Abstract
Chronic myeloid leukemia (CML) is a myeloproliferative disease caused by the BCR–ABL1 tyrosine kinase (TK). The development of TK inhibitors (TKIs) revolutionized the treatment of CML patients. However, TKIs are not effective to those at advanced phases when amplified BCR–ABL1 levels and increased genomic instability lead to secondary oncogenic modifications. Wiskott–Aldrich syndrome protein (WASP) is an important regulator of signaling transduction in hematopoietic cells and was shown to be an endogenous inhibitor of the c-ABL TK. Here, we show that the expression of WASP decreases with the progression of CML, inversely correlates with the expression of BCR–ABL1 and is particularly low in blast crisis. Enforced expression of BCR–ABL1 negatively regulates the expression of WASP. Decreased expression of WASP is partially due to DNA methylation of the proximal WASP promoter. Importantly, lower levels of WASP in CML advanced phase patients correlate with poorer overall survival (OS) and is associated with TKI response. Interestingly, enforced expression of WASP in BCR–ABL1-positive K562 cells increases the susceptibility to apoptosis induced by TRAIL or chemotherapeutic drugs and negatively modulates BCR–ABL1-induced tumorigenesis in vitro and in vivo. Taken together, our data reveal a novel molecular mechanism that operates in BCR–ABL1-induced tumorigenesis that can be used to develop new strategies to help TKI-resistant, CML patients in blast crisis (BC).
Similar content being viewed by others
Main
Chronic myeloid leukemia (CML) is a hematological malignancy caused by the chromosome translocation t(9:22) (q34;q11), which originates the oncogene BCR–ABL1. BCR–ABL1 is a protein composed of multiple domains involved in the initiation of a complex signaling transduction cascade.1, 2 The BCR portion of the oncoprotein contains a domain with serine and threonine kinase activity and regions that bind to Src homology-2 (SH2) domains, whereas the c-ABL portion contains SH3, SH2 and the tyrosine kinase (SH1) domains, in addition to four proline-rich domains, a nuclear translocation signal, a DNA- and an actin-binding domains.3 Binding of BCR–ABL1 to actin filaments was shown to contribute to transformation. Importantly, the tyrosine kinase (TK) activity is necessary for the transformation potential of BCR–ABL13, 4 Particularly, BCR–ABL1 TK activity induces a strong resistance to both intrinsic and extrinsic pathways of apoptosis,5 for instance, by inducing overexpression of BCL-XL6 and MCL-17 and downregulating TRAIL.8 Therefore, it is not surprising that the development of specific inhibitors directed to the TK catalytic site of BCR–ABL1, such as imatinib mesylate, revolutionized the treatment of CML patients.9
The evolution of CML comprehends three stages: chronic, accelerated and blast phases. In chronic phase (CP), the bulk of leukemic stem cells remains capable of undergoing differentiation, leading to the excessive production of mature granulocytes. In the accelerated phase (AP), differentiation becomes arrested, and the severe disease phenotype is caused by the proliferation of immature blasts, which increases in the BC.3, 10 Mechanisms of imatinib resistance are multifactorial and include point mutations within the BCR–ABL kinase domain,11, 12 BCR–ABL gene amplification,11 decreased intracellular drug availability13 and activation of alternative signaling oncogenic pathways.14, 15 In the advanced phases, patients present a stronger resistance to imatinib or other second- and third-generation TK inhibitors (TKIs),16 indicating an unmet need for improved therapeutic approaches in CML treatment.
BCR–ABL1 is the indispensable genetic modification responsible for CML,3, 17, 18 but other secondary19 genetic and epigenetic changes were also shown to contribute to malignancy, including resistance to TKIs.20 Although poorly understood, disturbance of the hematopoietic compartment is an evident phenomenon in CML that includes failure in differentiation of the stem cells in the bone marrow21 and inefficient function of mature leukocytes.22, 23, 24 Interestingly, these events are similar to those observed in patients with Wiskott–Aldrich syndrome (WAS), an immunodeficiency caused by the dysfunction of WAS protein (WASP).25 In addition, CML patients in BC also share symptoms with WAS patients, such as spontaneous bleeding, petechiae, ecchymosis, thrombocytopenia, enlargement of the spleen and increased susceptibility to infections.26, 27, 28 WASP is an adaptor protein exclusively expressed in hematopoietic cells capable of promoting actin polymerization.29 Important to our work, WASP was shown to act as an endogenous inhibitor of the TKs LCK, FYN and c-ABL.30, 31, 32
Here, we show that expression of WASP inversely correlates with BCR–ABL1 levels and the progression of the disease in CML patients. BCR–ABL1 downregulates WASP in part by epigenetic modification of its proximal promoter. Most importantly, downregulation of WASP contributes to the resistance to apoptosis and to BCR–ABL1-induced tumorigenesis. Therefore, WASP may serve as a molecular marker of prognosis, as well as a potential target for combined antitumor therapies for CML.
Results
WASP is downregulated in BCR–ABL1-positive cell lines and CML patients by a mechanism that involves epigenetic modification
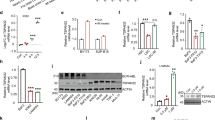
Initially, we sought to investigate the expression of WASP in CML patients and in BCR–ABL1-positive cell lines. We found that PBMC from CML patients expressed significantly lower levels of WASP, comparing with healthy donors (Figure 1a). CML patients in the CP presented lower levels of WASP and its expression was decreased during the progression of the disease to accelerated and blast phases (Figure 1b). Importantly, PBMC from patients resistant to TKI exhibited significantly lower levels of WASP compare with patients responsive to TKI (patients who achieved the complete cytogenetic remission (CCyR) and major molecular remission (MMR) after treatment with imatinib and dasatinib). Patients responsive to TKI were no different from either healthy individuals or CML patients at diagnosis (Figure 1b). These data indicate that WASP expression is linked to CML patient’s response to TKIs therapy and to BCR–ABL1 levels. Moreover, we detected that patients who presented secondary resistance to TKI therapy (patients who achieved but subsequently lose relevant response) have higher WASP expression in comparison with patients who showed primary resistance (patients who did not reach a benchmark response) (Supplementary Figure 1). Twenty-one CML patients resistant to TKIs were tested for BCR–ABL mutation; 9 of them were positive for Y253H (2); M244V (2); T315I (2); F317L (1); H396R (1) e G250E/Y253H (1) mutations. There was no association with mutation status and WASP expression levels (P=0.97; Supplementary Figure 2).
