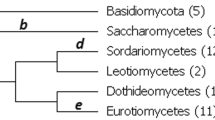
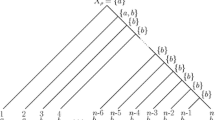
Summary
A maximum likelihood method for inferring protein phylogeny was developed. It is based on a Markov model that takes into account the unequal transition probabilities among pairs of amino acids and does not assume constancy of rate among different lineages. Therefore, this method is expected to be powerful in inferring phylogeny among distantly related proteins, either orthologous or parallogous, where the evolutionary rate may deviate from constancy. Not only amino acid substitutions but also insertion/deletion events during evolution were incorporated into the Markov model. A simple method for estimating a bootstrap probability for the maximum likelihood tree among alternatives without performing a maximum likelihood estimation for each resampled data set was developed. These methods were applied to amino acid sequence data of a photosynthetic membrane protein,psbA, from photosystem II, and the phylogeny of this protein was discussed in relation to the origin of chloroplasts.
Similar content being viewed by others
References
Aldrich J, Cherney B, Merlin E (1986) Sequence of the chloroplast-encodedpsbA gene for the QB polypeptide of alfalfa. Nucleic Acids Res 14:9537
Bishop MJ, Friday AE (1985) Evolutionary trees from nucleic acid and protein sequences. Proc R Soc Lond B 226:271–302
Bishop MJ, Friday AE (1987) Tetrapod relationships: the molecular evidence. In: Patterson C (ed) Molecules and morphology in evolution: conflict or compromise? Cambridge University Press, Cambridge, pp 123–139
Britten RJ (1986) Rates of DNA sequence evolution differ between taxonomic groups. Science 231:1393–1398
Burger-Wiersma T, Veenhuis M, Korthals HJ, Van de Wiel CCM, Mur LR (1986) A new prokaryote containing chlorophylls a and b. Nature 320:262–264
Cavalier-Smith T (1982) The origins of plastids. Biol J Linn Soc 17:289–306
Chakraborty R (1977) Estimation of time of divergence from phylogenetic studies. Can J Genet Cytol 19:217–223
Clarke B (1970) Selective constraints on amino-acid substitutions during the evolution of proteins. Nature 228:159–160
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. In: Dayhoff MO (ed) Atlas of protein sequence structure, vol 5, suppl 3. National Biomedical Research Foundation, Washington DC, pp 345–352
Erickson JM, Rahire M, Rochaix J-D (1984)Chlamydomonas reinhardii gene for the 32,000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J 3:2753–2762
Felsenstein J (1978) Cases in which parsimony and compatibility methods will be positively misleading. Syst Zool 27:401–410
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1983a) Methods for inferring phylogenies: a statistical view. In: Felsenstein J (ed) Numerical taxonomy. Springer-Verlag, Berlin, pp 315–334
Felsenstein J (1983b) Statistical inference of phylogenies. J R Stat Soc A 146:246–272
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Golden SS, Brusslan J, Haselkorn R (1986) Expression of a family ofpsbA genes encoding a photosystem II polypeptide in the cyanobacteriumAnacystis nidulans r2. EMBO J 5:2789–2798
Hasegawa M, Kishino H (1989) Confidence limits on the maximum-likelihood estimate of the hominoid tree from mitochondrial-DNA sequences. Evolution 43:672–677
Hasegawa M, Yano T (1984) Maximum likelihood method of phylogenetic inference from DNA sequence data. Bull Biomet Soc Jpn 5:1–7
Hasegawa M, Iida Y, Yano T, Takaiwa F, Iwabuchi M (1985) Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J Mol Evol 22:32–38
Hasegawa M, Kishino H, Yano T (1987) Man's place in Hominoidea as inferred from molecular clocks of DNA. J Mol Evol 26:132–147
Hasegawa M, Kishino H, Yano T (1988) Phylogenetic inference from DNA sequence data. In: Matusita K (ed) Statistical theory and data analysis II: Proceedings of Second Pacific Area Statistical Conference. North-Holland, Amsterdam, pp 1–13
Hendy MD, Penny D (1989) A framework for the quantitative study of evolutionary trees. Syst Zool 38:297–309
Jeffreys AJ, Wilson V, Thein SL (1985) Hypervariable ‘minisatellite’ regions in human DNA. Nature 314:67–73
Kikuno R, Hayashida H, Miyata T (1985) Rapid rate of rodent evolution. Proc Jpn Acad B 61:153–156
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kishino H, Hasegawa, M (1989) Evolution of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Lynch M (1988) Estimation of relatedness by DNA fingerprinting. Mol Biol Evol 5:584–599
Meyer TE, Cusanovich MA, Kamen MD (1986) Evidence against use of bacterial amino acid sequence data for construction of all-inclusive phylogenetic trees. Proc Natl Acad Sci USA 83:217–220
Miller KR, Jacob JS (1989) OnProchlorothrix. Nature 338:303–304
Morden CW, Golden SS (1989a)psbA genes indicate common ancestry of prochlorophytes and chloroplasts. Nature 337:382–385
Morden CW, Golden SS (1989b)psbA genes indicate common ancestry of prochlorophytes and chloroplasts (Corrigendum). Nature 339:400
Mulligan B, Schultes N, Chen L, Bogorad L (1984) Nucleotide sequence of a multiple-copy gene for the B protein of photosystem II of a cyanobacterium. Proc Natl Acad Sci USA 81:2693–2697
Nei M, Tateno Y (1978) Nonrandom amino acid substitution and estimation of the number of nucleotide substitutions in evolution. J Mol Evol 11:333–347
Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S, Inokuchi H, Ozeki H (1986) Chloroplast gene organization deduced from complete sequence of liverwortMarchantia polymorpha chloroplast DNA. Nature 322:572–574
Osiewacz HD, McIntosh L (1987) Nucleotide sequence of a member of thepsbA multigene family from the unicellular cyanobacteriumSynechocystis 6803. Nucleic Acids Res 15:10585
Penny D (1989) What, if anything, isProchloron?, Nature 337:304–305
Penny D, Hendy MD, Henderson IM (1987) Reliability of evolutionary trees. Cold Spring Harbor Symp Quant Biol 52:857–862
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Sinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5:2043–2049
Turner S, Burger-Wiersma T, Giovannoni SJ, Mur LR, Pace NR (1989) The relationship of a prochlorophyteProchlorothrix hollandica to green chloroplasts. Nature 337:380–382
Wu C-I, Li W-H (1985) Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci USA 82:1741–1745
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kishino, H., Miyata, T. & Hasegawa, M. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J Mol Evol 31, 151–160 (1990). https://doi.org/10.1007/BF02109483
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02109483