Expression analysis of four pseudo-response regulator (PRR) genes in Chrysanthemum morifolium under different photoperiods
- Published
- Accepted
- Received
- Academic Editor
- Savithramma Dinesh-Kumar
- Subject Areas
- Genetics, Molecular Biology, Plant Science
- Keywords
- Chrysanthemum, Circadian clock, PRR, Flower bud differentiation, Gene expression
- Copyright
- © 2019 Wang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Expression analysis of four pseudo-response regulator (PRR) genes in Chrysanthemum morifolium under different photoperiods. PeerJ 7:e6420 https://doi.org/10.7717/peerj.6420
Abstract
Genes encoding pseudo-response regulator (PRR) proteins play significant roles in plant circadian clocks. In this study, four genes related to flowering time were isolated from Chrysanthemum morifolium. Phylogenetic analysis showed that they are highly homologous to the counterparts of PRRs of Helianthus annuus and named as CmPRR2, CmPRR7, CmPRR37, and CmPRR73. Conserved motifs prediction indicated that most of the closely related members in the phylogenetic tree share common protein sequence motifs, suggesting functional similarities among the PRR proteins within the same subtree. In order to explore functions of the genes, we selected two Chrysanthemum varieties for comparison; that is, a short-day sensitive Zijiao and a short-day insensitive Aoyunbaixue. Compared to Aoyunbaixue, Zijiao needs 13 more days to complete the flower bud differentiation. Evidence from spatio-temporal gene expression patterns demonstrated that the CmPRRs are highly expressed in flower and stem tissues, with a growing trend across the Chrysanthemum developmental process. In addition, we also characterized the CmPRRs expression patterns and found that CmPRRs can maintain their circadian oscillation features to some extent under different photoperiod treatment conditions. These lines of evidence indicated that the four CmPRRs undergo circadian oscillation and possibly play roles in regulating the flowering time of C. morifolium.
Introduction
Circadian clocks have evolved as a timekeeping molecular mechanism that enable organisms to predict and anticipate these periodic changes in their surrounding environment, for example, light/dark cycles and temperature oscillations (Nohales & Kay, 2016). The flowering time of plants is regulated by circadian clocks. In the photoperiodic response, light has two distinct functions: a resetting cue for circadian clock and a day-length signal. Light receptors can sense and transfer the light signal to biological circadian clock that impacts rhythmic outputs of downstream genes. These rhythm-regulated genes eventually activate or inhibit the expression of meristem genes and floral organs formation genes, which lead to the regulation of plant flowering. In Arabidopsis, the transcription and translation feedback loop, comprising the evening-expressed TIMING OF CAB EXPRESSION 1 (TOC1) gene, the morning-expressed CIRCADIAN CLOCK ASSOCIATED 1/LATE ELONGATED HYPOCOTYL (CCA1/LHY) gene and the evening complex EARLY FLOWERING 3 gene, is one of the core components of the oscillator (Alabadí et al., 2001; Hazen et al., 2005; Li et al., 2011; Perales & Más, 2007; Nusinow et al., 2012). Additional transcriptional repressors/co-repressors PSEUDO-RESPONSE REGULATORS 5 (PRR5), PRR7, and PRR9, TOPLESS, LUX ARRHYTHMO (LUX), and activators/co-activators REVEILLE (RVE) 4, RVE6, NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED (LNK) 1, LNK2 are necessary in establishing the circadian clock (Cha et al., 2017; Farré et al., 2005; Kamioka et al., 2016; Hsu & Harmer, 2014).
Pseudo-response regulator represents a set of PRR proteins, which plays a significant role in circadian rhythm, light signal transduction, and flowering regulation. In Arabidopsis, many studies have reported the importance of PRR genes in the circadian clock (Eriksson et al., 2003; Farré et al., 2005; Ito et al., 2009; Kaczorowski & Quail, 2003; Michael et al., 2003; Norihito et al., 2005; Para et al., 2007; Prunedapaz et al., 2009; Salomé & Robertson McClung, 2005; Yoko et al., 2003). In Arabidopsis, the F box protein ZEITLUPE directly interacts with the pseudo-receiver (PR) domain of PRR5 and targets PRR5 for degradation by 26S proteasomes in the circadian clock and in early photomorphogenesis (Kiba et al., 2007). PRR5, PRR7, and PRR9 act as transcriptional repressors of CCA1 and LHY. The repressor activities of these proteins account for their roles in the interlocking feedback loop of the circadian clocks (Nakamichi et al., 2010). TOC1 can repress direct targets through the CCT motif, and the repression activity is in the PR domain of the protein (Gendron et al., 2012). PRR5, through binding to the CCT motifs of its target genes, is involved in flowering-time regulation, hypocotyl elongation and cold-stress (Nakamichi et al., 2012). Evidence from reverse genetics indicated that several other PRR genes also have the function of changing the length of the circadian cycle (McClung, 2006). SbPRR37 can inhibit the flowering process of Sorghum under the long sunshine condition, with its expression level influenced by the photoperiod (Murphy et al., 2011). PRR37 down-regulates Hd3a expression to suppress rice flowering under long-day conditions (Koo et al., 2013). In long-day plants such as barley and wheat, AtPRR7 homologous gene PHOTOPERIOD 1 plays a critical role in photoperiod response and flowering regulation (Boden et al., 2015; Turner et al., 2005). OsPRR73, a rice PRR gene co-expressed with Hdl protein gene OsBBX29, is involved in regulating the flowering process through nucleic acid metabolism pathway (Iniesta et al., 2008; Murakami et al., 2007).
Similarly to the PRRs, GIGANTEA (GI), a large plant-specific protein family, which lacks well-characterized functional domains, is also required for circadian timekeeping (Gould et al., 2006; Martin-Tryon, Kreps & Harmer, 2007). GI is involved not only in the central oscillator but also in light input and photoperiodic flowering output pathways (Gould et al., 2006; Martin-Tryon, Kreps & Harmer, 2007; Mishra & Panigrahi, 2015). GI genes can be repressed in the morning by CCA1 and LHY (Lu et al., 2012), both of which in turn appear to be induced by GI (Martin-Tryon, Kreps & Harmer, 2007). In addition, TOC1 and EC (evening complex) both contribute to GI repression (Huang et al., 2012; Mizuno et al., 2014).