WASP is downregulated in CML patients and BCR–ABL1-positive cell lines. (a) The expression of WASP is suppressed in CML patients comparing with healthy donors (HD). The relative expression of WASP in PBMC was determined by real-time PCR using GAPDH as housekeeping gene (**P<0.01, comparing with HD). Values are plotted as 2−ΔΔCt. 't' student was used as statistical test. (b) The downregulation of WASP was observed in advanced phases of the disease (AP and BC). (P<0.0001, comparing with HD). Patients in CCyR showed normal WASP levels, according to the healthy donor control group (P<0.001, comparing with BP). Analysis of variance (ANOVA) and Bonferronni posttest were used to statistical analysis. (c) The expression of WASP negatively correlates with BCR–ABL1 expression in CML patients (P<0.0001). Spearman test was used with linear regression. (d) Western blot showing the expression of WASP in cell lines derived from different leukemia types. BCR–ABL1-negative cell lines express high levels of WASP, whereas BCR–ABL1-positive cells express discrete or undetectable WASP. Actin was used as loading control. (e) Stable expression of BCR–ABL1 was induced in HL-60 and Jurkat cell lines by retroviral infection, resulting in downregulation of WASP both at the mRNA and protein levels. GAPDH was used as housekeeping control for qPCR. Western blots show the increased numbers of tyrosine phosphorylated proteins in BCR–ABL-positive HL-60 and Jurkat cell lines, and the complete WASP silencing after BCR–ABL1 expression. Actin protein was used as loading control
In addition, the expression levels of WASP and BCR–ABL1 were inversely correlated (Figure 1c), suggesting that BCR–ABL1 could potentially be responsible for WASP downregulation. In order to investigate this possible cause–effect relationship, we evaluated WASP protein expression in cell lines derived from BCR–ABL1-negative leukemia (HL-60, Jurkat, SKW6.4, THP-1 and U937) and BCR–ABL1-positive CML patients (K562, BV173, LAMA-84 and KCL22). Unlike the BCR–ABL1-negative cells, WASP was strongly downregulated in BCR–ABL1-positive cell lines (Figure 1d) further supporting that BCR–ABL1 is a negative regulator of WASP. Our hypothesis was confirmed by transducing BCR–ABL1 in the HL-60 and Jurkat cell lines, thereby producing HL-60.BCR–ABL1 and Jurkat.BCR–ABL1 cells. Enforced expression of BCR–ABL1 induced a strong suppression of WASP, observed at both mRNA and protein levels (Figure 1e). Efficacy of BCR–ABL1 transduction was confirmed by immunoblot using primary antibodies against c-ABL/BCR–ABL1 (to verify its expression) or to phosphotyrosine (to verify its activity) (Figure 1e). These results show that expression of BCR–ABL1 inhibits WASP at both mRNA and protein levels.
As most of BCR–ABL1-mediated signal transduction is dependent on its TK activity, we tested whether treatment with imatinib could reinstate the expression of WASP in BCR–ABL1-positive cells. To our surprise, the inhibition of the TK activity of BCR–ABL1 did not result in WASP re-expression in K562, BV173 or HL-60.BCR–ABL1 cells (Figure 2a), even in the presence of the dual calpain/proteasome inhibitor N-acetyl-l-leucyl-l-leucyl-l-norleucinal (Figure 2b), as protein levels of WASP is also tightly controlled by degradation via calpain-mediated proteolysis.33 These results suggest the emergence of an additional mechanism that maintains WASP inhibited independently of constant BCR–ABL1 TK activity.
WASP downregulation is not rescued by imatinib or calpain inhibitor and involves epigenetic modification. (a and b) Cell lines were treated for 24 h with 10 μM IM and/or the calpain inhibitor N-acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN), and western blot was performed to reveal the expression of WASP, WIP and the status of BCR–ABL1-TK activity. Actin was used as loading control. (c) Scheme of the distal and proximal promoters of WASP. (d) In silico analysis using UCSC Genome Browser public data (ww.ucsc.edu) show the presence of CpG sites in the promoters of WASP (green boxes). (e) CpG methylation status in the proximal promoter of WASP was evaluated. Black and white circles: methylated and unmethylated CpG sites
DNA methylation at CpG sites localized in the promoter region is a well-known mechanism to stably suppress gene expression.34 As hematopoietic-specific expression of the WASP gene was shown to be driven by a promoter located upstream to the TSS35 (Figures 2c and d), we measured the DNA methylation levels at CpGs dinucleotide in a 400 bp CpG island positioned at this promoter region by bisulfite sequencing. We found an inverse correlation between DNA methylation of WASP promoter and gene expression. CML cell lines with suppressed WASP, such as BV173 and K562, presented the higher levels of DNA methylation: 99% and 77%, respectively. KCL22 (which express low levels of WASP) presented intermediated levels of DNA methylation: 54% (Figure 2e). In contrast, THP-1, a BCR–ABL1-negative cell line, which expresses high amounts of WASP, presented low level of DNA methylation: 22% (Figure 2e).
Azacytidine (5-AZA) has presented significant therapeutic results for patients with acute myeloid leukemia and myelodysplastic syndromes, by inducing the demethylation of CpG sites and the consequent upregulation of silenced genes.36, 37 Treatment of CML cell lines with 1μM 5-AZA alone restored the levels of WASP expression in LAMA-84, BV173 (greatly) and KCL22 (partially) cell lines, confirming that CpG methylation is indeed one epigenetic mechanism involved in BCR–ABL1-induced WASP suppression (Figure 3). Interestingly enough, 5-AZA failed to restore WASP levels in K562 cells, although it restored RASSF1A and GST control genes, suggesting the existence of a further mechanism responsible for downregulation of WASP in this cell line. Taken together, our data suggest that BCR–ABL1 inhibits the expression of WASP via multiple molecular mechanisms.
