Abstract
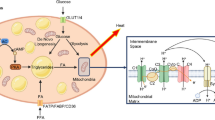
The lipid component of milk provides the critical nutritional source for generating both energy and essential nutrients to the growth of the newborn. Three types of substrate are utilized to synthesize milk triacylglycerides (TAG): dietary fat, fatty acids mobilized from adipose tissue stores, and lipids synthesized de novo synthesis from glucose and other dietary precursors, a process often referred to as de novo lipogenesis. The utilization of these various sources for TAG synthesis by the mammary epithelial cells is influenced by both the stage of lactation and the diet. From studies of gene expression in FVB mice, we observed that genes for β-oxidation of fatty acids are downregulated along with the expression of Acyl-CoA thioesterase 1 (ACOT1). As a control mechanism we propose that during pregnancy ACOT1 provides a supply of cytoplasmic free fatty acids which increase the activation of PPARγ. Ligand-induced activation of the PPAR/RXR transcription factor complex by free fatty acids, upregulates expression of genes required for β-oxidation of fatty acids. The fall in ACOTs at secretory activation may facilitate the switch to lipogenesis perhaps mediated by activation of the LXR/RXR transcription factor complex. The response to changes in the supply of dietary lipids, on the other hand, is likely to be mediated by SREBP1, possibly acting through modulation of Spot 14. Stability of SREBP1 may be enhanced by a significant increase in Akt at secretory activation. These regulatory pathways may be critical to the production of milk with a balanced TAG composition to support neonatal development of the newborn.






Similar content being viewed by others
Abbreviations
- ACBP:
-
acyl-CoA binding protein
- ACOT:
-
acyl-CoA thioesterase
- ADRP:
-
adipocyte differentiation regulated protein or adipophillin
- AGPAT:
-
acyl-glycerol-3-phosphate-acyl transferase
- ATP:
-
adenosine triphosphosphate
- ER:
-
endoplasmic reticulum
- ELOVL:
-
elongation of very long chain fatty acids
- FABP:
-
fatty acid binding protein
- FABP-FA:
-
fatty acid bound fatty acid binding protein
- FAD:
-
Δ5-fatty acid desaturase
- FASN:
-
fatty acid synthase
- FFA:
-
free fatty acid
- GK:
-
glycerol kinase
- GPAT:
-
glycerol-3-phosphate acyltransferase
- HSL:
-
hormone sensitive lipase
- INSIG:
-
insulin induced gene
- L2:
-
day 2 of lactation
- L9:
-
day 9 of lactation
- LACS:
-
long chain acyl-CoA synthase
- LC-PUFA:
-
long chain polyunsaturated fatty acid
- LPL:
-
lipoprotein lipase
- LXR:
-
liver X receptor
- MAG:
-
monoacylglycerol
- MCFA:
-
medium chain fatty acid
- mTOR:
-
mammalian target of Rapamycin
- MUFA:
-
monounsaturated fatty acid
- P17:
-
day 17 of pregnancy
- n-SREBP:
-
nuclear sterol response element binding protein
- PPAR:
-
peroxidase proliferators-activated receptor
- PPRE:
-
PPAR response element
- PUFA:
-
polyunsaturated fatty acid
- RXR:
-
retinoic X receptor
- S14:
-
Spot 14
- SCAP:
-
SREBP cleavage activating protein
- SCD:
-
stearoyl-coenzyme A desaturase
- SREBP:
-
sterol response element binding protein
- TAG:
-
triacyl glycerol
- VLDL:
-
very low density lipoprotein
Reference
Neville MC. Lactation and its hormonal control. In: Neill JD, editor. Knobil and Neils physiology of reproduction. 3rd ed. New York: Elsevier; 2006. p. 2993–3053.
Voet D, Voet JG. Biochemistry: biomolecules, mechanism of enyme action and metabolism , vol 1. 3rd ed. Hoboken: Wiley; 2004.
Larque ELVI, Demmelmair HANS, Koletzko BERT. Perinatal supply and metabolism of long-chain polyunsaturated fatty acids: importance for the early development of the nervous system. Ann NY Acad Sci 2002;967:299–310.
