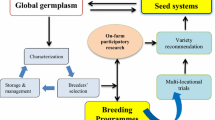
Abstract
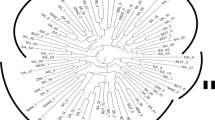
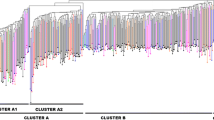
Kersting’s groundnut (Macrotyloma geocarpum (Harms) Maréchal & Baudet) is an important source of protein and essential nutrients that contribute to food security in West Africa. However, the crop is still underexploited by the populations and under-researched by the scientific community. This study aimed to investigate the genetic diversity and population structure of 217 Kersting’s groundnut accessions from five origins using 886 DArTseq markers. Gene diversity was low and ranged from 0.049 to 0.064. The number of private alleles greatly varied among populations (42–192) and morphotypes (40–339). Moderate to very high levels of selfing and inbreeding were observed among populations (s = 56–85%, FIS = 0.389–0.736) and morphotypes (s = 57–82%, FIS = 0.400–0.691). Moreover, little to very high genetic differentiations were observed among populations (0.006 ≤ FIS ≤ 0.371) and morphotypes (0.029 ≤ FIS ≤ 0.307). Analysis of molecular variance partitioned 38.5% of the genetic variation among and 48.7% within populations (P < 0.001). Significant isolation by distance was detected between populations (R2 = 0.612, P = 0.011) and accessions (R2 = 0.499, P < 0.001). Discriminant analysis of principal components and neighbour joining consistently distinguished eight distinct clusters. These data provide a global picture of the existing genetic diversity for Kersting’s groundnut and will guide the choice of breeding strategies to increase production.




Similar content being viewed by others
Availability of data and material
The datasets supporting the conclusions of this article are included within the article.
References
Achigan Dako EG, Vodouhè SR (2006) Macrotyloma geocarpum (Harms) Maréchal and Baudet. Brink M, Belay G (Editeurs) PROTA 1
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5(4):384–387
Adu-Gyamfi R, Dzomeku IK, Lardi J (2012) Evaluation of growth and yield potential of genotypes of Kersting’s groundnut (macrotyloma geocarpum harms) in Northern Ghana. Int Res J Agric Sci Soil Sci 2(12):509–515
Adu-Gyamfi R, Fearon J, Bayorbor T, Dzomeku I, Avornyo V (2011) The Status of Kersting’s Groundnut (Macrotyloma Geocarpum [Harms] Marechal and Baudet) An Underexploited Legume in Northern Ghana. Outlook Agric 40(3):259–262
Agoyi EE, N’danikou S, Kafoutchoni M, Ayena M, Sodedji FA, Agbahoungba S, Sossou HS, Vodouhe R, Assogbadjo AE (2019a) Kersting’s Groundnut [Macrotyloma geocarpum (Harms) Maréchal & Baudet] crop attracts more field pests and diseases than reported before. Agric Res Technol Open Access J 21(5)
Agoyi EE, Tumuhairwe JB, Chigeza G, Tukamuhabwa P, Diers BW (2019b) First attempt to identify and map QTLs associated with promiscuous nodulation In Soybean. bioRxiv:688028
Ajala SO, Olayiwola MO, Ilesanmi OJ, Gedil M, Job AO, Olaniyan AB (2019) Assessment of genetic diversity among low-nitrogen-tolerant early generation maize inbred lines using SNP markers. South Afr J Plant Soil 36(3):181–188
Akohoue F, Achigan-Dako EG, Coulibaly M, Sibiya J (2019) Correlations, path coefficient analysis and phenotypic diversity of a West African germplasm of Kersting’s groundnut [Macrotyloma geocarpum (Harms) Maréchal & Baudet]. Genet Resour Crop Evol 66(8):1825–1842
Akohoue F, Achigan-Dako EG, Sneller C, Van Deynze A, Sibiya J (2020) Genetic diversity, SNP-trait associations and genomic selection accuracy in a west African collection of Kersting’s groundnut [Macrotyloma geocarpum (Harms) Maréchal & Baudet]. PLoS ONE 15(6):e0234769