5-AZA treatment restored the levels of WASP expression in some but not all CML cell lines. (a) Treatment of CML cell lines with 1 μM 5-AZA alone restored the levels of WASP expression in LAMA-84, KCL22 and BV173, but not in K562 cells. The relative expression of RASSF1A and GST was used as positive controls for 5-AZA treatment. Two way Anova (Analysis of variance) and Bonferroni post test were used to determine statistic significance among the groups. *P < 0.05, **P < 0.01, ***P < 0.001
Lower WASP expression in PBMC from CML patients in advanced phases correlates with poor OS and re-expression of WASP negatively modulates BCR–ABL1-induced tumorigenesis in vitro and in vivo
Next, we investigated the biological relevance of decreased levels of WASP for CML patients. We analyzed the relationship between the levels of WASP with the OS of the 31 newly diagnosed CML patients and in 23 CML patients at AP and BC. For this analysis, we divided the patients in two groups according to the median of WASP expression. Patients with WASP levels below the median were called WASP BM, and patients with WASP levels above the median, were called WASP AM. There was no significant difference in OS according to WASP expression at CML diagnosis (Supplementary Figure 3A). In addition, we observed a tendency of a lower expression of WASP in high-risk Sokal patients (Supplementary Figure 3B). Interestingly, when the OS was calculated just for CML patients in advanced phases, resistant to TKIs and with significantly lower levels of WASP compared with healthy donors, we observed that milder suppression of WASP (levels above median – WASP low) correlated with longer OS, whereas strong suppression of this gene (levels below median – WASP very low) correlated with poorer OS (Figure 4b). Altogether, these data suggest that the downregulation of WASP by BCR–ABL1 may be relevant to the CML prognosis, particularly for patients in advanced phases of disease.
Lower levels of WASP in CML patients at advanced phases may indicate a worse OS and WASP re-expression negatively modulates BCR–ABL1-induced tumorigenesis. (a) The relative WASP expression values obtained from patients in AP and BP were divided in two groups according to the median: WASP very low for patients exhibiting a strong WASP downregulation (lower than median), and WASP low for patients presenting a mild WASP suppression (higher than median). (b) Patients with higher levels of WASP (grouped in WASP low) presented longer OS (median=61.75 months) comparing with the patients in WASP very low group. Kaplan–Meier was used as statistical test. (c) K562 cell line was transduced with lentivirus vector to induce the re-expression of WASP (eGFP) or WIP (mCherry). (d) K562.WT or re-expressing WASP or WIP were plated and the number of cells was counted in Neubauer chamber according to the time point in the graph. ***P<0.001 comparing K562.WASP with K562.WT. ##P<0.01 comparing K562.WASP with K562.WIP. (e) K562.WT was co-cultured with K562.WASP in three different initial proportions and the frequency of each population was assessed 7 days later by flow cytometry. (f) K562.WIP (mCherry-positive) and K562.WASP (eGFP-positive) were subcutaneously injected in the right and left flanks of the BALB/c nude mice, respectively, and the development of the solid tumors was followed and imaging of the tumors was assessed at 21st day. The tumors were collected to determine volume (g) and weight (h). Student's t-test was used for statistical analysis. *P<0.05
Therefore, we investigated the consequences of re-expression of WASP on in vitro and in vivo models of BCR–ABL1-induced tumorigenesis. Using lentiviral transduction, we enforced the expression of eGFP-WASP or mCherry-WIP (as a control) in K562 cells, which lacks both WASP and WIP (WASP-interacting protein) (Figure 4c). First of all, the expression of WIP, a chaperone for WASP that prevents its targeting by calpain, thus increasing WASP half-life,33 did not result in increased expression of WASP in K562 cells, as expected after our results showing WASP promoter is silenced by DNA methylation in these cells (Figure 2e), as described above. Growth of tumor cells comprehends one important aspect of tumorigenesis and it is directly associated to the balance of proliferation and cell death in vitro and in vivo. In line with this, enforced expression of WASP decreased the growth of K562 cells compared with K562.WT and K562.WIP (Figure 4d). We also performed a mixed culture assay with 25%/75%, 50%/50% and 75%/25% of K562.WT (eGFP-negative) and K562.WASP (eGFP-positive), respectively. After 7 days of co-culture, the frequency of the cell populations was determined by flow cytometry. Under all the conditions tested, the relative percentage of K562.WT to K562.WASP increased with respect to the initial input (Figure 4e). In combination, these results suggest that re-expression of WASP decreases the robustness of the BCR–ABL1-positive cells, impacting its ability to grow in vitro (Figures 4c–e).
We next tested the impact of the re-expression of WASP in K562 cell growth in vivo, using a xenogeneic murine model of tumor growth. K562.WASP (eGFP-positive) and K562.WIP (mCherry-positive) were subcutaneously injected in the left and right flanks of BALB/c nude mice, respectively, and the growth of the tumors was followed up to 21 days. In agreement with the in vitro data, the expression of WASP in K562 drastically inhibited the in vivo tumor development, as assessed by in vivo imaging of the whole animal (Figure 4f), and by measuring the volume and weight of extracted tumors (Figures 4g and h).
Taken together, our data demonstrate that the expression of WASP has a negative impact on BCR–ABL1-induced tumorigenesis both in vitro and in vivo.
WASP sensitizes BCR–ABL1-positive cells to TRAIL- and chemotherapy-induced apoptosis and interferes with BCR–ABL1 tumorigenic activity
To complete our study, we investigated the possible mechanisms involved in the negative impact of WASP on BCR–ABL1-mediated tumorigenesis. In fact, we observed a consistently increased 'spontaneous' apoptosis in cultures of K562.WASP cells compared with K562.WT (Figure 5a). This could be related to the fact that expression of WASP in K562 cells resulted in upregulation of the death receptors DR4 and DR5, and also their ligand TRAIL (Figure 5b). In fact, when we culture both cell lines in the presence of the chimeric TRAIL inhibitor, TRAIL-R2/Fc, we observed a reduction of PI-positive cells only in K562.WASP cell samples, and to the level found in K562.WT cultures (Figure 5a).