Schwertfeger KL, McManaman JL, Palmer CA, Neville MC, Anderson SM. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J Lipid Res 2003;44:1100–12.
Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia 2003;8:287–307.
Rudolph MC, McManaman JL, Phang T, Russel T, Kominsky DM, Serkova N, Anderson SM, Neville MC. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genomics 2006;28:323–36.
Insull W Jr, Hirsch J, James T, Ahrens EH. The fatty acids of human milk. II Alterations produced by manipulations of caloric balance and exhange of dietary fats. J Clin Invest 1958;38:443–50.
Neville MC, Picciano MF. Regulation of milk lipid synthesis and composition. Annu Rev Nutr 1997;17:159–84.
Jensen DR, Bessesen DH, Etienne J, Eckel RH, Neville MC. Distribution and source of lipoprotein lipase inmouse mammary gland. J Lipid Res 1991;32:733–42.
Ntambi JM, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res 2004;43:91–104.
Pegorier JP, May CL, Girard J. Control of gene expression by fatty acids. J Nutr 2004;134:2444S–9S.
Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med 2006;57:313–29.
Ide T, Shimano H, Yoshikawa T, Yahagi N, memiya-Kudo M, Matsuzaka T, Nakakuki M, Yatoh S, Iizuka Y, Tomita S, Ohashi K, Takahashi A, Sone H, Gotoda T, Osuga Ji, Ishibashi S, Yamada N. Cross-talk between peroxisome proliferator-activated receptor (PPAR) {alpha} and Liver X Receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol Endocrinol 2003;17:1255–67.
Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev 2000;14:2831–8.
Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev 2000;14:2819–30.
Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha áand delta. Proc Natl Acad Sci USA 1997;94:4312–7.
Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha áandágamma. Proc Natl Acad Sci USA 1997;94:4318–23.
Brauweiler A, Lorick KL, Lee JP, Tsai YC, Chan D, Weissman AM, Drabkin HA, Gemmill RM. RING-dependnet tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene. Oncogene 2007;26:2263–71.
Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, Zeng C, Erickson PF, Hooper JE, Drabkin HA. The TRC8 hereditary kidney gene suppresses growth and functions wiht VHL in a common pathway. Oncogene 2002;21:3507–16.
Kuramochi Y, Takagi-Sakuma M, Kitahara M, Emori R, Asaba Y, Sakaguchi R, Watanabe T, Kuroda J, Hiratsuka K, Nagae Y, Suga T, Yamada J. Characterization of mouse homolog of brain acyl-CoA hydrolase: molecular cloning and neuronal localization. Mol Brain Res 2002;98:81–92.
Kuramochi Y, Nishimura Si, Takagi-Sakuma M, Watanabe T, Kuroda J, Hiratsuka K, Nagae Y, Suga T, Yamada J. Immunohistochemical localization of acyl-CoA hydrolase/thioesterase multigene family members to rat epithelia. Histochem Cell Biol 2002;117:211–217.
Yamada J, Kuramochi Y, Takoda Y, Takagi M, Suga T. Hepatic induction of mitochondrial and cytosolic acyl-coenzyme a hydrolases/thioesterases in rats under conditions of diabetes and fasting. Metabolism 2003;52:1527–9.
Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5′-AMP activated protein kinase activator, 5-aminoimidazole-4-carboxamide-1-[beta]-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun 2001;287:562–7.
Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha á(PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA 1999;96:7473–8.
Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 2003;100:12027–32.
Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 2004;6:839–48.
Goldstein JL, Bose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell 2006;24:35–46.
Rawson RB. The SREBP pathway [mdash] insights from insigs and insects. Nat Rev Mol Cell Biol 2003;4:631–40.
Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 2002;109:1125–31.
Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest 1997;99:838–45.
Du X, Kristiana I, Wong J, Brown AJ. Involvement of Akt in ER-to-golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol Biol Cell 2006;17:2735–45.
Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metaboliusmand beyond. Curr Opin Cell Biol 2007;19:215–22.
Gimpl G, Burger K, Fahrenholz F. A closer look at the cholersterol sensor. Trends Biochem Sci 2002;27:596–9.