Chrysanthemum morifolium, one of the most important global cut-flowers and pot plants, is a typical short-day plant that is distributed worldwide (Teixeira da Silva, 2003; Liu et al., 2016). Breeders have attempted to develop more Chrysanthemum varieties with different flowering times and colors which promote C. morifolium as an ideal material to study the light inducible flowering system (Teixeira da Silva, 2003; Liu et al., 2016). Several flowering-related genes, such as COL1, COL5, ClFT1, and ClFT2, have been cloned from C. lavandulifolium and transferred into Arabidopsis (Fu, 2014). For instance, research indicated that transgenic Arabidopsis overexpressing COL1 and COL5 are both bolting and flowering earlier than wild-type strains. ClFT1 and ClFT2 may play opposite roles in the flowering process, as ClFT1 transgenic Arabidopsis is flowering earlier than the control, while ClFT2 transgenic lines are flowering later compared to the control (Fu, 2014). Transgenic Chrysanthemum with overexpression of CsFTL3 is able to bloom normally even under long-day conditions (Atsushi et al., 2012). However, there are limited studies on the PRR genes of C. morifolium. In this study, we compared the flower bud differentiation process between short-day sensitive C. morifolium varieties Zijiao and short-day insensitive Aoyunbaixue and found that Zijiao needs 13 more days than Aoyunbaixue to complete the flower bud differentiation. We then cloned four putative PRR genes (CmPRR2, CmPRR7, CmPRR37, and CmPRR73) from C. morifolium, and showed their phylogenetic relationship related to PRRs from Arabidopsis and other species. Expression patterns of these four CmPRR genes under different photoperiod conditions and across different flower bud differentiation stages were also analyzed to understand their functions in regulating flowering time or circadian rhythms of C. morifolium.
Materials and Methods
Cultivation of Chrysanthemum morifolium variety
The short-day sensitive C. morifolium variety Zijiao and insensitive variety Aoyunbaixue were cultivated in the greenhouse of Shanxi Agricultural University under normal growth conditions with 60–70% relative humidity, 14/10-h light/dark cycles, and a constant temperature of 25 ± 2 °C.
Gene cloning and phylogenetic analysis
Tender leaves were harvested from Aoyunbaixue, frozen immediately in liquid nitrogen, and stored at −70 °C for RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR). Primers targeting the UTR (Untranslated Region) region were designed and used to amplify the coding sequences of CmPRR2, CmPRR7, CmPRR37, and CmPRR73 according to the RNA-seq data (Table 1). RT-PCR products were then purified and sent to BGI Shenzhen (Shenzhen, China) for DNA sanger sequencing to confirm the identity of the fragments. Non-reverse transcribed RNA was used as the negative control to judge amplified DNA is from mRNA not genomic DNA.
| Genes | Primers (5′-3′) | cDNA length (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| PRR2 | AGCTATGGTTTGCACTGCGAAC | CAGCATTAACGAGAGCTGCTGAT | 1,605 |
| PRR7 | GTTGATGAGGAGTGTTGGTGT | CAAGAGCTCTGAGTTCCACTTC | 1,017 |
| PRR37 | GGTTAATGAAGAGTGTTGGAGTG | TCAGTAGTCCGACAAACCGA | 2,019 |
| PRR73 | TCGATGACTAGTAGCAGCAGAG | ACTAGACATCAGCAGCATTAGCG | 1,941 |
Nine PRR protein sequences from Arabidopsis and 18 from other related species were blasted and downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/gene/). Multiple sequence alignments of PRR proteins were performed using Clustal X 1.83 (Jeanmougin et al., 1998). Unrooted phylogenetic trees were constructed with MEGA 7.0.21 using the neighbor joining (NJ) methods and the bootstrap test carried out with 1,000 iterations (Kumar, Stecher & Tamura, 2016). Pairwise gap deletion mode was used to ensure the divergent domains could contribute to the topology of the NJ tree (Kumar, Stecher & Tamura, 2016). Conserved motifs in PRR proteins were also detected using the program MEME version 5.0.2 (Bailey et al., 2009). MEME was run with the following parameters: any number of repetitions, 10 maximum motifs, and between 6 and 50 residues for the optimum motif widths.
RNA extraction and RTq-PCR analysis
Total RNA of each sample material was extracted using the Column Plant RNAout Kit (Tiandz, Beijing, China) followed by cDNA synthesis using PrimeScipt™RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. Quality and quantity of RNA were determined by agarose gel electrophoresis and a NanoDrop 2000c Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). RTq-PCR was performed on an ABI 7,500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using the SYBR Premix Ex Taq™ kit (Takara, Dalian, China) according to the manufacture’s instructions. Primers used for RTq-PCR are shown in Table 2. The amplification curve was generated after analyzing the raw data and the cycle threshold value was calculated based on the fluorescence threshold of 0.01 (Bustin et al., 2009; Gu et al., 2011; Bustin & Mueller, 2005). The average expression levels of Chrysanthemum house-keeping genes Actin and UBC were used as the internal control (Table 2). The relative expression level of target genes in different samples was calculated using 2−ΔΔCt method, defined as ΔΔCt = (Ct-target—Ct-control)2–(Ct-target—Ct-control)1 (Gu et al., 2011; Bustin & Mueller, 2005). RTq-PCR was carried out with three technical and three biological replicates per sample.