WASP sensitizes BCR–ABL1-positive cells to TRAIL- and chemotherapy-induced apoptosis. (a) K562.WT and K562.WASP were cultured in the presence or absence of TRAIL-R2/Fc to prevent TRAIL-mediated apoptosis and PI uptake was assesed 48 h later. (b) qPCR shows the upregulation of DR4, DR5 and TRAIL after re-expression of WASP in K562 cells. (c) K562.WT and K562.WASP were treated with different concentrations of IM and cell death was assessed by the hypodiploid DNA content (Sub G0) (*P<0.05; **P<0.01; ***P<0.001). (d) Cell lines were treated with camptothecin, actinomycin D and ultra violet (UV) radiation and cell death was assessed after 48 h by hypodiploid DNA content. Two way ANOVA was used for statistical analysis. ***P<0.001
As TRAIL was shown to sensitize BCR–ABL1-positive cells to imatinib mesylate,38 we tested whether the expression of WASP in K562 cells rendered them more sensitive to imatinib. Indeed, imatinib treatment at a dose range achievable in patients resulted in higher frequency of apoptotic K562.WASP cells compared with K562.WT (Figure 5c). Finally, we tested whether the expression of WASP could also sensitize K562 cells to chemotherapy-induced apoptosis. Similarly, K562.WASP cells were more susceptible to apoptosis induced by camptothecin, actinomycin D and UV irradiation than K562.WT cells (Figure 5d). Therefore, increased levels of apoptosis appear to account for the tumor-suppressor activity mediated by WASP. These results indicate that the expression of WASP sensitizes BCR–ABL1-positive cells to apoptosis.
Discussion
The development of TKI was a turning point for the treatment of CML. However, its efficiency depends on the early diagnosis, early administration and continuing treatment. In addition, part of the CML patients do not respond to imatinib or second- or third-generation TKIs.16 Therefore, the discovery of novel signaling pathways associated with BCR–ABL1-induced tumorigenesis is still a matter of importance for patients with BCR–ABL1-positive leukemia. We show here for the first time that WASP may serve as a molecular marker of prognosis, as well as a potential target for combined antitumor therapies for CML.
Initially, we demonstrated that BCR–ABL1 inhibits the expression of WASP both at mRNA and protein levels. Particularly important is the fact that downregulation of WASP seems to take place mostly at the transition of chronic to advanced phases of the disease. At this point, increased genomic instability may result in additional genetic abnormalities that are discretely acquired with no significant changes in the clinical aspects of disease. This includes no relevant increase in blast numbers, making it difficult to recognize the molecular evolution of CML patients.16, 39, 40 The observed inhibition of WASP expression coincided exactly with this transition, as patients in CP still express WASP at levels similar to healthy individuals, whereas patients in AP and BC has significantly less WASP (Figure 1b). Therefore, monitoring the expression of WASP could be useful as a molecular marker of progression of CML and may help in future therapeutic decisions, such as early switch to more potent TKI or to use combining chemotherapies.
Besides its potential use as a biomarker for AP/BC, WASP suppression seems to have a role in the severity of the disease. First, as mentioned above, WASP levels become significantly lower in advanced phases of CML (AP+BC). Most importantly, by dividing these two groups of patients according to their (diminished) expression of WASP, we observed that those with very low levels of WASP (below the median) had a significantly shorter OS than those with low levels (above the median), independently of the stage of the disease (Figures 4a and b). Interestingly, another member of the WASP family, N-WASP, has been implicated in the progression of breast and colorectal cancer.41, 42, 43 Similar to our results, the authors showed a strong correlation between low levels of N-WASP and shorter OS/disease-free survival. However, they linked the increased severity of the disease to an augmented effect of the absence of N-WASP on tumor invasiveness/metastasis, which clearly does not apply to our case, because of the leukemic nature of CML.
The enforced expression of WASP in K562 cells revealed the biological consequences of WASP suppression in CML. Our data demonstrate that the strong resistance to apoptosis conferred by BCR–ABL1 to leukemic cells was attenuated by the introduction of a WASP-expressing construct. Besides inducing TRAIL-dependent spontaneous apoptosis, with participation of the increased expression of DR4 and DR5, recovery of WASP turned K562 cells sensitive to different chemotherapeutic drugs, as well as to imatinib. These data suggest that re-activation of WASP could be an interesting therapeutic strategy in CML, especially in combination with imatinib or other TKI. Interestingly, the deficiency of WASP expression has already been reported to induce or to facilitate apoptosis in other contexts.44, 45, 46
In order to explore this possibility, we investigated the mechanisms behind WASP downregulation in CML. To our surprise, treatment of CML cell lines with imatinib did not show any sign of recovery of WASP, suggesting that the inhibition of WASP by BCR–ABL1 is either a kinase-independent event or may undergo a secondary layer of regulation that does not depend on the constant kinase activity of BCR–ABL1. The existence of kinase-independent mechanisms derived from the expression of BCR–ABL1 has been described.47 For instance, BCR–ABL1 can bind and activate HCK independently of its catalytic activity.48 Moreover, the expression of BCR–ABL1, but not its kinase activity, was shown to be important to the survival of quiescent CML stem cells.49 In line with this, we have shown that robust pharmacological inhibition of BCR–ABL1 TK, as assessed by the disappearance of phosphotyrosine-containing proteins, does not interfere with the strong resistance to apoptosis observed in BCR–ABL1-positive cell lines, at least for 24 h, when levels of BCL-XL were significantly reduced.50, 51
The stability of WASP suppression in all tested CML cell lines led us to investigate the existence of an epigenetic inhibitory mechanism. Indeed, we found that a CpG island located at WASP promoter was strongly methylated in BV173 and K562 cell lines, implicating DNA methylation as a novel mechanism for WASP inhibition.
