Abstract
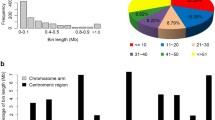
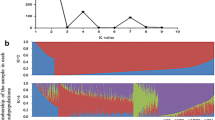
A set of 89 near-isogenic lines (NILs) of maize was created using marker-assisted selection. Nineteen genomic regions, identified by restriction fragment length polymorphism loci and chosen to represent portions of all ten maize chromosomes, were introgressed by backcrossing three generations from donor line Tx303 into the B73 genetic background. NILs were genotyped at an additional 128 simple sequence repeat loci to estimate the size of introgressions and the amount of background introgression. Tx303 introgressions ranged in size from 10 to 150 cM, with an average of 60 cM. Across all NILs, 89% of the Tx303 genome is represented in targeted and background introgressions. The average proportion of background introgression was 2.5% (range 0–15%), significantly lower than the expected value of 9.4% for third backcross generation lines developed without marker-assisted selection. The NILs were grown in replicated field evaluations in two years to map QTLs for flowering time traits. A parallel experiment of testcrosses of each NIL to the unrelated inbred, Mo17, was conducted in the same environments to map QTLs in NIL testcross hybrids. QTLs affecting days to anthesis, days to silking, and anthesis-silk interval were detected in both inbreds and hybrids in both environments. The testing environments differed dramatically for drought stress, and different sets of QTLs were detected across environments. Furthermore, QTLs detected in inbreds were typically different from QTLs detected in hybrids, demonstrating the genetic complexity of flowering time. NILs can serve as a valuable genetic mapping resource for maize breeders and geneticists.






Similar content being viewed by others
References
Beavis WD (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, FL, pp 145–162
Bolaños J, Edmeades GO (1996) The importance of the anthesis-silking interval in breeding for drought tolerance in tropical maize. Field Crops Res 48:65–80
Brouwer DJ, St Clair DA (2004) Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor Appl Genet 108:628–638
Brownie C, Bowman DT, Burton JW (1993) Estimating spatial variation in analysis of data from yield trials: a comparison of methods. Agron J 85:1244–1253
Byrne PF, McMullen MD, Snook ME, Musket TA, Theuri JM, Widstrom NW, Wiseman BR, Coe EH (1996) Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc Natl Acad Sci USA 93:8820–8825
Canady MA, Meglic V, Chetelat RT (2005) A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48:685–697
Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A (2004) Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168:2169–2185
Cowen NM (1988) The use of replicated progenies in marker-based mapping of QTL’’s. Theor Appl Genet 75:857–862
Eduardo I, Arús P, Monforte AJ (2005) Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor Appl Genet 112:139–148
Edwards MD, Helentjaris T, Wright S, Stuber CW (1992) Molecular-marker-facilitated investigations of quantitative trait loci in maize. 4. Analysis based on genome saturation with isozyme and restriction fragment length polymorphism markers. Theor Appl Genet 83:765–774
Edwards MD, Stuber CW, Wendel JF (1987) Molecular-marker-facilitated investigations of quantitative-trait loci in maize. I. Numbers, genomic distribution, and types of gene action. Genetics 116:113–125
Ellis THN (1986) Restriction fragment length polymorphism markers in relation to quantitative characters. Theor Appl Genet 72:1–2
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennelli in the cultivatred tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Gonzalo M, Vyn TJ, Holland JB, McIntyre LM (2006) Mapping density response in maize: a direct approach for testing genotype and treatment interactions. Genetics 173:331–348
Graham GI, Wolff DW, Stuber CW (1997) Characterization of a yield quantitative trait locus on chromosome five of maize by fine mapping. Crop Sci 37:1601–1610
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Holland JB (2004) Implementation of molecular markers for quantitative traits in breeding programs—challenges and opportunities. In: Fischer T, Turner N, Angus J, McIntyre L, Robertson M, Borrell A, Lloyd D (eds) New directions for a diverse planet: proceedings for the 4th international crop science congress. Brisbane, Australia. Online: http://www.cropscience.org.au/icsc2004/symposia/3/4/203_hollandjb.htm
