Identification of genes encoding ALMT and MATE transporters as candidate aluminum tolerance genes from a typical acid soil plant, Psychotria rubra (Rubiaceae)
- Published
- Accepted
- Received
- Academic Editor
- Joseph Gillespie
- Subject Areas
- Ecology, Genomics, Plant Science
- Keywords
- Psychotria, Plant, Acid soils, Transcriptome, MATE, ALMT
- Copyright
- © 2019 Iguchi et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Identification of genes encoding ALMT and MATE transporters as candidate aluminum tolerance genes from a typical acid soil plant, Psychotria rubra (Rubiaceae) PeerJ 7:e7739 https://doi.org/10.7717/peerj.7739
Abstract
To understand how tropical plants have adapted to acid soils, we analyzed the transcriptome of seedlings of Psychotria rubra, a typical species found on acid soils. Using RNA-seq, we identified 22,798 genes, including several encoding proteins of the Al3+-activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) families. Molecular phylogenetic analysis of ALMTs and MATEs revealed the grouping of those from P. rubra, which may be useful to select targets for elucidating the molecular basis of P. rubra adaptation to acid soils in the future. The transcriptome datasets obtained in this study would help us to further understand the physiological and ecological aspects of soil adaptation of Psychotria species.
Introduction
Understanding how plants adapt to various soils is essential in plant biology (Hiradate, Ma & Matsumoto, 2007) because plants are sessile and need to grow roots in settled soils. Adaptation to acid soils is an important issue because acid soils cover a considerable part of Earth’s arable land and prevent agriculture of most plants (Von Uexküll & Mutert, 1995). In acid soils, aluminum is toxic to root tip growth, and various aspects, from molecular to physiological, of the mechanisms of aluminum toxicity have been explored in detail (reviewed in Ma, 2007).
Proteins of the Al3+-activated malate transporter (ALMT) and multidrug and toxic compound extrusion (MATE) families are likely involved in plant adaptation to acid soils (Delhaize, Gruber & Ryan, 2007; Delhaize, Ma & Ryan, 2012; Ma, 2007; Ma, Chen & Shen, 2014). ALMTs and MATEs release organic acids (malate and citrate, respectively), which bind Al3+ and detoxify it. ALMTs and MATEs related to aluminum tolerance have been identified in model and agricultural plants (wheat: Sasaki et al., 2004; barley: Delhaize et al., 2004; Furukawa et al., 2007; maize: Maron et al., 2010; Arabidopsis: Hoekenga et al., 2006; Liu et al., 2009), but the composition of those families in non-model wild plants has hardly been explored.
Psychotria (Rubiaceae) is a highly diversified genus comprising more than 1,600 species distributed in all tropical and some subtropical regions (Hamilton, 1989; Davis et al., 2001; Razafimandimbison et al., 2014). Because Psychotria species adapt to several types of soils (e.g., soils with high concentrations of nickel; Merlot et al., 2014), the genus is an ideal target to use to understand how adaptation of wild plants to different types of soils has evolved. In this study, we report ALMTs and MATEs of P. rubra, which grows on acid soils (Miyawaki, 1989).
Materials and Methods
Sampling, RNA extraction, RNA-seq library preparation, and sequencing
Seeds of P. rubra were collected on Mt. Nago-dake, in the north of Okinawa Island, and seedlings were grown in a greenhouse of the National Institute of Technology, Okinawa College (Fig. 1). RNA was extracted from the seedlings (three–five cm height) using an RNeasy Plant Mini kit (Qiagen, Hilden, Germany). An RNA-seq library was prepared using a TruSeq Stranded mRNA Sample Prep Kit (Illumina, San Diego, CA, USA). The library was sequenced (100-bp paired-end reads) on an Illumina HiSeq 2500 platform. The above procedures on RNA-seq were outsourced to Hokkaido System Science Corporation, Japan.
Figure 1: Psychotria rubra.