| Genes | Primers (5′-3′) | |
|---|---|---|
| Forward | Reverse | |
| Actin | CCAAAAGCCAATCGTGAGAAG | CACCATCACCAGAATCCAACA |
| UBC | TCTCGCTTGTCCGGTTTGTG | ACCTTGGGTGGCTTGAATGG |
| CmPRR2 | GTGAGGGCAGACAACGAAGA | TTCTCGAGGAATTCGACCGC |
| CmPRR73 | GGGATGACGATGAGAGCACC | ACGGATAACGAGGCCACAAG |
| CmPRR7 | TTGTTCAGTGGGAGCGGTTT | ATAGCCGCAATTACGGAGCA |
| CmPRR37 | AGCATCCTTCCCATTCACCC | CAAGTTCCGCCAGAAGAGGA |
| CmCOL1 | GTGTCCCGGTTATGCCTATTTC | CGCTGCTTCGTCTTCTTCTTC |
| CmCOL5 | GTTCTTGTGTCTCGCGTGTG | AGCATCAGCCTTACACGTCA |
| COL9Y | TTGGTGGTGCCGAATCTTCA | CCTTCCATAGCGGGTTGGTT |
| CmCOL13 | TACCCAAGAACGGGAGACCG | GCAAAGCGACCTCTGATCCT |
| COL14Y | ACAGCTAACGTGAGCAGCAT | TTCTGCAACAATAGGCCAGC |
| CmCOL15 | TGAAACCGTCGACAGAGGTG | GTCGTTCTGCTCCTGTCTCC |
| COL16Y | CCACGACCAAGGAGCAAATAC | ACACGGGCACTAAAGGATACAAA |
| CmCOL20 | TGTCCAGCTGACGATGCTTT | GAAACTGCGCCTGAACCAAA |
| CmGI1-F | ATGGATAGCGGTGACGAACC | GCCTCCCCCATTAGATACGC |
| CmGI2-F | GCGAAAATACCGATGCCACC | TAGCTGAAGTTCGCAGGCAA |
| CmGI3-F | GAGTTGGTTCACCACCGCTA | ACCAGCGGAAGTAGTCATGC |
| CmFT-F | ACAGGAGCACAGTTTGGTCA | ACCCAATTGCCGGAATAGCA |
Expression patterns of CmPRRs across different flower bud differentiation stages of Zijiao and Aoyunbaixue
In the early stage of vegetative growth, mature leaves at the 3rd and 4th nodes were harvested every 3 days at 9:00 am from respective Zijiao and Aoyunbaixue, in order to study the expression patterns of CmPRR2, CmPRR7, CmPRR37, and CmPRR73 across nine different flower bud differentiation stages (Vegetative growth stage, Inflorescence primordial differentiation stage, Initial stage of involucres primordial differentiation, Final stage of involucres differentiation, Initial stage of ligulae flower differentiation, Final stage of ligulae flower differentiation, Initial stage of corolla formation, Secondary stage of corolla formation, and Finish stage of corolla formation). For comparison, other genes related to circadian clock, including CmGI1, CmGI2, CmGI3, CmCOL1, CmCOL5, CmCOL9, CmCOL13, CmCOL14, CmCOL15, CmCOL16, CmCOL20, and CmFT, were also used in the study (Table 2). All of the leaf samples were stored at −70 °C for RNA isolation and RTq-PCR analysis. Terminal buds of Zijiao and Aoyunbaixue were cut every 3 days in addition to the leaf tissue harvested. Then the bud materials were fixed with FAA (Formalin–acetic acid–alcohol) solution for 1 week and used for paraffin section operation to observe the flower bud differentiation process under the microscope (Gerlach, 1969).
Expression pattern of CmPRRs in Zijiao across different developmental stages
Mature leaves at the 3rd and 4th nodes were harvested at 9:00 am from Zijiao at the respective stages of flower bud undifferentiation, squaring, visible bud color, initial flowering, full-blossom, and the senescing flowers. Materials were collected and stored at −70 °C for RNA isolation and gene expression analysis using RTq-PCR.
Expression patterns of CmPRRs in Zijiao different tissues
Since CmPRRs are highly expressed at the senescing flower stage (will be addressed below), we harvested roots, stems, leaves, and flowers from Zijiao at 9:00 am at this stage, in order to study the tissue-specific expression of the CmPRRs using RTq-PCR.
Expression patterns of CmPRRs in Zijiao under different photoperiod conditions
Long- or short-day treatments
Stem cuttings from Zijiao were propagated in the green house with 60–70% relative humidity, 14/10-h light/dark cycles and a constant temperature of 25 ± 2 °C. 1 month later, at least 50 strong and healthy seedlings were selected and planted in flowerpots for long-day (LD; 16/8-h light/dark cycles) and short-day (SD; 8/16-h light/dark cycles) treatments, respectively. After 2 weeks, leaves at the 3rd and 4th nodes were harvested every 3 h within 24 h for gene expression analysis using RTq-PCR. Materials collection was carried out with three biological replications at every time point.
Night-break treatment
After 2 weeks of short-day treatment, seedlings were further treated with night-break; that is, light was on at 15 h, was off at 17 h, and lasted for 2 h. After that, leaves at the 3rd and 4th nodes were harvested every 3 h within 24 h for gene expression pattern analysis. Materials collection was carried out with three biological replications at every time point.
Continuous light or dark treatment
After being treated with the long-day or short-day photoperiod for 2 weeks, seedlings were transferred to continuous light or dark conditions. Then leaves at the 3rd and 4th nodes were harvested every 3 h within 48 h for gene expression analysis. Period length and relative amplitude were estimated using program FFT-NLLS (Millar et al., 1995; Plautz et al., 1997). Materials collection was carried out with three biological replications at every time point.
Statistical analysis
Statistical analysis methods included single variable analysis and multiple variable analysis. Single variable analysis was used to compare the CmPRRs gene expression in Zijiao across different development stages (FU, SQ, VB, IF, FB, SF), and to compare gene expression level in Zijiao in different tissues (Flowers, Leaves, Stems, Roots). The student t-test was applied for this purpose. Multivariate analysis was employed to study the relationships and interactions between the two variables: different Chrysanthemum varieties (Zijiao and Aoyunbaixue) and different flower bud differentiation stages. The multiple linear regression modeling followed by the analysis of variance was performed to examine the significance of one variable after adjustment for another variable. In addition, the errors from both technical and biological replications were illustrated using standard error (SE) method. Data represents the mean ± SE of the experiments. All statistical analyses were conducted using the package corrplot in R (R Core Team, 2018).