Re-expression of WASP in the BCR–ABL1-positive cells hampers both the in vitro and in vivo tumorigenic potential, and increases sensitivity to imatinib and to chemotherapeutic drugs, providing a rationale for the development of new therapeutic strategies aiming to restore the levels of WASP in BCR–ABL1 leukemia. Our data suggest that one interesting strategy is combining Azacytidine, an FDA-approved DNA demethylating agent, to increase WASP expression with TKIs, such as imatinib. Another potential strategy is using gene therapy approaches to correct WASP expression. Interesting enough, γ-retroviral and HIV-based lentiviral vectors designed to correct WASP levels are already in place for patients with WAS.52, 53, 54
One final consequence of our findings is related to proposed immunotherapy using dendritic cells isolated from CML patients.55, 56, 57 Although there are evidences that the immune system can potentially eliminate CML cells, particularly imatinib-resistant BCR–ABL1-positive stem cells, a note of caution should be added to the use of autologous dendritic cells, as patients in advanced phase of CML express low levels of WASP and, therefore, are likely to present impaired immunological synapses and consequent deficiency in antigen presentation. In fact, using a retrovirus-induced murine model of CML, Mumprecht and collaborators have shown that BCR–ABL-expressing DCs do not efficiently induce specific CTL immune response.22, 23, 24
Taken together, we described a new tumor-suppressor activity for WASP and demonstrated that its expression in CML interferes with BCR–ABL1-mediated signaling transduction, increases the sensitivity of leukemic cells to apoptosis and is associated with a better prognosis of CML patients. Therefore, the strategies aimed to recover the expression of WASP, which are currently being tested for WAS patients,58 may be potentially considered as a therapeutic approach for some CML patients.
Materials and Methods
Patients and controls
WASP expression was analyzed in 32 healthy individual and 85 CML patients (62 CP, 11 AP and 12 crisis blastic phase) treated at the Euryclides de Jesus Zerbini Transplantation Hospital and the Hematology and Hemotherapy Center, University of Campinas, State of São Paulo, Brazil, between April 2003 and October 2010. One patient was diagnosed in August 2015. Thirty-nine patients were responsive to TKIs therapy (imatinib or dasatinib) and 46 were resistant. Most of patients from responsive groups presented a major molecular response after dasatinib therapy. The demographic and clinical features of evaluated patients are shown at Tables 1 and 2 (Supplementary Data).
The study was approved by the local research ethics committees and patients gave written informed consent for their participation, in accordance with the Declaration of Helsinki. Demographic and disease characteristics were collected at baseline. CML diagnosis of the patients enrolled in this study was confirmed by the demonstration of Philadelphia chromosome in conventional cytogenetics and/or BCR–ABL1 detection by RT-PCR. Hematologic, cytogenetic and molecular responses were redefined according to the European LeukemiaNet 2013 recommendations.
Peripheral blood mononuclear cells from patients and controls were isolated according to standard protocol with the Ficoll–Hypaque 1077 density technique.
Cell lines
All cell lines K562, LAMA-84, KCL22, BV173, Jurkat, HL-60, SKW6.4, THP-1 and U937 were cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum, 25 mM Hepes, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. HL-60 and K562 cells were obtained from ATCC (Manassas, VA, USA). LAMA-84 was kindly provided by Dr. James Griffin (Dana-Farber Cancer Institute, Boston, MA, USA). HL-60.BCR–ABL1 cells were derived from wild-type HL-60 by retroviral transfection with pSRαMSVp185BCR–ABL1tkneo, as previously described.51, 52 K562 were infected with lentivirus vectors to induce expression of WASP.GFP and WIP.mchery,59 and sorted using FACS ARIA (Becton-Dickinson, Franklin Lakes, NJ, USA) to obtain high purity cell lines.
Reagents
For western blot, we used anti-WASP (Santa Cruz Biotechnologies, Dallas, TX, USA), anti-WIP (Santa Cruz Biotechnologies), anti-c-ABL OP-20 (Oncogene Research Products, La Jolla, CA, USA), anti-Actin (Sigma Aldrich, St. Louis, MO, USA). TRAIL-R2/Fc protein was generously provided by Dr. Henning Walczak (Imperial College, London, UK).
Quantitative PCR
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were determined spectrophotometrically by measuring fluorescence at 260 and 280 nm. Three micrograms of RNA was reverse transcribed into cDNA using Superscript III (Invitrogen) transcription reagents according to the manufacturer’s instructions. After obtaining the cDNA, gene expression was quantified by quantitative PCR using Platinum SYBRGreen Kit (Invitrogen) in Mx3005P detector equipment (Stratagene, Santa Clara, CA, USA). The following primers were used: BCR–ABL 5′-TGGGTCCAGCGAGAAGGTT-3′ (forward) and 5′-GCATTCCGCTGACCATCAAT-3′ (reverse); GAPDH 5′-GGAGAAGGCTGGGGCTCAT-3′ (forward) and 5′-TCCTTCCACGATACCAAAGTT-3′ (reverse); TRAIL 5′-AAGGCTCTGGGCCGCAAAATAAAC-3′ (forward) and 5′-CCAACTAAAAAGGCCCCGAAAAA-3′ (reverse); DR4 5′-GTACGCCCTGGAGTGACATC-3′ (forward) and 5′-CCTCGTAGGAGACCCAAGC-3′ (reverse); DR5 5′-CTAGCTCCCCAGCAGAGAGT-3′ (forward) and 5′-GTGGTGCAGGGACTTAGCTC-3′ (reverse). WASP 5′-GGCTGGTCGGCTGCTCTGGGAACA-3′ (forward) and 5′-GGTGGTGGGGGTAGCTGGCGTCTGT-3′ (reverse). Results were given as relative expression represented as 2−ΔΔCt.
Western blot
Protein samples were resolved under reducing conditions as previously described.45 Separated proteins were transferred onto polyvinylidene difluoride membranes and reactions were detected with a suitable secondary antibody conjugated to horseradish peroxidase (Jackson Laboratory, Bar Harbor, ME, USA and Amersham, Arlington, IL, USA) using enhanced chemiluminescence (Pierce, Rockford, IL, USA).
Treatments
Cell lines were treated with imatinib mesylate (0.1, 1 and 10 μM), actinomycin D (5 μM), camptothecyn (100 μM) and UV.C. (400 J/cm2) for 48 h and cell death was determined by flow cytometry. For TRAIL pathway blockage, cell lines were treated with 5 μg/ml of TRAIL-R2/Fc for 48 h.