Sundqvist A, goechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCFFbw7. Cell Metabolism 2005;1:379–91.
Lee JN, Gong Y, Zhang X, Ye J. Proteasomal degradation of ubiquitinated Insig proteins is determined by serine residues flanking ubiquitinated lysines. Proc Natl Acad Sci USA 2006;103:4958–63.
memiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga Ji, Harada K, Gotoda T, Sato R, Kimura S, Ishibashi S, Yamada N. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J Lipid Res 2002;43:1220–35.
Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab 2002;13:444–51.
Yang ZZ, Tschopp O, Bauudy A, Dümmler B, Hynx D, Hemmings BA. Physiological functions of protein kinase B/Akt. Biochem Soc Trans 2004;32:350–4.
DeBerardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem 2006;281:37372–80.
Anderson SM, Rudolph MC, McManaman JL, Neville MC. Secretory activation in the mammary gland: it’s not just about milk protein synthesis. Breast Cancer Res 2007;9:204–17.
Summers SA, Kao AW, Kohn AD, Backus GS, Roth RA, Pessin JE, Birnbaum MJ. The role of glycogen synthase kinase 3β in insulin-stimulated glucose metabolism. J Biol Chem 2000;274:17934–40.
Boxer RB, Stairs DB, Dugan KD, Notarfrancesco KL, Portocarrero CP, Keister BA, Belka GK, Cho H, Rathmell JC, Thompson CB, Birnbaum MJ, Chodosh LA. Isoform-specific requirement for Akt1 in the developmental regulation of cellular metabolism during lactation. Cell Metab 2006;4:475–90.
Porstmann T, Griffiths B, CHung Y-L, Delpuech O, Griffiths JR, Downward J, Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 2005;24:6465–81.
Seelig S, Liaw C, Towle H, Oppenheimer J. Thyroid hormone attenuates and augments hepatic gene expression at a pretranslational level. Proc Natl Acad Sci USA 1981;78:4733–7.
Kinlaw WB, Tron P, Friedmann AS. Nuclear localization and hepatic zonation of rat “spot 14” protein: immunohistochemical investigation employing anti-fusion protein antibodies. Endocrinology 1992;131:3120–2.
Kinlaw WB, Church JL, Harmon J, Mariash CN. Direct evidence for a role of the “Spot 14” Protein in the Regulation of Lipid Synthesis. J Biol Chem 1995;270:16615–8.
Zhu Q, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, Mariash CN. The spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology 2005;146:3343–50.
Brown S, Maloney M, Kinlaw W. “Spot 14” protein functions at the pretranslational level in the regulation of hepatic metabolism by thyroid hormone and glucose. J Biol Chem 1997;272:2163–6.
Blackman B, Russell T, Nordeen S, Medina D, Neville M. Claudin 7 expression and localization in the normal murine mammary gland and murine mammary tumors. Breast Cancer Res 2005;7:R248–55.
Stein T, Morris J, Davies C, Weber-Hall S, Duffy MA, Heath V, Bell A, Ferrier R, Sandilands G, Gusterson B. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res 2004;6:R75–91.
Wan Y, Saghatelian A, Chong LW, Zhang CL, Cravatt BF, Evans RM. Maternal PPARγ protects nursing neonates by suppressing the production of inflammatory milk. Genes Dev 2007;21:1895–1908.
Acknowledgements
We especially thank Dr. Yamada, Department of Clinical Biochemistry at Tokyo University of Pharmacy and Life Sciences, for kindly providing reagents and for his informative correspondence. We also acknowledge Lisa Litzenberger for her assistance with the figures presented in this review. The authors would also like to thank the members of the Mammary Gland Biology Program Project Grant at the University of Colorado Health Sciences Center for discussions and sharing of ideas. Research in our labs (MCN and SMA) is supported by PO1-HD38129.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rudolph, M.C., Neville, M.C. & Anderson, S.M. Lipid Synthesis in Lactation: Diet and the Fatty Acid Switch. J Mammary Gland Biol Neoplasia 12, 269–281 (2007). https://doi.org/10.1007/s10911-007-9061-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-007-9061-5