(A) Shrub on Mount Nago-dake, Okinawa, Japan. (B) Seedlings.De novo assembly and annotation of transcriptome sequences
FASTQ files were filtered, the reads with poor-quality bases (Q < 20) and those shorter than 20-bp were excluded, and adapter sequences were removed in cutadapt v. 1.9.1 software (Martin, 2011). PCR duplicates that arose during library preparation were removed by ConDeTri software (Smeds & Künstner, 2011). The remaining paired-end reads were assembled in Trinity v. 2.8.4 software (Grabherr et al., 2011) with the default options. Open reading frames of >150 amino acids were identified by using TransDecoder software (Haas et al., 2013) and the results of BLASTP searches within the Swiss-Prot database (e-value <1e−5). Redundant amino acid sequences were removed in CD-HIT v. 4.7 software (−c 0.95; Li & Godzik, 2006). The remaining sequences were used as queries in BLASTP searches (e-value <1e−5) of Arabidopsis thaliana sequences (TAIR10; Swarbreck et al., 2007) and selected sequences assumed to be P. rubra itself. Basic information on sequences was obtained in SeqKit v. 0.9.3 software (Shen et al., 2016). Sequence data are accessible under the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (accession: DRA008339). The raw data has been also available at Figshare (https://doi.org/10.6084/m9.figshare.7848425.v2).
| Number of contigs | 131,578 |
| Total bases (bp) | 161,583,381 |
| Longest contig length (bp) | 16,195 |
| Shortest contig length (bp) | 185 |
| Average contig length (bp) | 1,228 |
| N50 | 2,007 |
| Gene name | Sequence ID | % identity | e-value | Bit score | Annotate description (Swiss-prot) | Accession no. (Swiss-prot) | e-value (Swiss-prot) |
|---|---|---|---|---|---|---|---|
| TaALMT1 | Prub_08163 | 45.966 | 1.05E−110 | 334 | Aluminum-activated malate transporter 2 (AtALMT2) | Q9SJE8 | 3.67E−166 |
| TaALMT1 | Prub_08164 | 49.477 | 4.78E−90 | 277 | Aluminum-activated malate transporter 2 (AtALMT2) | Q9SJE8 | 1.12E−133 |
| TaALMT1 | Prub_10125 | 43.902 | 1.19E−77 | 251 | Aluminum-activated malate transporter 12 (AtALMT12) (Quick anion channel 1) | O49696 | 0 |
| TaALMT1 | Prub_08553 | 36.111 | 3.39E−63 | 214 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 0 |
| TaALMT1 | Prub_16333 | 33.333 | 6.06E−57 | 197 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 0 |
| TaALMT1 | Prub_03305 | 51.19 | 5.72E−53 | 176 | Aluminum-activated malate transporter 10 (AtALMT10) | O23086 | 3.21E−59 |
| TaALMT1 | Prub_02168 | 33.772 | 4.74E−23 | 98.6 | Putative aluminum-activated malate transporter 3 (AtALMT3) | Q9LPQ8 | 1.74E−74 |
| TaALMT1 | Prub_02171 | 27.801 | 1.10E−22 | 98.6 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 1.99E−141 |
| TaALMT1 | Prub_16334 | 39.655 | 1.03E−19 | 86.3 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 6.62E−51 |
| TaALMT1 | Prub_08405 | 30.108 | 2.45E−19 | 88.6 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 9.01E−62 |
| TaALMT1 | Prub_08554 | 51.852 | 6.42E−12 | 63.2 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 3.20E−27 |
| TaALMT1 | Prub_02170 | 39.062 | 4.77E−11 | 62 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 7.28E−32 |
| TaALMT1 | Prub_02169 | 30 | 2.57E−07 | 51.6 | Aluminum-activated malate transporter 4 (AtALMT4) | Q9C6L8 | 1.57E−73 |
| TaALMT1 | Prub_02167 | 38.129 | 3.26E−07 | 50.8 | Aluminum-activated malate transporter 9 (AtALMT9) | Q9LS46 | 3.96E−26 |
| HvAACT1 | Prub_03062 | 57.752 | 0 | 537 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 0 |
| HvAACT1 | Prub_17943 | 55.955 | 0 | 536 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 0 |
| HvAACT1 | Prub_17944 | 59.432 | 0 | 531 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 0 |