Results
Gene cloning and amino acids sequence analysis
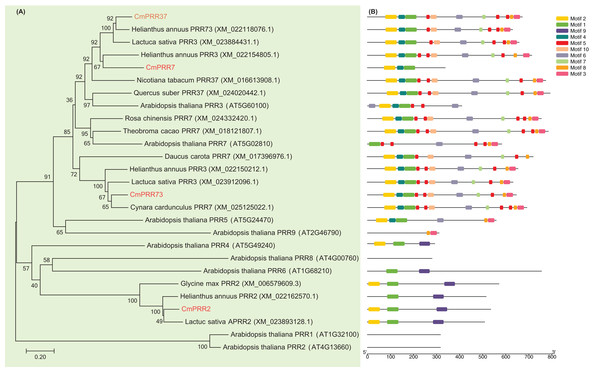
The cDNA fragments of CmPRR2, CmPRR7, CmPRR37, and CmPRR73 were amplified using RT-PCR from Chrysanthemum Aoyunbaixue according to the RNA sequencing pre-test (SI.1, SI.2). In order to examine the phylogenetic relationships among the PRR protein sequences from Chrysanthemum and other related species, we constructed unrooted trees based on alignments with the full-length protein sequences. Evidences from the sequence comparisons indicated that the Chrysanthemum PRRs are highly homologous to the counterparts from other species, especially the Asteraceae plant Helianthus annuus (Fig. 1A). For instance, CmPRR2 shares high homology with the PRR2 from H. annuus (XM_022162570.1), Glycine max (XM_006579609.3), and Lactuca sativa (XM_023893128.1) (Fig. 1A). CmPRR7 has high similarity with protein TaPRR7 from Theobroma cacao (XM_018121807.1), HaPRR3 from H. annuus (XM_022154805.1), and NtPRR37 from Nicotiara tabacum (XM_016613908.1). CmPRR37 shares 82%, 83%, and 77% sequence identity with protein HaPRR73 (XM_022118076.1), LsPRR3 (XM_023884431.1), and QsPRR37 (XM_024020442.1), respectively. CmPRR73 also shows high similarity to HaPRR3 (XM_022150212.1) and LsPRR3 (XM_023912096.1). However, we found that all of the four CmPRR proteins have a relatively low homology with PRR proteins from Arabidopsis (Fig. 1A).
Figure 1: Phylogenetic relationships and motif composition of PRRs.
(A) Multiple alignment of full-length amino acids of PRRs. (B) Schematic representation of conserved motifs in the PRR proteins.To further reveal the diversification of the PRR genes, putative motifs were predicted by the program MEME, and 10 distinct motifs were identified (Fig. 1B). Details of the 10 putative motifs are shown in Supplementary Information 3. Most of the closely related members in the phylogenetic tree have common motif compositions, suggesting functional similarities among the PRR proteins within the same subtree (Fig. 1). All of the CmPRR2, CmPRR7, CmPRR37, and CmPRR73 contain the conserved Motif1 and Motif2 in the N-terminal of amino acid sequence (Fig. 1B). Moreover, CmPRR37 and CmPRR73 share high consistency in the motifs harbored (Fig. 1B).
Observation of flower bud differentiation process
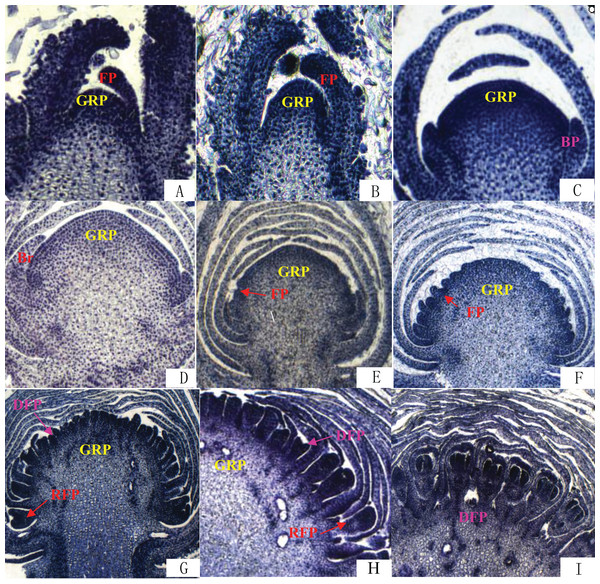
The differentiation of Chrysanthemum buds is divided into two processes: inflorescence differentiation and floret differentiation. The former contains such stages as vegetative growth, inflorescence primordial differentiation, involucres differentiation, and ligulae flower differentiation. The latter mainly has the corolla formation stage. On the basis of the results of paraffin section (Fig. 2), we compared the time of flower bud differentiation between Zijiao and Aoyunbaixue (Table 3). The flower bud of Zijiao started to differentiate after 9 days of vegetative growth, and floret differentiate initiated after 54 days of vegetative growth (Table 3). The entire flowering process lasted for 64 days. In contrast, Aoyunbaixue flower bud differentiation started after 15 days of vegetative growth, and floret differentiate initiated after 39 days of vegetative growth (Table 3). The entire flowering process took 51 days. Compared to Aoyunbaixue, Zijiao needs 13 more days to complete the flower bud differentiation. In addition, the initiation of flower bud differentiation in Zijiao is 38 days later compared to that of Aoyunbaixue.
Figure 2: Flower bud differentiation observation using paraffin section.