Assessment of cell death
Apoptosis was quantified using the fluorescence-activated cell sorter (FACS) Calibur flow cytometer (Becton-Dickinson), by analysis of DNA content as described.51 We also evaluated cell death by propidium iodide (PI) incorporation for dead cells using flow cytometry. The results represent the average±S.D. in triplicate samples. Every experiment was repeated at least three times.
Cell growth analysis and proliferation assays
For absolute count of cells, 4 × 104 cells K562.WT, K562.WASP, K562.WIP and K562.WASP.WIP were cultured in six-well plates with 1 ml of medium, and after each 24 h, 0.5 ml of fresh medium was added to avoid starvation and cellular stress. The absolute number of the cells was counted in Neubauer chamber each 24 h during 5 consecutive days. For the co-culture assays, 2 × 105 cells K562.WT (GFP negative) and K562.WASP (GFP positive) were co-cultured in three different proportions: 25%:75%; 50%:50%; 75%:25%, respectively. The cells were homogenized by pipetting, half part was discarded and the culture received fresh medium every 24 h. After 7 days of assay, the frequency of the populations was evaluated by flow cytometry and compared with day 0. For proliferation assay, K562.WT and K562.WASP were stained with Violet Cell Trace (Molecular Probes, Invitrogen, Carlsbad, CA, USA) and cultured for 4 days, and the proliferation rate was determined by fluorescence decay using flow cytometry analysis. The results represent the average±S.D. in triplicate samples. Every experiment was repeated three times.
Animals and in vivo model of K562-induced solid tumors
Five million K562.WASP.gfp and K562.WIP.mcherry cells were subcutaneously injected in the left and right superior flanks of 6–8-week-old, female BALB/c nude mice, respectively. The tumor growth was followed during 21 days, when the animals were killed and the tumors were removed to determine weight and volume. Images of the animals were collected at day 18 after inoculation, and tumors were analyzed through detection of gfp and mcherry fluorescence using IVIS Spectrum Pre-clinical In Vivo Imaging System (Perkin Elmer, Waltham, MA, USA). Groups of at least 6 mice were used in each experiment, which was repeated three times. All animals were housed at our animal facility at the Institute of Biomedical Sciences, University of Sao Paulo (ICB-USP). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Brazilian National Council of Animal Experimentation (http://www.cobea.org.br/). The protocols were approved by the Animal Ethics Committee of the ICB-USP.
Methylation analysis in WASP promoter
One μg of genomic DNA from each cell line was bisulfite converted using the Zymo EZ DNA methylation kit (Zymo Research, Orange, CA, USA), according to the manufacturer’s instructions. To analyze the DNA methylation status of individual DNA molecules, we amplified by PCR a 400 bp region upstream to WAS TSS after the bisulfite conversion. The PCR fragments were cloned into the pCR2.1 vector using the TOPO-TA cloning kit (Invitrogen). Individual colonies were screened for the insert and the region of interest was sequenced using M13 primers as previously described.60
5’Azacytidine treatment and induction of DNA demethylation
The cell lines K562, LAMA-84, KCL22 and BV173 were treated with 1 μM of Azacytidine for 120 h. The supplemented medium was refreshed every 24 h in order to maintain the drug concentration during the experiment. Cells were harvested and RNA was obtained. Real-time PCR was performed to investigate the recovery of WASP after DNA demethylation. Glutatione S-tranferase-pi 1 (GSTP1) and Ras association domain family member 4 (RASSF4), which are described as suppressed genes by CpG methylation in CML cell lines61, 62, 63 were used as control of the efficacy of the treatment, and GAPDH as the housekeeping control.
Statistical analysis
The probability of OS of 31 CML newly diagnosed patients (clinical and laboratorial features at Table 3,Supplementary Data) according to WASP expression levels was calculated from CML diagnosis until death or last follow-up using the Kaplan–Meier method and the log-rank test, using IBM SPSS software, version 21 (IBM, Armonk, NY, USA). We also calculated the OS of 23 CML patients in advanced phases (AP and BP) resistant to IM 400 mg daily according to WASP expression levels.
WASP gene expression analysis among healthy individuals and patients' different groups were performed by using the GraphPad Prism software version 7 (GraphPad Software Inc., La Jolla, CA, USA).
References
Ahmed W, Van Etten RA . Signal transduction in the chronic leukemias: implications for targeted therapies. Curr Hematol Malign R ep 2013; 8: 71–80.
Wong S, Witte ON . The BCR-ABL story: bench to bedside and back. Ann Rev Immunol 2004; 22: 247–306.
Ren R . Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005; 5: 172–183.
Pendergast AM, Quilliam LA, Cripe LD, Bassing CH, Dai Z, Li N et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell 1993; 75: 175–185.
Amarante-Mendes GP, Naekyung Kim C, Liu L, Huang Y, Perkins CL, Green DR et al. Bcr-Abl exerts its antiapoptotic effect against diverse apoptotic stimuli through blockage of mitochondrial release of cytochrome C and activation of caspase-3. Blood 1998; 91: 1700–1705.
Amarante-Mendes GP, McGahon AJ, Nishioka WK, Afar DE, Witte ON, Green DR . Bcl-2-independent Bcr-Abl-mediated resistance to apoptosis: protection is correlated with up regulation of Bcl-xL . Oncogene 1998; 16: 1383–9130.
Aichberger KJ, Mayerhofer M, Krauth MT, Skvara H, Florian S, Sonneck K et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood 2005; 105: 3303–3311.
De Carvalho DD, Binato R, Pereira WO, Leroy JM, Colassanti MD, Proto-Siqueira R et al. BCR-ABL-mediated upregulation of PRAME is responsible for knocking down TRAIL in CML patients. Oncogene 2011; 30: 223–233.
Carroll M, Ohno-Jones S, Tamura S, Buchdunger E, Zimmermann J, Lydon NB et al. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood 1997; 90: 4947–4952.
Radich JP . The biology of chronic myelogenous leukemia progression: who, what, where, and why? Hematol Oncol Clin North Am 2011; 25: 967–980.
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001; 293: 876–880.
Pagnano KB, Bendit I, Boquimpani C, De Souza CA, Miranda EC, Zalcberg I et al. BCR-ABL mutations in chronic myeloid leukemia treated with tyrosine kinase inhibitors and impact on survival. Cancer Invest 2015; 33: 451–458.