Akohoue F, Sibiya J, Achigan-Dako EG (2018) On-farm practices, mapping, and uses of genetic resources of Kersting’s groundnut [Macrotyloma geocarpum (Harms) Maréchal et Baudet] across ecological zones in Benin and Togo. Genet Resour Crop Evol 66(1):195–214
Alam M, Neal J, O’Connor K, Kilian A, Topp B (2018) Ultra-high-throughput DArTseq-based silicoDArT and SNP markers for genomic studies in macadamia. PLoS ONE 13(8):e0203465
Amujoyegbe B, Obisesan I, Ajayi A, Aderanti F (2007) Disappearance of Kersting's groundnut (Macrotyloma geocarpum (Harms) Marechal and Baudet) in south-western Nigeria: an indicator of genetic erosion. Plant Genetic Resources Newsletter (Bioversity International/FAO)
Assogba P, Ewedje EB, Dansi A, Loko Y, Adjatin A, Dansi M, Sanni A (2015) Indigenous knowledge and agro-morphological evaluation of the minor crop Kersting’s groundnut (Macrotyloma geocarpum (Harms) Maréchal et Baudet) cultivars of Benin. Genet Resour Crop Evol 63(3):513–529
Bampuori AH (2007) Effect of Traditional Farming Practices on the Yield of Indigenous Kersting’s Groundnut (Macrotyloma geocarpum Harms) Crop in the Upper West Region of Ghana. J Dev Sustain Agric 2(2):128–144
Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Vale G, Cattivelli L (2016) Next generation breeding. Plant Sci 242:3–13. https://doi.org/10.1016/j.plantsci.2015.07.010
Bayorbor TB, Dzomeku IK, Avornyo VK, Opoku-Agyeman MO (2010) Morphological variation in Kersting’s groundnut (Kerstigiella geocarpa Harms) landraces from northern Ghana. Agric Biol J N Am 1(3):290–295
Campa A, Murube E, Ferreira JJ (2018) Genetic diversity, population structure, and linkage disequilibrium in a spanish common bean diversity panel revealed through genotyping-by-sequencing. Genes 9(11):518
Campoy JA, Lerigoleur-Balsemin E, Christmann H, Beauvieux R, Girollet N, Quero-García J, Dirlewanger E, Barreneche T (2016) Genetic diversity, linkage disequilibrium, population structure and construction of a core collection of Prunus avium L. landraces and bred cultivars. BMC Plant Biol 16(1):1–15
Chen W, Hou L, Zhang Z, Pang X, Li Y (2017) Genetic diversity, population structure, and linkage disequilibrium of a core collection of Ziziphus jujuba assessed with genome-wide SNPs developed by genotyping-by-sequencing and SSR markers. Front Plant Sci 8:575
Chikwendu JN (2015) Comparative evaluation of chemical composition of fermented ground bean flour (Kerstingella geocarpa), cowpea flour (Vigna unguiculata) and commercial wheat flour (Triticum spp.). Pak J Nutr 14:218–224
Cortés AJ, Blair MW (2018) Genotyping by sequencing and genome-environment associations in wild common bean predict widespread divergent adaptation to drought. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00128
Dansi A, Vodouhe R, Azokpota P, Yedomonhan H, Assogba P, Adjatin A, Loko YL, Dossou-Aminon I, Akpagana K (2012) Diversity of the neglected and underutilized crop species of importance in Benin. ScientificWorldJournal 2012:932947. https://doi.org/10.1100/2012/932947
Dray S, Dufour A-B (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22(4):1–20
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits? Am Nat 169(5):662–672
Echendu A, Obizoba I, Ngwu E, Anyika J (2009) Chemical composition of ground bean based cocoyam, yam and plantain pottage dishes and roasted ground bean. Pak J Nutr 8(11):1786–1790
Egea LA, Mérida-García R, Kilian A, Hernandez P, Dorado G (2017) Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology “genotyping-by-sequencing” platform (DArTseq). Front Genet 8:98
Eltaher S, Sallam A, Belamkar V, Emara HA, Nower AA, Salem KF, Poland J, Baenziger PS (2018) Genetic diversity and population structure of F3: 6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front Genet 9:76