(A) Vegetative growth stage; (B) inflorescence primordial differentiation stage; (C) initial stage of involucres primordial differentiation stage; (D) final stage of involucres differentiation; (E) initial stage of ligulae flower differentiation; (F) final stage of ligulae flower differentiation; (G) initial stage of corolla formation; (H) secondary stage of corolla formation; (I) finish stage of corolla formation; Br, Bract; BP, Bract primordium; DFP, Disk primordium; FP, Floret primordium; GRP, Growth point; RFP, Ray floret primordium. For paraffin section operation to observe the flower bud differentiation process under the fluorescence microscope. The paraffin section operation was performed with about 10 buds per samples.| Stage | Start time | Last time (Days) | ||
|---|---|---|---|---|
| Zijiao | Aoyunbaixue | Zijiao | Aoyunbaixue | |
| A-Vegetative growth stage | 14, June | 30, April | 9 | 15 |
| B-Inflorescence primordial differentiation stage | 23, June | 15, May | 18 | 10 |
| C1-Initial stage of involucres primordial differentiation stage | 11, July | 25, May | 21 | 10 |
| C2-Final stage of involucres differentiation | 2, August | 4, June | 6 | 4 |
| D1-Initial stage of ligulae flower differentiation | 8, August | 8, June | 3 | 7 |
| D2-Final stage of ligulae flower differentiation | 11, August | 15, June | 3 | 7 |
| E1-Initial stage of corolla formation | 14, August | 22, June | 6 | 3 |
| E2-Secondary stage of corolla formation | 20, August | 25, June | 6 | 15 |
| E3-Finish stage of corolla formation | 26, August | 9, July | ||
Expression patterns of CmPRRs in Zijiao and Aoyunbaixue flower bud differentiation stages
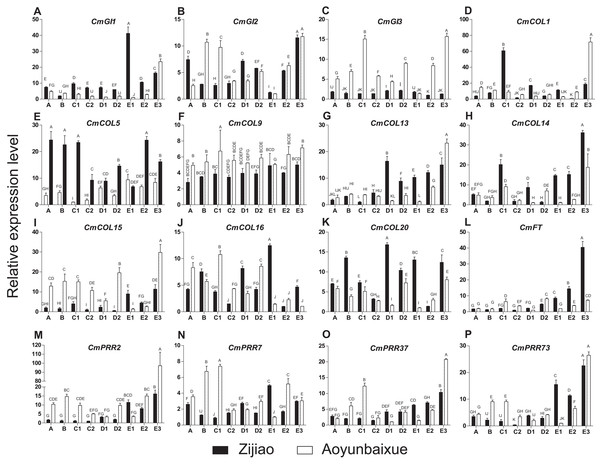
We compared gene expression patterns between short-day sensitive Zijiao and short-day insensitive Aoyunbaixue. Four genes, including CmPRR2, CmPRR7, CmPRR37, and CmPRR73, were overexpressed in Aoyunbaixue at stage E3 compared to Zijiao (Fig. 3). Before the E3 stage, expression levels of CmPRR2 were relatively low in both Zijiao and Aoyunbaixue (Fig. 3). The expression of CmPRR7 was significantly different between Zijiao and Aoyunbaixue across the entire flower bud differentiation stages except the E3 stage. It reached to the peak at stage D1 in Aoyunbaixue, while in Zijiao the peak displayed at stage E1 (Fig. 3). CmPRR37 and CmPRR73 showed similar gene expression patterns, except that their expression levels were relatively higher in Zijiao than that in Aoyunbaixue at stages D1, D2, E1, and E2, and vice versa at the other stages (Fig. 3). In addition, expression levels of other circadian clock related genes, including CmGI1, CmGI2, CmGI3, CmCOL1, CmCOL5, CmCOL9, CmCOL13, CmCOL14, CmCOL15, CmCOL16, CmCOL20, and CmFT, were identified using RTq-PCR. In general, CmCOL14, CmCOL20, and CmFT genes showed opposite expression patterns with the CmPRRs gene at stage E3. However, genes, includingCmGI1, CmGI3, CmCOL1, CmCOL5, CmCOL13, and CmCOL15, displayed a similar expression pattern with the CmPRRs (Fig. 3). In addition, there was no significant difference between Zijiao and Aoyunbaixue in the expression of CmCOL9 during the flower bud differentiation.
Figure 3: Expression pattern of circadian clock related genes across Zijiao and Aoyunbaixue different flower bud differentiation stages.
A-Vegetative growth stage; B-Inflorescence primordial differentiation stage; C1-Initial stage of involucres primordial differentiation stage; C2-Final stage of involucres differentiation; D1-Initial stage of ligulae flower differentiation; D2-Final stage of ligulae flower differentiation; E1-Initial stage of corolla formation; E2-Secondary stage of corolla formation; E3-Finish stage of corolla formation. Capital letters (A–P) indicate whether the samples are significantly different from each other. Data represents the mean ± SE of three technical and three biological repeats per sample.Expression patterns of CmPRRs in Zijiao across different developmental stages
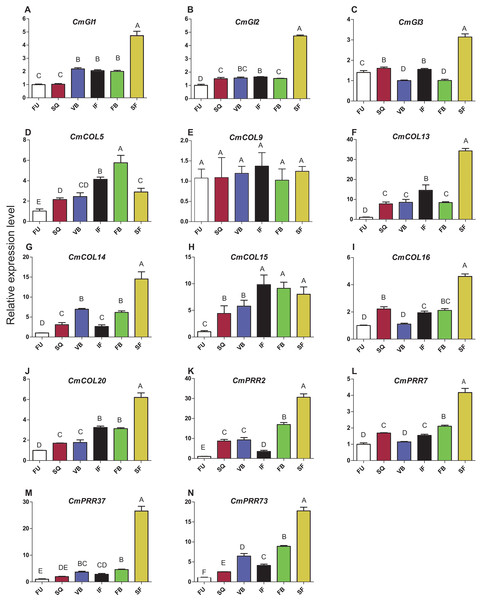
Expression patterns of circadian clock genes, including CmPRR2, CmPRR7, CmPRR37, and CmPRR73, were analyzed across developmental stages of Zijiao (Fig. 4). In general, expression levels of CmPRR2, CmPRR7, CmPRR37, and CmPRR73 showed a growing trend across Chrysanthemum developmental process. The expression of CmPRR2 and CmPRR73 showed a brief decrease at the initial flowering stage; in contrast, the expression of CmPRR7 went down at the stage of visible bud color (Fig. 4). At squaring stage, the expression level of CmPRR2 is almost five times that of the other three CmPRRs (Fig. 4). At the visible bud color stage, the expression level of CmPRR73 was twice that of CmPRR37, and expression level of CmPRR37 was three times that of CmPRR7 (Fig. 4). However, other genes, including CmGI1 and CmCOL20, exhibited a gradual increase in expression across the entire developmental stages of Zijiao, which was consistent with the expression of CmPRR37. In addition, the expression patterns of CmGI2, CmGI3, CmCOL13, CmCOL14, and CmCOL16 were similar to that of CmPRR2, CmPRR7, and CmPRR73 (Fig. 4).
