Abstract
The oomycete pathogen Plasmopara viticola (Berk. et Curt.) Berl. et de Toni is the causing agent of the destructive downy mildew disease in grapevine. Despite the advances towards elucidation of grapevine resistance mechanisms to downy mildew, increased knowledge of the biological and genetic components of the pathosystem is important to design suitable breeding strategies. Previously, a cDNA microarray approach was used to compare two Vitis vinifera genotypes Regent and Trincadeira (resistant and susceptible to downy mildew, respectively) in field conditions. The same cDNA microarray chip was used to confirm field-based results and to compare both genotypes under greenhouse conditions at 0, 6, and 12 h post-inoculation with P. viticola. Results show that when comparing both cultivars after pathogen inoculation, there is a preferential modulation of several defense, signaling, and metabolism associated transcripts in Regent. Early transcriptional changes are discussed in terms of genetic background and resistance mechanism. This study is the first to directly compare resistant and susceptible cultivars responses as early as 6 hpi with P. viticola, providing several candidate genes potentially related to the expression of resistance traits.


Similar content being viewed by others
Abbreviations
- cDNA:
-
Complementary DNA
- EST:
-
Expressed sequence tag
- NCBI:
-
National Center for Biotechnology Information
- MIPS:
-
Munich information center for protein sequences
- hpi:
-
Hours post-inoculation
References
Akkurt M, Welter L, Maul E, Topfer R, Zyprian E (2007) Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L. and Vitis sp.). Mol Breed 19:103–111
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Anonymous (2000) Description list of varieties—grapes 2000. Landburg Verlag, Hannover
Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J, Harris M, Hill D, Issel-Tarver L, Kasarskis A, Lewis S, Matese J, Richardson J, Ringwald M, Rubin G, Sherlock G, Consortium GO (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G et al (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genetics 120:163–176
Bilgin D, Zavala J, Zhu J, Clough S, Ort D, DeLucia E (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33(10):1597–1613
Bisson L, Waterhouse A, Ebeler S, Walker M, Lapsley J (2002) The present and future of the international wine industry. Nature 418:696–699
Blasi P, Blanc S, Wiedemann-Merdinoglu S, Prado E, Rühl EH, Mestre P, Merdinoglu D (2011) Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor Appl Genet 123:43–53
Blein J, Coutos-Thevenot P, Marion D, Ponchet M (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defense mechanisms. Trends Plant Sci 7(7):293–296
Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92
Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl M, Shipley G, Vandesompele J, Wittwer C (2009a) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Bustin S, Vandesompele J, Pfaffl M (2009b) Standardization of qPCR and RT-qPCR. Genet Eng Biotechnol 29(14):40–43
Coffeen W, Wolpert T (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16:857–873
Dalbo M, Ye G, Weeden N, Steinkellner H, Sefc K, Reisch B (2000) A gene controlling sex in grapevines placed on a molecular marker-based genetic map. Genome 43(2):333–340
Diez-Navajas A, Wiedemann-Merdinoglu S, Greif C, Merdinoglu D (2008) Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 98:776–780
Doddapaneni H, Lin H, Walker M, Yao J, Civerolo E (2008) VitisExpDB: a database resource for grape functional genomics. BMC Plant Biol 8:23
Espinoza C, Vega A, Medina C, Schlauch K, Cramer G, Arce-Johnson P (2007) Gene expression associated with compatible viral diseases in grapevine cultivars. Funct Integr Genomics 7:95–110
FAOSTAT (2010) Food and Agriculture Organization of the United Nations (FAOSTAT). http://apps.fao.org
Figueiredo A, Fortes A, Ferreira S, Sebastiana M, Choi Y, Sousa L, Acioli-Santos B, Pessoa F, Verpoorte R, Pais M (2008) Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J Exp Bot 59:3371–3381