Fatokun C, Girma G, Abberton M, Gedil M, Unachukwu N, Oyatomi O, Yusuf M, Rabbi I, Boukar O (2018) Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci Rep 8(1):1–10
Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. R package version 004-22 10
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487. https://doi.org/10.1155/2015/431487
Hamrick J, Godt M, Murawski D, Loveless M (1991) Correlations between species traits and allozyme diversity: implications for conservation biology. Genet Conserv Rare Plants 3:30
Hamrick JL, Godt MW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci 351(1345):1291–1298
Hassani SMR, Talebi R, Pourdad SS, Naji AM, Fayaz F (2020) In-depth genome diversity, population structure and linkage disequilibrium analysis of worldwide diverse safflower (Carthamus tinctorius L.) accessions using NGS data generated by DArTseq technology. Mol Biol Rep 47(3):2123–2135
Hepper F (1963) Plants of the 1957–58 West African Expedition: II. The Bambara Groundnut (Voandzeia subterranea) and Kersting’s Groundnut (Kerstingiella geocarpa) Wild in West Africa. Kew Bull 16(3):395–407
Iquira E, Humira S, François B (2015) Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol 15(1):5
Isidro J, Jannink J-L, Akdemir D, Poland J, Heslot N, Sorrells ME (2015) Training set optimization under population structure in genomic selection. Theor Appl Genet 128(1):145–158
Islam AF, Blair MW (2018) Molecular characterization of Mung bean germplasm from the USDA core collection using newly developed KASP-based SNP markers. Crop Sci 58(4):1659–1670
Jaiswal SK, Mohammed M, Dakora FD (2019) Microbial community structure in the rhizosphere of the orphan legume Kersting’s groundnut [Macrotyloma geocarpum (Harms) Marechal & Baudet]. Mol Biol Rep 46(4):4471–4481
Jarquín D, Kocak K, Posadas L, Hyma K, Jedlicka J, Graef G, Lorenz A (2014) Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom 15(1):740
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405
Kabbaj H, Sall AT, Al-Abdallat A, Geleta M, Amri A, Filali-Maltouf A, Belkadi B, Ortiz R, Bassi FM (2017) Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front Plant Sci 8:1277
Kafoutchoni KM, Agoyi EE, Dassou GH, Sossou HS, Ayi S, Glèlè CA, Adomou AC, Yédomonhan H, Agbangla C, Assogbadjo AE (2021) Reproductive biology, phenology, pollen viability and germinability in Kersting’s groundnut (Macrotyloma geocarpum (Harms) Maréchal & Baudet, Fabaceae). S Afr J Bot 137:440–450
Kafoutchoni KM, Agoyi EE, Sossou HS, Agbahoungba S, Agbangla C, Assogbadjo AE (2021) Impacts of sociodemographic factors on the production system of the orphan legume Kersting’s groundnut (Macrotyloma geocarpum) and prioritisation of production constraints in Benin. Environ Dev Sustain (in press)
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In: Data production and analysis in population genomics. Humana Press, Totowa, NJ, pp 67–89
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9(1):29. https://doi.org/10.1186/1746-4811-9-29
Kumar S, Ambreen H, Variath MT, Rao AR, Agarwal M, Kumar A, Goel S, Jagannath A (2016) Utilization of molecular, phenotypic, and geographical diversity to develop compact composite core collection in the oilseed crop, safflower (Carthamus tinctorius L.) through maximization strategy. Front Plant Sci 7:1554
Mensah AG, Gyan AD, Yeboah AM (2016) An assessment on variation among genotypes of kersting’s groundnut in Northern Ghana. Adv Agric Agric Sci 2(7):149–155
Mergeai G (1993) Influence des facteurs sociologiques sur la conservation des ressources phytogénétiques. Le cas de la lentille de terre (Macrotyloma geocarpum (Harms) Marechal & Baudet) au Togo. Bull des Recherches Agronomiques de Gembloux 28(4):487–500