Kaeppler SM (1997) Quantitative trait locus mapping using sets of near-isogenic lines: relative power comparisons and technical considerations. Theor Appl Genet 95:384–392
Kao C-H, Zeng Z-B, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Knapp SJ (1991) Using molecular markers to map multiple quantitative trait loci: models for backcross, recombinant inbred, and doubled haploid progeny. Theor Appl Genet 81:333–338
Knapp SJ, Bridges WC (1990) Using molecular markers to estimate quantitative trait locus parameters: power and genetic variances for unreplicated and replicated progeny. Genetics 126:769–777
Koester RP, Sisco PH, Stuber CW (1993) Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci 33:1209–1216
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Littell RC, Milliken GA, Stroup WA, Wolfinger RD (1996) SAS system for mixed models. SAS Institute Inc., Cary, NC
Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsattelites. Genetics 165:2117–2128
Melchinger AE, Utz HF, Schon CC (1998) Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveal low power of QTL detection and large bias in estimates of QTL effects. Genetics 149:383–403
Mikel MA, Dudley JW (2006) Evolution of North American dent corn from public to proprietary germplasm. Crop Sci 46:1193–1205
Monforte AJ, Tanksley SD (2000) Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in an L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43:803–813
Muehlbauer GJ, Staswick PE, Specht JE, Graef GL, Shoemaker RC, Keim P (1991) RFLP mapping using a set of near-isogenic lines in the soybean [Glycine max (L.) Merr]. Theor Appl Genet 81:189–198
Paran I, Kesseli R, Michelmore R (1991) Identification of restriction-fragment-length-polymorphism and random amplified polymorphic DNA markers linked to downy mildew resistance genes in lettuce, using near-isogenic lines. Genome 34:1021–1027
Ramsay LD, Jennings DE, Bohuon E JrR, Arthur AE, Lydiate DJ, Kearsey MJ, Marshall DF (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39:558–567
Riede CR, Anderson JA (1996) Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci 36:905–909
Senior ML, Murphy JP, Goodman MM, Stuber CW (1998) Utility of SSRs for determining genetic similarities and relationships in maize using an agarose gel system. Crop Sci 38:1088–1098
Sharopova N, McMullen MD, et al (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48:463–481
Steel RGD, Torrie JH (1980) Principles and procedures of statistics, 2nd edn. McGraw-Hill, New York
Stuber CW (1998) Case history in crop improvement: yield heterosis in maize. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, FL, pp 197–206
Stuber CW, Polacco M, Senior ML (1999) Synergy of empirical breeding, marker-assisted selection, genomics, and genetic engineering to increase crop yield potential. Crop Sci 39:1571–1583
Tallury SP, Goodman MM (1999) Experimental evaluation of the potential of tropical germplasm for temperate maize improvement. Theor Appl Genet 98:54–61
Tanksley D, Medina-Filho H, Rick CM (1982) Use of naturally-occurring enzyme variation to detect and map genes controlling quantitative traits in an interspecific backcross of tomato. Heredity 49:11–25
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tarter JA, Goodman MM, Holland JB (2003) Testcross performance of semiexotic inbred lines derived from Latin American maize accessions. Crop Sci 43:2272–2278
Vroh Bi I, McMullen MD, et al (2006) Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46:12–21
Yu ZH, Mackill DJ, Bonman JM, Tanksley SD (1991) Tagging genes for blast resistance in rice via linkage to RFLP markers. Theor Appl Genet 81:471–476
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nature Reviews Genetics 2:983–989
Zhao W, Canaran P, et al (2006) Panzea: a database and resource for molecular and functional diversity in the maize genome. Nucl Acids Res 34:D752–D757
Acknowledgments
Research funded in part by United States Department of Agriculture National Research Initiative Competitive Grants Program Award No. 2001-35301-10601. We thank Wayne Dillard, David Rhyne, and Brooke Peterson for technical assistance; Stella Salvo for help in preparing Fig. 2; and Drs. Luciana Carlini Garcia and Martin Bohn, and several anonymous reviewers for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Bohn.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Szalma, S.J., Hostert, B.M., LeDeaux, J.R. et al. QTL mapping with near-isogenic lines in maize. Theor Appl Genet 114, 1211–1228 (2007). https://doi.org/10.1007/s00122-007-0512-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0512-6