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards K, Topfer R, Zyprian E (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Gene 108(3):501–515
Fung R, Gonzalo M, Fekete C, Kovacs L, He Y, Marsh E, McIntyre L, Schachtman D, Qiu W (2008) Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 146:236–249
Galet P (1977) Apoplexie. In: Les maladies et les parasites de la vigne, Vol. I (pp. 409–430) Imp. Paysan du Midi, Montpellier
Godoy A, Lazzaro A, Casalongue C, San Segundo B (2000) Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci 152(2):123–134
Golldack D, Vera P, Dietz K (2003) Expression of subtilisin-like serine proteases in Arabidopsis thaliana is cell-specific and responds to jasmonic acid and heavy metals with developmental differences. Physiol Plant 118(1):64–73
Gutha L, Casassa L, Harbertson J, Naidu R (2010) Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol 10:187
Hofius D, Tsitsigiannis D, Jones J, Mundy J (2007) Inducible cell death in plant immunity. Semin Cancer Biol 17:166–187
Hren M, Nikolic P, Rotter A, Blejec A, Terrier N, Ravnikar M, Dermastia M, Gruden K (2009) ‘Bois noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics 10:460
Jorda L, Coego A, Conejero V, Vera P (1999) Genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem 274:2360–2365
Jurges G, Kassemeyer H-H, Dürrenberger M, Duggelin M, Nick P (2009) The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol 11(6):886–898
Kong HY, Lee SC, Hwan BK (2001) Expression of pepper cyclophilin gene is differentially regulated during the pathogen infection and abiotic stress conditions. Physiol Mol Plant Pathol 59:189–199
Kortekamp A, Zyprian E (2003) Characterization of Plasmopara-resistance in grapevine using in vitro plants. J Plant Physiol 160:1393–1400
Kortekamp A, Welter L, Vogt S, Knoll A, Schwander F, Topfer R, Zyprian E (2008) Identification, isolation and characterization of a CC-NBS-LRR candidate disease resistance gene family in grapevine. Mol Breed 22(3):421–432
Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J (2005) The TIGR gene indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33:D71–D74
Lee J, Park S, Kim J, Lee S, Park Y, Cheong G, Hahm K, Lee S (2007) Molecular and functional characterization of a cyclophilin with antifungal activity from Chinese cabbage. Biochem Bioph Res Co 353(3):672–678
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25:402–408
Lukasik E, Takken F (2009) STANDing strong, resistance proteins instigators of plant defense. Curr Opin Plant Biol 12:427–436
Malacarne G, Vrhovsek U, Zulini L, Cestarol A, Stefanini M, Mattivi F, Delledonne M, Velasco R, Moser C (2011) Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol 11:114
Merdinoglu D, Wiedemann-Merdinoglu S, Coste P, Dumas V, Haetty S, Butterlin G, Greif C (2003) Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hort 451–456.
Palloix A, Ayme V, Moury B (2009) Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol 183:190–199
Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, Bellin D, Di Gaspero G, Testolin R, Merdinoglu D, Mestre P (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10:147
Polesani M, Desario F, Ferrarini A, Zamboni A, Pezzotti M, Kortekamp A, Polverari A (2008) CDNA-AFLP analysis of plant and pathogen genes expressed in grapevine infected with Plasmopara viticola. BMC Genomics 9:142
Polesani M, Bortesi L, Ferrarini A, Zamboni A, Fasoli M, Zadra C, Lovato A, Pezzotti M, Delledonne M, Polverari A (2010) General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics 11:117–133
Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29(1):159–164
Richard S, Lapointe G, Rutledge R, Seguin A (2000) Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol 41(8):982–987
Rotter A, Camps C, Lohse M, Kappel C, Pilati S, Hren M, Stitt M, Coutos-Thevenot P, Moser C, Usadel B, Delrot S, Gruden K (2009) Gene expression profiling in susceptible interaction of grapevine with its fungal pathogen Eutypa lata: extending MapMan ontology for grapevine. BMC Plant Biol 9:104
Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes H (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32:5539–5545