Figure 4: Expression pattern of circadian clock genes in Zijiao different development stages.
(A) Relative expression level of CmGI1; (B) Relative expression level of CmGI2; (C) Relative expression level of CmGI3; (D) Relative expression level of CmCOL5; (E) Relative expression level of CmCOL9; (F) Relative expression level of CmCOL13; (G) Relative expression level of CmCOL14; (H) Relative expression level of CmCOL15; (I) Relative expression level of CmCOL16; (J) Relative expression level of CmCOL20; (K) Relative expression level of CmPRR2; (L) Relative expression level of CmPRR7; (M) Relative expression level of CmPRR37; (N) Relative expression level of CmPRR73. AFU, Flower bud undifferentiated stage; SQ, squaring stage; VB, visible bud color stage; IF, initial flowering stage; FB, full blossom stage; SF, senescing flower stage. Letters in (A–N) indicate whether the samples are significantly different from each other. Data represents the average ± SE of three technical and three biological repeats per sample.Expression patterns of CmPRRs in different tissues of Zijiao
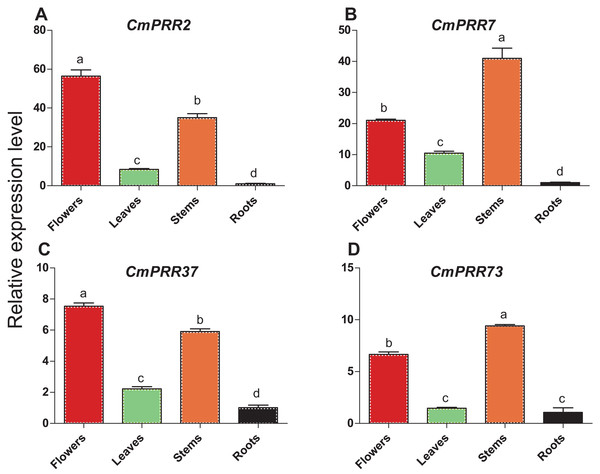
Expression patterns of CmPRR2, CmPRR7, CmPRR37, and CmPRR73 in the respective root, stem, leave, and flower tissues of Zijiao were analyzed. These four genes were expressed in all of the tissues tested, with high expression level in flowers and stems, and the lowest level in roots (Fig. 5). CmPRR2 and CmPRR37 displayed similar expression patterns to that of CmPRR7 and CmPRR73. It is notable that the expression of CmPRR7 in roots was significantly lower than that in the leaf; but the expression of CmPRR73 in roots was no significantly different from that in leaves.
Figure 5: Expression pattern of CmPRRs in different tissues of Zijiao.
(A) Relative expression level of CmPRR2; (B) Relative expression level of CmPRR7; (C) Relative expression level of CmPRR37; (D) Relative expression level of CmPRR73. Letters a–d indicate whether the samples are significantly different from each other. Data represents the average ± SE of three technical and three biological repeats per sample.Expression patterns of CmPRRs in Zijiao under different photoperiod conditions
Long- or short-day treatments
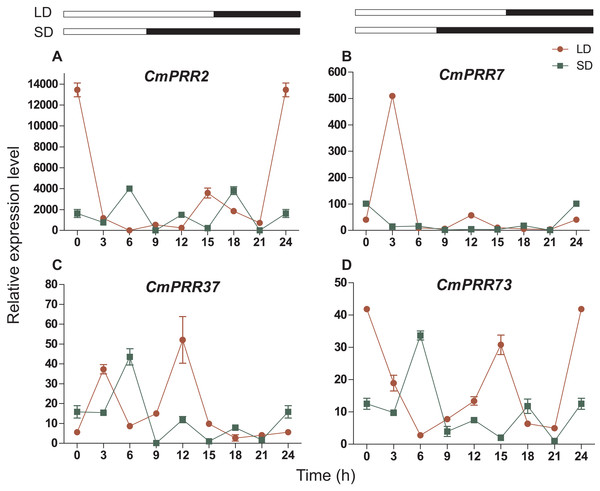
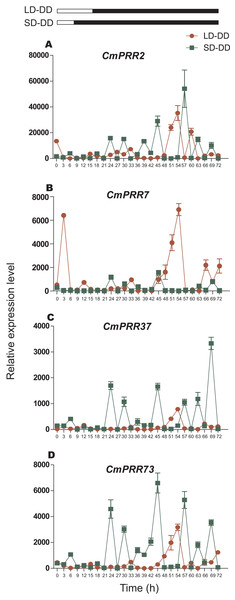
The CmPRR gene responded differentially to long- and short-day treatments. Expression of CmPRR2 was similar to that of CmPRR73 under both long- and short-day conditions; that is, the expression peaks of the two genes coincide with one another, and their expression levels were higher under long-day condition, compared to the short-day condition (Fig. 6). When challenged with long-day, both CmPRR2 and CmPRR73 expressed the highest level at the start of light and the end of darkness; In contrast, under the short-day condition, the peak of expression occurred at 6 h in the light phase. Under the long-day condition, expression level of CmPRR7 started to increase upon the light, reached to the peak after 3 h, and then declined (Fig. 6). Under short-day condition, however, its expression generally continued to decline upon light (Fig. 6). Nevertheless, expression of CmPRR7 showed a rebound at the end of dark phase in both long- and short-day conditions. Compared to the dark phase, CmPRR37 expressed at a higher level in the light phase under both long- and short-day conditions, with the peak occurred before the end of light treatment (Fig. 6).
Figure 6: Expression pattern of CmPRRs of Zijiao in long-day and short-day photoperiod conditions.
