S, Krugman T, Saranga Y . (2008). Genetic structure of wild emmer wheat populations as reflected by transcribed versus anonymous SSR markers. Genome 51: 187–195.
Postel S, Küfner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A et al. (2010). The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Euro J Cell Biol 89: 169–174.
Pritchard JK, Stephens M, Donnelly P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
R Development Core Team. (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Raymond M, Rousset F . (1995). Genepop (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Reed DH, Frankham R . (2003). Correlation between fitness and genetic diversity. Conser Biol 17: 230–237.
Rice WR . (1989). Analyzing table of statistical tests. Evolution 43: 223–249.
Räsänen K, Hendry AP . (2008). Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecol Lett 11: 624–626.
Siame L, Chu HT, Carcaillet J, Bourles D, Braucher R, Lu WC et al. (2007). Glacial retreat history of Nanhuta Shan (north-east Taiwan) from preserved glacial features: the cosmic ray exposure perspective. Quat Sci Rev 26: 2185–2200.
Steinwand BJ, Kieber JJ . (2010). The role of receptor-like kinases in regulating cell wall function. Plant Physiol 153: 479–484.
Storz JF . (2005). Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol 14: 671–688.
Taiwan Forestry Bureau. (1995) The Third Survey of Forest Resources and Land Use in Taiwan. Taiwan Forestry Bureau, Department of Agriculture: Taipei, Taiwan. In Chinese.
Tan XX, Li Y, Ge XJ . (2009). Development and characterization of eight polymorphic microsatellites for Rhododendron simsii Planch (Ericaceae). Conser Genet 10: 1553–1555.
Tsuda Y, Ide Y . (2005). Wide-range analysis of genetic structure of Betula maximowicziana, a long-lived pioneer tree species and noble hardwood in the cool temperature zone of Japan. Mol Ecol 14: 3929–3941.
Turk B, Turk D, Salvesen GS . (2002). Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr Pharm Des 8: 1623–1637.
Turk V, Turk B, Turk D . (2001). Lysosomal cysteine proteases: facts and opportunities. EMBO J 20: 4629–4633.
van Oosterhout C, Hutchinson WF, Willis DPM, Shipley P . (2004). MICRO_CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Von Wettberg EJ, Remington DL, Schmitt J . (2008). Partitioning adaptive differentiation across a patchy landscape: shade avoidance traits in Impatiens capensis. Evolution 62: 654–667.
Wilson GA, Rannala B . (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191.
Wu S-H, Hwang C-Y, Lin T-P, Chung J-D, Cheng Y-P, Hwang S-Y . (2006). Contrasting phylogeographic patterns of two closely related species Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. J Biogeogr 33: 936–947.
Acknowledgements
WE thank Mr Ji-Sheng Wu of the Taiwan Forest Research Institute for his assistance with sample collection. This study was supported by the National Science Council to S-YH (grant no. NSC100-2621-B-003-001), Executive Yuan, Taiwan. Funding for a graduate studentship to Y-CH and a postdoctoral associateship to C-TC and C-YC by the National Science Council is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Hsieh, YC., Chung, JD., Wang, CN. et al. Historical connectivity, contemporary isolation and local adaptation in a widespread but discontinuously distributed species endemic to Taiwan, Rhododendron oldhamii (Ericaceae). Heredity 111, 147–156 (2013). https://doi.org/10.1038/hdy.2013.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2013.31
Keywords
This article is cited by
-
Projecting the potential distribution and analyzing the bioclimatic factors of four Rhododendron subsect. Tsutsusi species under climate warming
Journal of Forestry Research (2023)
-
Population genetic variation characterization of the boreal tree Acer ginnala in Northern China
Scientific Reports (2020)
-
Demographic history and adaptive synonymous and nonsynonymous variants of nuclear genes in Rhododendron oldhamii (Ericaceae)
Scientific Reports (2020)
-
Postglacial range expansion and the role of ecological factors in driving adaptive evolution of Musa basjoo var. formosana
Scientific Reports (2017)
-
Population genetic structure and post-LGM expansion of the plant bug Nesidiocoris tenuis (Hemiptera: Miridae) in China
Scientific Reports (2016)