| HvAACT1 | Prub_17945 | 55.894 | 3.61E−168 | 485 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 0 |
| HvAACT1 | Prub_03064 | 63.372 | 4.58E−147 | 429 | Protein DETOXIFICATION 43 (AtDTX43) (Multidrug and toxic compound extrusion protein 43) (MATE protein 43) (Protein FERRIC REDUCTASE DEFECTIVE 3) (AtFRD3) (Protein MANGANESE ACCUMULATOR 1) | Q9SFB0 | 1.12E−160 |
| HvAACT1 | Prub_03541 | 41.322 | 1.43E−120 | 368 | Protein DETOXIFICATION 45, chloroplastic (AtDTX45) (Multidrug and toxic compound extrusion protein 45) (MATE protein 45) | Q9SVE7 | 0 |
| HvAACT1 | Prub_18165 | 39.506 | 2.75E−114 | 350 | Protein DETOXIFICATION 45, chloroplastic (AtDTX45) (Multidrug and toxic compound extrusion protein 45) (MATE protein 45) | Q9SVE7 | 0 |
| HvAACT1 | Prub_03065 | 54.4 | 6.52E−55 | 186 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 3.65E−69 |
| HvAACT1 | Prub_03063 | 53.441 | 4.31E−52 | 179 | Protein DETOXIFICATION 42 (AtDTX42) (Aluminum-activated citrate transporter) (AtMATE) (FRD-like protein) (Multidrug and toxic compound extrusion protein 42) (MATE protein 42) | Q9SYD6 | 1.00E−66 |
| HvAACT1 | Prub_14684 | 43.541 | 4.14E−37 | 137 | Protein DETOXIFICATION 44, chloroplastic (AtDTX44) (Multidrug and toxic compound extrusion protein 44) (MATE protein 44) | Q84K71 | 4.41E−94 |
| HvAACT1 | Prub_14685 | 38.174 | 2.44E−22 | 96.7 | Protein DETOXIFICATION 44, chloroplastic (AtDTX44) (Multidrug and toxic compound extrusion protein 44) (MATE protein 44) | Q84K71 | 4.59E−53 |
| HvAACT1 | Prub_18629 | 23.25 | 7.67E−14 | 73.6 | Protein DETOXIFICATION 46, chloroplastic (AtDTX46) (Multidrug and toxic compound extrusion protein 46) (MATE protein 46) (Protein EDS5 HOMOLOGUE) | Q8W4G3 | 0 |
Extraction of ALMTs and MATEs of P. rubra and molecular phylogenetic analysis
We searched for P. rubra ALMTs and MATEs by BLASTP (e-value <1e−5) using sequences of TaALMT1 (UniProt database ID: Q76LB1) and HvAACT1 (UniProt ID: A7M6U2; this was the first MATE identified in barley (Furukawa et al., 2007) as queries). We then performed BLASTP searches against the Swiss-Prot database (e-value <1e−5) with each ALMT and MATE of P. rubra and selected the top 10 hits for each. The amino acid sequences of ALMTs and MATEs from P. rubra and related sequences from Swiss-Prot and other studies (Dreyer et al., 2012; Liu et al., 2016) were aligned in MAFFT v. 7.407 software (Katoh & Standley, 2013). We excluded four P. rubra ALMT sequences (Prub_02169, Prub_02171, Prub_08405, Prub_08554) from the following analysis because of poor alignment. We selected only plant MATEs, including HvAACT1 and those from (Liu et al., 2016), for the following analysis. Neighbor-joining trees of ALMTs and MATEs were constructed in MEGA7 software (Kumar, Stecher & Tamura, 2016) with the following settings: Poisson model, Uniform rates, and Pairwise deletion. To evaluate the confidence of phylogenetic trees, bootstrap tests were performed with 1,000 replicates.
Results and Discussion
Our RNA-seq analysis of P. rubra yielded 57,110,261 paired-end reads, of which 53,994,410 remained after filtering. De novo assembly of the remaining reads resulted in 131,578 contigs (Table 1), in which we found 24,687 non-redundant amino acid sequences; 22,798 of them were expected to originate from P. rubra itself as indicated by BLASTP analysis of TAIR10 (the remaining 1,889 sequences were almost no-hit in Swiss-Prot database or included those from microorganisms, etc). Among these 22,798 sequences, 19,701 ones were hit against Swiss-Prot database, and gene ontology (GO) numbers were found in 1,0348 ones. From these sequences, we found 14 ALMTs and 12 MATEs (Table 2).