White DL, Saunders VA, Dang P, Engler J, Venables A, Zrim S et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood 2007; 110: 4064–4072.
Dai Y, Rahmani M, Corey SJ, Dent P, Grant S . A Bcr/Abl-independent, Lyn-dependent form of imatinib mesylate (STI-571) resistance is associated with altered expression of Bcl-2. J Biol Chem 2004; 279: 34227–34239.
Silveira RA, Fachel AA, Moreira YB, De Souza CA, Costa FF, Verjovski-Almeida S et al. Protein-coding genes and long noncoding RNAs are differentially expressed in dasatinib-treated chronic myeloid leukemia patients with resistance to imatinib. Hematol 2014; 19: 31–41.
Radich JP . Chronic myeloid leukemia 2010: where are we now and where can we go? Hematology/the Education Program of the American Society of Hematology. Am Soc Hematol 2010; 2010: 122–128.
Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G . Structural organization of the bcr gene and its role in the Ph' translocation. Nature 1985; 315: 758–761.
Gross AW, Ren R . Bcr-Abl has a greater intrinsic capacity than v-Abl to induce the neoplastic expansion of myeloid cells. Oncogene 2000; 19: 6286–6296.
Soverini S, de Benedittis C, Mancini M, Martinelli G . Mutations in the BCR-ABL1 kinase domain and elsewhere in chronic myeloid leukemia. Clin Lymphoma Myeloma Leuk 2015; 15 (Suppl): S120–S128.
Machova Polakova K, Koblihova J, Stopka T . Role of epigenetics in chronic myeloid leukemia. Curr Hematol Malig Rep 2013; 8: 28–36.
Eaves C, Cashman J, Eaves A . Defective regulation of leukemic hematopoiesis in chronic myeloid leukemia. Leukemia Res 1998; 22: 1085–1096.
Radhika V, Thennarasu S, Naik NR, Kumar A, Advani SH, Bhisey AN . Granulocytes from chronic myeloid leukemia (CML) patients show differential response to different chemoattractants. Am J Hematol 1996; 52: 155–164.
Chang WC, Fujimiya Y, Casteel N, Pattengale P . Natural killer cell immunodeficiency in patients with chronic myelogenous leukemia. III. Defective interleukin-2 production by T-helper and natural killer cells. Int J Cancer 1989; 43: 591–597.
Mumprecht S, Claus C, Schurch C, Pavelic V, Matter MS, Ochsenbein AF . Defective homing and impaired induction of cytotoxic T cells by BCR/ABL-expressing dendritic cells. Blood 2009; 113: 4681–4689.
Massaad MJ, Ramesh N, Geha RS . Wiskott-Aldrich syndrome: a comprehensive review. Ann N Y Acad Sci 2013; 1285: 26–43.
Ariga T . Wiskott-Aldrich syndrome; an x-linked primary immunodeficiency disease with unique and characteristic features. Allergol Int 2012; 61: 183–189.
Albert MH, Notarangelo LD, Ochs HD . Clinical spectrum, pathophysiology and treatment of the Wiskott-Aldrich syndrome. Curr Opinion Hematol 2011; 18: 42–48.
Blundell MP, Worth A, Bouma G, Thrasher AJ . The Wiskott-Aldrich syndrome: the actin cytoskeleton and immune cell function. Dis Markers 2010; 29: 157–175.
Matalon O, Reicher B, Barda-Saad M . Wiskott-Aldrich syndrome protein—dynamic regulation of actin homeostasis: from activation through function and signal termination in T lymphocytes. Immunol Rev 2013; 256: 10–29.
Banin S, Truong O, Katz DR, Waterfield MD, Brickell PM, Gout I . Wiskott-Aldrich syndrome protein (WASp) is a binding partner for c-Src family protein-tyrosine kinases. Curr Biol 1996; 6: 981–988.
Scott MP, Zappacosta F, Kim EY, Annan RS, Miller WT . Identification of novel SH3 domain ligands for the Src family kinase Hck. Wiskott-Aldrich syndrome protein (WASP), WASP-interacting protein (WIP), and ELMO1. J Biol Chem 2002; 277: 28238–28246.
Schulte RJ, Sefton BM . Inhibition of the activity of SRC and Abl tyrosine protein kinases by the binding of the Wiskott-Aldrich syndrome protein. Biochemistry 2003; 42: 9424–9430.
Chou HC, Anton IM, Holt MR, Curcio C, Lanzardo S, Worth A et al. WIP regulates the stability and localization of WASP to podosomes in migrating dendritic cells. Curr Biol 2006; 16: 2337–2344.
Kelly TK, De Carvalho DD, Jones PA . Epigenetic modifications as therapeutic targets. Nature Biotech 2010; 28: 1069–1078.
Petrella A, Doti I, Agosti V, Giarrusso PC, Vitale D, Bond HM et al. A 5' regulatory sequence containing two Ets motifs controls the expression of the Wiskott-Aldrich syndrome protein (WASP) gene in human hematopoietic cells. Blood 1998; 91: 4554–4560.
Estey EH . Epigenetics in clinical practice: the examples of azacitidine and decitabine in myelodysplasia and acute myeloid leukemia. Leukemia 2013; 27: 1803–1812.
Schmelz K, Wagner M, Dorken B, Tamm I . 5-Aza-2'-deoxycytidine induces p21WAF expression by demethylation of p73 leading to p53-independent apoptosis in myeloid leukemia. Int J Cancer 2005; 114: 683–695.
Nimmanapalli R, Porosnicu M, Nguyen D, Worthington E, O'Bryan E, Perkins C et al. Cotreatment with STI-571 enhances tumor necrosis factor alpha-related apoptosis-inducing ligand (TRAIL or apo-2 L)-induced apoptosis of Bcr-Abl-positive human acute leukemia cells. Clin Cancer Res 2001; 7: 350–357.
Melo JV, Barnes DJ . Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 2007; 7: 441–453.
Perrotti D, Jamieson C, Goldman J, Skorski T . Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest 2010; 120: 2254–2264.