Mogga M, Sibiya J, Shimelis H, Lamo J, Yao N (2018) Diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PloS one 13(6):e0198012
Mondini L, Noorani A, Pagnotta MA (2009) Assessing plant genetic diversity by molecular tools. Diversity 1(1):19–35
Morris J (2008) Macrotyloma axillare and M. uniflorum: descriptor analysis, anthocyanin indexes, and potential uses. Genet Resour Crop Evol 55(1):5–8
Ndjiondjop M-N, Semagn K, Gouda AC, Kpeki SB, Dro Tia D, Sow M, Goungoulou A, Sie M, Perrier X, Ghesquiere A (2017) Genetic variation and population structure of Oryza glaberrima and development of a mini-core collection using DArTseq. Front Plant Sci 8:1748
Ndjiondjop MN, Semagn K, Sow M, Manneh B, Gouda AC, Kpeki SB, Pegalepo E, Wambugu P, Sié M, Warburton ML (2018) Assessment of genetic variation and population structure of diverse rice genotypes adapted to lowland and upland ecologies in Africa using SNPs. Front Plant Sci 9:446
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70(12):3321–3323
Nemli S, Aşçioğul TK, Ateş D, Eşiyok D, Tanyolac MB (2017) Diversity and genetic analysis through DArTseq in common bean (Phaseolus vulgaris L.) germplasm from Turkey. Turk J Agric For 41(5):389–404
O’Leary SJ, Puritz JB, Willis SC, Hollenbeck CM, Portnoy DS (2018) These aren’t the loci you’e looking for: principles of effective SNP filtering for molecular ecologists. Mol Ecol 27(16):3193–3206
O’Connor K, Kilian A, Hayes B, Hardner C, Nock C, Baten A, Alam M, Topp B (2019) Population structure, genetic diversity and linkage disequilibrium in a macadamia breeding population using SNP and silicoDArT markers. Tree Genet Genomes 15(2):24
Pasquet RS, Mergeai G, Baudoin J-P (2002) Genetic diversity of the African geocarpic legume Kersting’s groundnut, Macrotyloma geocarpum (Tribe Phaseoleae: Fabaceae). Biochem Syst Ecol 30(10):943–952
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28(19):2537
Priyanka S, Sudhagar R, Vanniarajan C, Ganesamurthy K, Souframanien J (2019) Combined mutagenic ability of gamma ray and EMS in horsegram (Macrotyloma uniflorum (Lam) Verdc.). Electron J Plant Breed 10(3):1086–1094
Pusadee T, Jamjod S, Chiang Y-C, Rerkasem B, Schaal BA (2009) Genetic structure and isolation by distance in a landrace of Thai rice. Proc Natl Acad Sci 106(33):13880–13885
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Redjeki ES, Ho WK, Shah N, Molosiwa OO, Ardiarini N, Kuswanto MS (2020) Understanding the genetic relationships between Indonesian bambara groundnut landraces and investigating their origins. Genome 999:1–9
Ritland K (1990) Inferences about inbreeding depression based on changes of the inbreeding coefficient. Evolution 44(5):1230–1241
Robbana C, Kehel Z, Naceur B, Sansaloni C, Bassi F, Amri A (2019) Genome-wide genetic diversity and population structure of tunisian durum wheat landraces based on DArTseq technology. Int J Mol Sci 20(6):1352
Schröder S, Mamidi S, Lee R, McKain MR, McClean PE, Osorno JM (2016) Optimization of genotyping by sequencing (GBS) data in common bean (Phaseolus vulgaris L.). Mol Breed 36(1):6
Shaibu AS, Sneller C, Motagi BN, Chepkoech J, Chepngetich M, Miko ZL, Isa AM, Ajeigbe HA, Mohammed SG (2020) Genome-wide detection of SNP markers associated with four physiological traits in groundnut (Arachis hypogaea L.) mini core collection. Agronomy 10(2):192
Sharma S, Upadhyaya H, Varshney R, Gowda C (2013) Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00309
Singh S, Gupta S, Thudi M, Das RR, Vemula A, Garg V, Varshney R, Rathore A, Pahuja S, Yadav DV (2018) Genetic diversity patterns and heterosis prediction based on SSRs and SNPs in hybrid parents of pearl millet. Crop Sci 58(6):2379–2390