(A) Relative expression level of CmPRR2; (B) Relative expression level of CmPRR7; (C) Relative expression level of CmPRR37; (D) Relative expression level of CmPRR73. LD, Long-day condition; SD, short-day condition. Data represents the average ± SE of three technical and three biological repeats per sample.Night-break treatment
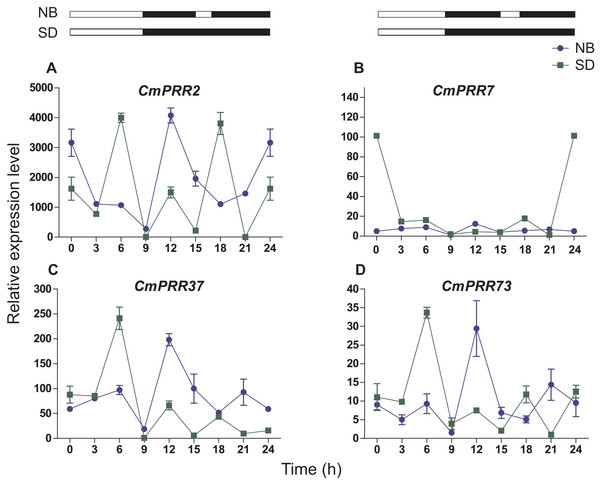
Under the night-break condition, expression levels of CmPRR2, CmPRR37, and CmPRR73 were similar to that challenged with the short-day, with the peak of expression levels coincided with one another (Fig. 7). In contrast, expression level of CmPRR7 was significantly reduced due to the night-break, compared to its expression under the short-day condition. When challenged with the night-break, the expression of CmPRR2, CmPRR37, and CmPRR73 genes decreased at 15–17 h, which was the opposite to that with the short-day treatment (Fig. 7). After 3-h night-break, the expression levels of CmPRR2, CmPRR37, and CmPRR73 started to rise, but still displayed a trend that was against that under the short-day condition (Fig. 7). In addition, CmPRR7 gene expression level was relatively stable at 15–17 h (Fig. 7).
Figure 7: Expression pattern of CmPRRs of Zijiao in night-break condition.
(A) Relative expression level of CmPRR2; (B) Relative expression level of CmPRR7; (C) Relative expression level of CmPRR37; (D) Relative expression level of CmPRR73. The light break occurred at 15 h, went off at 17 h, and lasted for 2 h. NB, Night-break condition; SD, short-day condition. Data represents the average ± SE of three technical and three biological repeats per sample.Continuous light
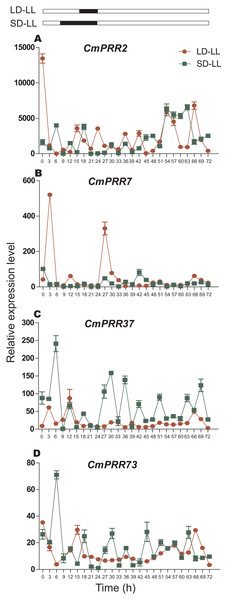
After switching from the long-day to the continuous light condition, expression of CmPRR2 and CmPRR7 still maintained the original circadian rhythm in the first 24 h, but the expression amplitude decreased (Fig. 8). Similarly, CmPRR37 and CmPRR73 also maintained a weak circadian rhythm. In the following 24 h, the four CmPRRs maintained a varied circadian rhythm (Fig. 8). After changing from the short-day to the continuous light condition, expression of CmPRR37 still kept the original circadian rhythm with the amplitude decreased in the first day (Fig. 8). Expression of CmPRR7 had a weak circadian rhythm (Fig. 8). CmPRR2 and CmPRR73 showed similar expression patterns with that under the long-day condition. In the following day, CmPRR7 hold a certain circadian rhythm, with a reduced amplitude.
Figure 8: Expression pattern of CmPRRs of Zijiao in continuous light condition.
(A) Relative expression level of CmPRR2; (B) Relative expression level of CmPRR7; (C) Relative expression level of CmPRR37; (D) Relative expression level of CmPRR73. LD, Long-day condition; SD, short-day condition; LL, continuous light condition. Data represents the average ± SE of three technical and three biological repeats per sample.Continuous dark
After switching from the long-day to the continuous dark condition, expression of CmPRR7, CmPRR37, and CmPRR73 genes still maintained a level of the original circadian rhythm in the first day, and the amplitude of CmPRR7 expression decreased (Fig. 9). In addition, expression of CmPRR2 gene exhibited only a moderate circadian rhythm. On the second day, expression of CmPRR7 showed its original circadian rhythm with an increased amplitude. Expression of CmPRR2, CmPRR37, and CmPRR73 held a certain level of circadian rhythm with enhanced amplitude and the rhythm. In addition, after changing from the short-day to the continuous dark condition, the four CmPRRs maintained a distinct circadian rhythm within the 48 h tested. The amplitude and rhythm of CmPRR2, CmPRR37, and CmPRR73 were increased.
Figure 9: Expression pattern of CmPRRs of Zijiao in continuous dark condition.
(A) Relative expression level of CmPRR2; (B) Relative expression level of CmPRR7; (C) Relative expression level of CmPRR37; (D) Relative expression level of CmPRR73. LD, Long-day condition; SD, short-day condition; DD, continuous dark condition. Data represents the average ± SE of three technical and three biological repeats per sample.Discussion
In the present study, we cloned four Chrysanthemum PRR genes and compared their full-length protein sequences with their counterparts from other related species. Our results indicated that PRR proteins from Chrysanthemum are highly homologous to those from the closely related Asteraceae plants H. annuus (Fig. 1A). However, we found that all of these four CmPRRs have a relatively low homology with PRR proteins from Arabidopsis (Fig. 1A). For instance, CmPRR2 shares 32% identity with PRR2 (AT4G13660) from Arabidopsis, whereas CmPRR7 has a relatively high identity (82%) with protein AT5G02810 (PRR7). Both CmPRR37 and CmPRR73 show 46% similarity to PRR3 (AT5G60100) of Arabidopsis, but the similarity of them compared to PRR7 of Arabidopsis is 53% and 37%, respectively. In addition, we predicted motifs of the four Chrysanthemum PRRs. They contain the conserved Motif1 and Motif2 in the N-terminal of amino acid sequence (Fig. 1B). As expected, closely related members in the phylogenetic tree share common motifs, suggesting functional similarities among the PRR proteins within the same subtree (Fig. 1). However, the biological significance of these putative motifs remains to be elucidated as they do not have homologs when searching against Pfam and simple modular architecture research tool databases (Finn et al., 2006; Letunic & Bork, 2018).