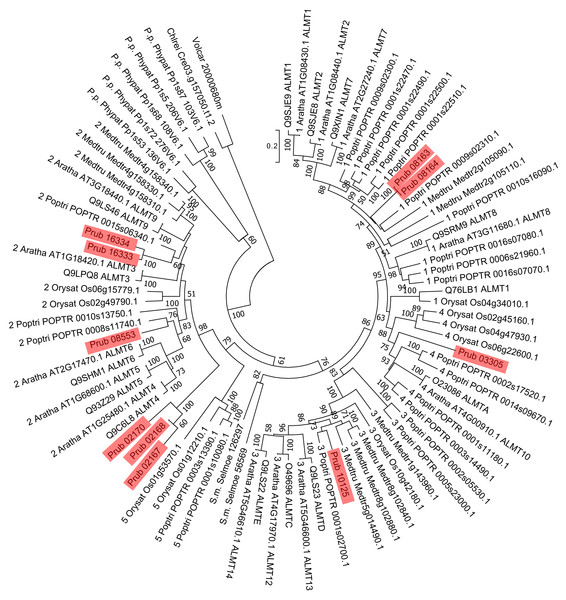
Figure 2: Phylogenetic tree of ALMTs of P. rubra and related proteins.
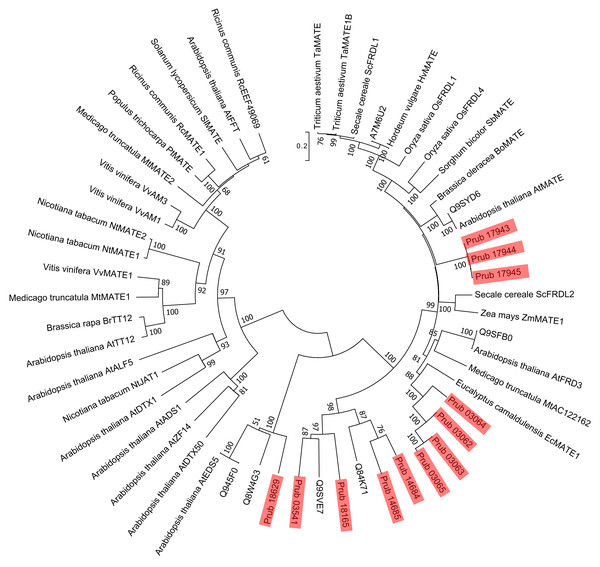
Sequences of P. rubra are shown using red shades. For sequences other than P. rubra ALMTs, the labels show the UniProt database ID and ALMT type, and ones from Dreyer et al. (2012). Number at each node is the bootstrap value.Figure 3: Phylogenetic tree of MATEs of P. rubra and related proteins.
Sequences of P. rubra are shown using red shades. For sequences other than P. rubra MATEs, the labels are UniProt database IDs, and ones from Liu et al. (2016). Number at each node is the bootstrap value.Using 14 ALMTs from P. rubra as queries in a BLASTP search against the Swiss-Prot database, we found 14 homologs, all of plant origin (mainly from Arabidopsis). Using a similar approach, we found 13 homologs of MATEs (six from Arabidopsis and seven from non-plant organisms). Molecular phylogenetic analysis did not detect P. rubra orthologs of TaALMT1 (UniProt ID: Q76LB1) (Fig. 2). ScALMT1 (accession number: ABA62397) from rye (Secale cereale) is the only known clear ortholog of TaALMT1 (Delhaize, Gruber & Ryan, 2007). ALMT1 of Arabidopsis (UniProt ID: Q9SJE9), encoded by an aluminum tolerance gene (Hoekenga et al., 2006), is clearly distinct from TaALMT1 (Delhaize, Gruber & Ryan, 2007). Thus, ALMTs related to aluminum tolerance may have multiple origins. Molecular phylogenetic analysis of MATEs revealed no clear orthologs of HvAACT1 (UniProt ID: A7M6U2) in P. rubra (Fig. 3).
Expression and functional analyses of ALMTs and MATEs of P. rubra would be useful for understanding their roles in soil adaptation of Psychotria (e.g., with and without Al treatment). Another aluminum tolerance mechanism of plants (different from releasing organic acids), aluminum accumulation, has been reported in several species of the Rubiaceae (Jansen et al., 2003). Genes related to this function are also good targets for future studies to explain the molecular basis of acid soil adaptation of P. rubra.
Conclusions
We succeeded in identifying transcriptome sequences including ALMTs and MATEs from P. rubra in this study. Comparative transcriptome analysis of several Psychotria species would help us to clarify the physiological and ecological aspects of diversification of this genus (e.g., adaptation to metalliferous soils; Merlot et al., 2014). In particular, Psychotria manillensis, which is closely related to P. rubra, is reportedly adapted to non-acid soils (Miyawaki, 1989). Thus, comparative analysis of P. rubra and P. manillensis should help to explain how soil adaptation–related genes are involved in adaptive evolution of Psychotria species.