Szczecińska M, Sramko G, Wołosz K, Sawicki J (2016) Genetic diversity and population structure of the rare and endangered plant species Pulsatilla patens (L.) Mill in East Central Europe. PLoS ONE 11(3):e0151730
Tamini Z (1997) Etude ethnobotanique et analyses morphophysiologiques du développement de la lentille de terre (Macrotyloma geocarpum (Harms) Marechal et Baudet. Université de Ouagadougou, Burkina-Faso
Varshney RK, Close TJ, Singh NK, Hoisington DA, Cook DR (2009) Orphan legume crops enter the genomics era! Curr Opin Plant Biol 12(2):202–210. https://doi.org/10.1016/j.pbi.2008.12.004
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91(11):1846–1862
Wright S (1978) Evolution and the genetics of populations, volume 4: variability within and among natural populations, 4th edn. University of Chicago Press, Chicago
Xiong H, Shi A, Mou B, Qin J, Motes D, Lu W, Ma J, Weng Y, Yang W, Wu D (2016) Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS ONE 11(8):e0160941
Yang X, Ren R, Ray R, Xu J, Li P, Zhang M, Liu G, Yao X, Kilian A (2016) Genetic diversity and population structure of core watermelon (Citrullus lanatus) genotypes using DArTseq-based SNPs. Plant Genet Resour 14(3):226–233
Yang Y, Pan Y, Gong X, Fan M (2010) Genetic variation in the endangered Rutaceae species Citrus hongheensis based on ISSR fingerprinting. Genet Resour Crop Evol 57(8):1239–1248
Yelome OI, Audenaert K, Landschoot S, Dansi A, Vanhove W, Silue D, Van Damme P, Haesaert G (2018) Analysis of population structure and genetic diversity reveals gene flow and geographic patterns in cultivated rice (O. sativa and O. glaberrima) in West Africa. Euphytica 214(11):215
Zavinon F, Adoukonou-Sagbadja H, Keilwagen J, Lehnert H, Ordon F, Perovic D (2020) Genetic diversity and population structure in Beninese pigeon pea [Cajanus cajan (L.) Huth] landraces collection revealed by SSR and genome wide SNP markers. Genet Resour Crop Evol 67(1):191–208
Acknowledgements
We are grateful to the different institutions that provided the accessions used in this study: IITA-Nigeria, INERA-Burkina-Faso, SARI-Ghana. Finally, we thank Dr. Mustapha Mohammed (Tshwane University of Technology, Pretoria, South Africa) for providing additional accessions from Ghana.
Funding
This research was funded by the World Academy of Sciences (TWAS), Grant number 18-238 RG/BIO/AF/AC_G-FR3240303667. We thank Carnegie Cooperation of New York for funding part of the work through the Regional Universities Forum for Capacity Building in Agriculture (RUFORUM) Grant number RU/2018/TQA/38.
Author information
Authors and Affiliations
Contributions
Conceptualization, Konoutan M. Kafoutchoni and Eric E. Agoyi; Data curation, Konoutan M. Kafoutchoni; Formal analysis, Konoutan M. Kafoutchoni; Funding acquisition, Konoutan M. Kafoutchoni, Symphorien Agbahoungba and Achille E. Assogbadjo; Project administration, Eric E. Agoyi; Resources, Eric E. Agoyi, Achille E. Assogbadjo and Clément Agbangla; Supervision, Achille E. Assogbadjo and Clément Agbangla; Writing – original draft, Konoutan M. Kafoutchoni; Writing – review & editing, Konoutan M. Kafoutchoni, Eric E. Agoyi and Symphorien Agbahoungba.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kafoutchoni, K.M., Agoyi, E.E., Agbahoungba, S. et al. Genetic diversity and population structure in a regional collection of Kersting’s groundnut (Macrotyloma geocarpum (Harms) Maréchal & Baudet). Genet Resour Crop Evol 68, 3285–3300 (2021). https://doi.org/10.1007/s10722-021-01187-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01187-4