Martin TA, Pereira G, Watkins G, Mansel RE, Jiang WG . N-WASP is a putative tumour suppressor in breast cancer cells, in vitro and in vivo, and is associated with clinical outcome in patients with breast cancer. Clin Exp Metastasis 2008; 25: 97–108.
Shekarabi M, Moore SW, Tritsch NX, Morris SJ, Bouchard JF, Kennedy TE . Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. J Neurosci 2005; 25: 3132–3141.
Escudero-Esparza A, Jiang WG, Martin TA . Claudin-5 is involved in breast cancer cell motility through the N-WASP and ROCK signalling pathways. J Exp Clin Cancer Res 2012; 31: 43.
Rawlings SL, Crooks GM, Bockstoce D, Barsky LW, Parkman R, Weinberg KI . Spontaneous apoptosis in lymphocytes from patients with Wiskott-Aldrich syndrome: correlation of accelerated cell death and attenuated Bcl-2 expression. Blood 1999; 94: 3872–3882.
Rengan R, Ochs HD, Sweet LI, Keil ML, Gunning WT, Lachant NA et al. Actin cytoskeletal function is spared, but apoptosis is increased, in WAS patient hematopoietic cells. Blood 2000; 95: 1283–1292.
Mateo V, Brown EJ, Biron G, Rubio M, Fischer A, Deist FL et al. Mechanisms of CD47-induced caspase-independent cell death in normal and leukemic cells: link between phosphatidylserine exposure and cytoskeleton organization. Blood 2002; 100: 2882–2890.
Ichim CV . Kinase-independent mechanisms of resistance of leukemia stem cells to tyrosine kinase inhibitors. Stem Cells Transl Med 2014; 3: 405–415.
Warmuth M, Bergmann M, Priess A, Hauslmann K, Emmerich B, Hallek M . The Src family kinase Hck interacts with Bcr-Abl by a kinase-independent mechanism and phosphorylates the Grb2-binding site of Bcr. J Biol Chem 1997; 272: 33260–33270.
Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest 2013; 123: 4144–4157.
Bueno-da-Silva AE, Brumatti G, Russo FO, Green DR, Amarante-Mendes GP . Bcr-Abl-mediated resistance to apoptosis is independent of constant tyrosine-kinase activity. Cell Death Differ 2003; 10: 592–598.
Amarante-Mendes GP, Jascur T, Nishioka WK, Mustelin T, Green DR . Bcr - Abl-mediated resistance to apoptosis is independent of PI 3-kinase activity. Cell Death Diff 1997; 4: 548–554.
Bosticardo M, Ferrua F, Cavazzana M, Aiuti A . Gene therapy for Wiskott-Aldrich syndrome. Curr Gene Ther 2014; 14: 413–421.
Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M et al. Gene therapy for Wiskott-Aldrich syndrome—long-term efficacy and genotoxicity. Sci Transl Med 2014; 6: 227ra233.
Hacein-Bey Abina S, Gaspar HB, Blondeau J, Caccavelli L, Charrier S, Buckland K et al. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA 2015; 313: 1550–1563.
Schurch CM, Riether C, Ochsenbein AF . Dendritic cell-based immunotherapy for myeloid leukemias. Front Immunol 2013; 4: 496.
Held SA, Heine A, Mayer KT, Kapelle M, Wolf DG, Brossart P . Advances in immunotherapy of chronic myeloid leukemia CML. Curr Cancer Drug Targets 2013; 13: 768–774.
Watanabe T, Hotta C, Koizumi S, Miyashita K, Nakabayashi J, Kurotaki D et al. The transcription factor IRF8 counteracts BCR-ABL to rescue dendritic cell development in chronic myelogenous leukemia. Cancer Res 2013; 73: 6642–6653.
Galy A, Thrasher AJ . Gene therapy for the Wiskott-Aldrich syndrome. Curr Opin Allergy Clin Immunol 2011; 11: 545–550.
Monypenny J, Chou HC, Banon-Rodriguez I, Thrasher AJ, Anton IM, Jones GE et al. Role of WASP in cell polarity and podosome dynamics of myeloid cells. Eur J Cell Biol 2011; 90: 198–204.
De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 2012; 21: 655–667.
Avramouli A, Tsochas S, Mandala E, Katodritou E, Ioannou M, Ritis K et al. Methylation status of RASSF1A in patients with chronic myeloid leukemia. Leukemia Res 2009; 33: 1130–1132.
Borde-Chiche P, Diederich M, Morceau F, Puga A, Wellman M, Dicato M . Regulation of transcription of the glutathione S-transferase P1 gene by methylation of the minimal promoter in human leukemia cells. Biochem Pharmacol 2001; 61: 605–612.
Fan J, Kodama E, Koh Y, Nakao M, Matsuoka M . Halogenated thymidine analogues restore the expression of silenced genes without demethylation. Cancer Res 2005; 65: 6927–6933.
Acknowledgements
WOP was a recipient of a PhD and BEPE fellowships from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP – Brazil) and financial support from Amigos da Oncologia e Hematologia (AmigoH), Hospital Israelita Albert Einstein, São Paulo, Brazil. MEZ received a PhD fellowship from FAPESP. FAC was a recipient of BPE FAPESP 2015/21237-4. BFR is a recipient of a PhD fellowship from CAPES. This work was supported by grants from FAPESP and from the Brazilian Research Council (CNPq – Brazil) to GPA-M and KBP, and from the Burton Myeloma Program to YC. Work in DDC’s laboratory is supported by grants from the Cancer Research Society, Canadian Cancer Society, CIHR, NSERC, Ontario Institute for Cancer Research (OICR) with funds from the province of Ontario, the Princess Margaret Cancer Foundation, and the University of Toronto McLaughlin Centre. KBBP has received speaker funding and advisory fees from Bristol-Meyers Squibb and Novartis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Publisher’s Note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pereira, W., De Carvalho, D., Zenteno, M. et al. BCR–ABL1-induced downregulation of WASP in chronic myeloid leukemia involves epigenetic modification and contributes to malignancy. Cell Death Dis 8, e3114 (2017). https://doi.org/10.1038/cddis.2017.458
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.458
This article is cited by
-
USP10 modulates the SKP2/Bcr-Abl axis via stabilizing SKP2 in chronic myeloid leukemia
Cell Discovery (2019)