Schwander F, Eibach R, Fechter I, Hausmann L, Zyprian E, Topfer R (2011) Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Gen. doi:10.007/s00122-011-1695-4
Soria-Guerra R, Rosales-Mendoza S, Chang S, Haudenshield J, Padmanaban A, Rodriguez-Zas S, Hartman G, Ghabrial S, Korban S (2010) Transcriptome analysis of resistant and susceptible genotypes of Glycine tomentella during Phakopsora pachyrhizi infection reveals novel rust resistance genes. Theor Appl Genet 120(7):1315–1333
Susnow N, Zeng L, Margineantu D, Hockenbery D (2009) Bcl-2 family proteins as regulators of oxidative stress. Semin Cancer Biol 19(1):42–49
Suzek B, Huang H, McGarvey P, Mazumder R, Wu C (2007) UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288
Takken FL, Albrecht M, Tameling WI (2006) Resistance proteins: molecular switches of plant defense. Curr Opin Plant Biol 9:383–390
Tornero P, Conejero V, Vera P (1996) Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proc Natl Acad Sci USA 6332–6337
Tornero P, Conejero V, Vera P (1997) Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem 272:14412–14419
Treutter D (2006) Significance of flavonoids in plant resistance: a review. Environ Chem Lett 4:147–157
van der Hoorn R, Jones J (2004) The plant proteolytic machinery and its role in defense. Curr Opin Plant Biol 7:400–407
Vitis International Variety Catalogue. http://www.vivc.de. Accessed 2 June 2011
Welter L, Gokturk-Baydar N, Akkurt M, Maul E, Eibach R, Topfer R, Zyprian E (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breed 20(4):359–374
Wu J, Zhang Y, Zhang H, Huang H, Folta K, Lu J (2010) Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol 10:16
Yan J, He C, Zhang H (2003) The BAG-family proteins in Arabidopsis thaliana. Plant Sci 165(1):1–7
Zeng Y, Yang T (2002) RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep 20: 417a–417e
Acknowledgements
This work was developed and supported within the frame of the project “Genomic Research-Assisted breeding for Sustainable Production of quality GRAPEs and WINE”, ERA-NET Plant Genomics granted research project (0313996A), ERA-PG/0004/2006, and by the Portuguese Foundation for Science and Technology with the fellowship SFRH/BPD/33281/2008 and SFRH/BPD/63641/2009.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Technical details of Array Design and Spotting provided as MIAME (PDF 135 kb).
Online Resource 2
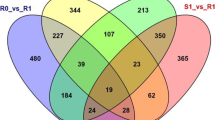
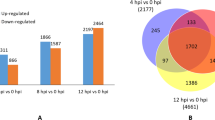
List of the unigenes differentially regulated in Regent and Trincadeira prior (0 h) and after inoculation with P. viticola (6 and 12 hpi). For each unigene is reported: (1) tentative consensus; (2) Genbank accession number obtained by sequence submission to dbEST; (3) expression profile at 0 h, 6 and 12 hpi; (4) description of putative function (if available) together with EST/TC and protein accession numbers obtained by nblast/nrblast analysis (March 2011) against the public databases: NCBI (Altschul et al. 1997), VitisEXPDB (Doddapaneni et al. 2008), DFCI Grape Gene Index (release 6.0) (Lee et al. 2005; Quackenbush et al. 2001), and UNIREF50 (Suzek et al. 2007) (PDF 161 kb).
OnlineResource 3
Transcript gene primer sequences and amplicon characteristics used for quantitative real-time PCR validation of the expression profiles of seven P. viticola-responsive genes following MIQE guidelines. Reaction efficiency calculated from the slope of the standard curve (Efficiency = −1 + 10(−1/slope)) is presented (PDF 110 kb).
Online Resource 4
Quantitative PCR efficiency plots. Median quantification cycle (Cq) values of each set of tenfold serial dilution plotted against the logarithm of cDNA concentration. Reaction efficiency is given by [10(1/−S) − 1] × 100%, where S represents the slope of the linear regression line (PDF 106 kb).
Rights and permissions
About this article
Cite this article
Figueiredo, A., Monteiro, F., Fortes, A.M. et al. Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct Integr Genomics 12, 379–386 (2012). https://doi.org/10.1007/s10142-012-0261-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-012-0261-8