Abstract
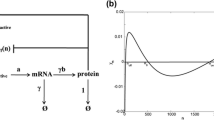
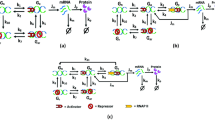
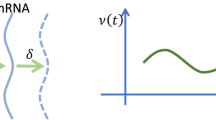
Intrinsic transcriptional noise induced by operator fluctuations is investigated with a simple spin-like stochastic model. The effects of transcriptional fluctuations in protein synthesis are probed by coupling transcription and translation by an amplificative interaction. In the presence of repression a new term contributes to the noise, which depends on the rate of mRNA production. If the switch decay time is small compared with the mRNA life time, the noise is also small. In general the damping of protein production by a repressive agent occurs linearly but fluctuations can show a maximum at intermediate repression. The discrepancy among the switch decay time, the mRNA degradation, and protein degradation is crucial for the repressive control in translation without large fluctuations. The noise profiles obtained here are in quantitative agreement with recent experiments.
Similar content being viewed by others
References
Abramowitz, M., Stegun, I.A.: Handbook of mathematical functions with formulas, graphs and mathematical tables. Nat. Bur. Standards Appl. Series, 55, U.S. Government Printing Office, Washington, D.C. (paperback edition published by Dover, New York) (1964)
Ackers G.K., Johnson A.D. and Shea M.A. (1982). Quantitative model for gene regulation by λ phage repressor. Proc. Natl. Acad. Sci. USA 79: 1129–1133
Becskei A. and Serrano L. (2000). Engineering stability in gene networks by autoregulation. Nature 405: 590–593
Berg O.G. (1978). A model for the statistical fluctuations of protein numbers in a microbial population. J. Theor. Biol. 71: 587–603
Bhalla U.S. and Iyengar R. (1999). Emergent properties of networks of biological signaling pathways. Science 283: 381–387
Blake W.J., Kaern M., Cantor C.R. and Collins J.J. (2003). Noise in eukaryotic gene expression. Nature 422: 633–637
Cook D.L., Gerber A.N. and Tapscott S.J. (1998). Modeling stochastic gene expression: Implications for haploinsufficiency. Proc. Natl. Acad. Sci. USA 95: 15641–15646
von Dassow G., Meir E., Munro E.M. and Odell G.M. (2000). The segment polarity network is a robust developmental module. Nature 406: 188–192
Elowitz M.B. and Leibler S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338
Gardner T.S., Cantor C.R. and Collins J.J. (2000). Construction of genetic toggle switch in Escherichia coli. Nature 403: 339–342
Gillespie D.T. (1977). Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81: 2340–2361
Hasty J., Pradines J., Dolnik M. and Collins J.J. (2000). Noise-based switches and amplifiers for gene expression. Proc. Natl. Acad. Sci. USA 97: 2075–2080
Hornos J.E.M., Schultz D., Innocentini G.C.P., Wang J., Walczak A.M., Onuchic J.N. and Wolynes P.G. (2005). Self-regulating gene: An exact solution. Phys. Rev. E 72: 051907
Innocentini, G.C.P., Hornos, J.E.M.: Stochastic gene expression: approaching the equilibrium (in preparation)
van Kampen N.G. (1992). Stochastic Processes in Physics and Chemistry. North-Holland, Amsterdam
Kennell D. and Riezman H. (1977). Transcription and translation initiation frequencies of the escherichia coli lac operon. J. Mol. Biol. 114: 1–21
Ko M.S.H. (1991). A stochastic model for gene induction. J. Theor. Biol. 153: 181–194
McAdams H.H. and Arkin A. (1997). Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA 94: 814–819
McAdams H.H. and Arkin A. (1999). Its a noisy business! genetic regulation at the nanomolar scale. Trends Genet. 15: 65–69
Monod J. and Jacob F. (1961). Genetic regulatory mechanisms in synthesis of protein. J. Mol. Biol. 3: 318–356
Ozbudak E.M., Thattai M., Kurtser I., Grossman A.D. and van Oudenaarden A. (2002). Regulation of noise in the expression of single gene. Nat. Genet. 31: 69–73
Paulsson J. (2004). Summing up the noise in gene networks. Nature 427: 415–418
Paulsson J., Berg O.G. and Ehrenberg M. (2000). Stochastic focusing: Fluctuation-enhanced sensitivity of intracellular regulation. Proc. Natl. Acad. Sci. USA 97: 7148–7153
Pedraza J.M. and van Oudenaarden A. (2005). Noise propagation in gene networks. Science 307: 1965–1969
Ptashne M. (1992). A Genetic Switch: Phage λ and Higher Organisms. Cell Press/Blackwell, Cambridge
van de Putte P. and Goosen N. (1992). Dna inversions in phages and bacteria. Trends Genet. 8: 457–462
Siegele D.A. and Hu J.C. (1997). Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci.USA 94: 8168–8172
Thattai M. and van Oudenaarden A. (2001). Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 98: 8614–8619
Walczak A.M., Sasai M. and Wolynes P.G. (2005). Self-consistent proteomic field theory of stochastic gene switches. Biophys. J. 88: 828–850
Author information
Authors and Affiliations
Corresponding author
Additional information
Work was supported by FAPESP and CNPq, Brazil.
Rights and permissions
About this article
Cite this article
Innocentini, G.C.P., Hornos, J.E.M. Modeling stochastic gene expression under repression. J. Math. Biol. 55, 413–431 (2007). https://doi.org/10.1007/s00285-007-0090-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-007-0090-x