Flower bud differentiation is the most important stage in the development of Chrysanthemum, which marks the transition from vegetative growth to reproductive growth. The entire process of flower bud differentiation varies by Chrysanthemum varieties. Previous studies by Yang et al. (2007) indicated that eight Japanese Chrysanthemum verities need 30–60 days to complete flower bud differentiation. In the present study, the flower bud differentiation process lasted for 64 days for Zijiao and 51 days for Aoyunbaixue (Fig. 2; Table 3). We observed that the flower bud differentiation started to speed up after the squaring stage in both Zijiao and Aoyunbaixue, which is similar to the trend observed in Dendranthema morifolium (Rong, 2006). Expression levels of circadian genes, including CmGI3, CmCOL1, CmCOL5, CmCOL9, CmCOL13, CmCOL15, CmCOL16, CmPRR2, CmPRR37, and CmPRR37, were higher in Aoyunbaixue, compared to that in Zijiao, suggesting that these genes may contribute to promote initiating floral meristem development. This is consistent with former studies that circadian clock genes including PRRs play essential roles within or close to the control of hypocotyl elongation and flowering time (Akinori et al., 2007; Ariadne et al., 2002; Eriko et al., 2002; Kaczorowski & Quail, 2003; Masaya, Takafumi & Takeshi, 2004; Makino et al., 2002; Takatoshi et al., 2007). However, other genes, including CmGI1, CmGI2, CmCOL14, CmCOL20, and CmFT, displayed the opposite expression pattern. These genes were highly expressed in Zijiao than that in Aoyunbaixue, suggesting their functions in inhibiting flowering (Fig. 3). Therefore, there exist two sets of circadian related genes and their interplay plays a significant role in regulating flowering time in Chrysanthemum. More work with knock-out mutants will help address the underlying molecular mechanisms.
During different stages of Chrysanthemum development, the circadian genes present different expression features in leaves. In this study, expression of CmCOL15 was the highest at the full-blossom stage and decreased in the senescing flower stage, suggesting that it is not involved in the process of senescence. Since CmPRR37 had a similar expression level to that of CmGI1 and CmCOL20, suggesting that it plays a significant role in the regulation of Chrysanthemum development (Fig. 4). The expression level of tobacco NtCO1 is significantly higher during the stage of squaring and vegetative growth, compared to the seedling stage (Lu et al., 2013). Seven CmCOL genes in the present study showed similar results (Fig. 4). In addition, we found that four CmPRRs were expressed in different tissues of Zijiao (Fig. 5). Interestingly, the CmPRRs had the highest expression level in flowers and stems, a moderate level in the leaves, and lowest level in the roots.
In the photoperiod pathway, plants perceive the light signal and produce the biorhythm to regulate the downstream flowering related gene expression, and then regulate the flowering process. In C. lavandulifolium, ClPRR1, ClPRR73, and ClPRR37 expression peaks appeared earlier under short-day condition, compared to long-day condition (Fu, 2014). Under long-day, SbPRR37 had expression peaks both in the morning and at night, while the peak at night disappeared under short-day condition (Murphy et al., 2011). The expression peak of CmPRR2, CmPRR7, and CmPRR73 under long-day appeared earlier, compared to the short-day condition. However, expression of CmPRR37 displayed the opposite pattern (Fig. 6). Previous studies indicated that the order of expression in five Arabidopsis PRRs is PRR9→PRR7→PRR5→PRR3→PRR1/TOC (Strayer et al., 2000). In comparison, five members from the PRR gene family were cloned from C. lavandulifolium, and the order of expression from morning to night in one day is ClPRR73/ ClPRR37→ClPRR5→ClPRR1 (Fu, Yang & Dai, 2014). In the present study, the order of the CmPRRs expression peaks under long day condition appears to be like this: CmPRR2/CmPRR73→CmPRR7→CmPRR37 (Fig. 6). Previous studies indicated that expression peaks of SbPRR37 in both morning and night disappeared when the Sorghum was placed into continuous dark condition (Murphy et al., 2011). We switched Zijiao from long- or short-day conditions to continuous dark and found that expression levels of the CmPRRs still maintained the original circadian oscillation, but the rhythmicity and amplitude enhanced (Fig. 9). This indicates that the CmPRRs can maintain their circadian oscillation features, which are subject to influence by external light. When compared with the results of continuous light, expression of the CmPRRs has increased in both rhythm and amplitude, which may be related to the regulation of the photo responsive element in CmPRRs (Figs. 8 and 9). Studies have found that giving short periods of light in the dark phase under short-day, which is equivalent to long-day condition, can significantly inhibit the flowering of short-day plants (Thomas & Vince-Prue, 1997). Results from dark interrupt in rice indicated that a significant flowering delay occurred after 10 min of light is given in the dark phase (Ishikawa et al., 2005). In the present study, the changes of expression patterns of the CmPRRs after the night-break may impact expression of the downstream genes of the CmPRRs and then change the flowering process of Chrysanthemum (Fig. 7).
Conclusion
In this study, four genes related to flowering time (CmPRR2C, mPRR7, CmPRR37, and CmPRR73) were isolated and cloned from C. morifolium. Sequences alignment and phylogenetic analysis showed that they are highly homologous with PRRs from the closely related Asteraceae plants H. annuus. In addition, conserved motifs in these PRR proteins were predicted, and we found that most of the closely related members in the phylogenetic tree contain common motif composition, which suggested the functional similarities among the PRR proteins within the same subtree. Spatio-temporal expression patterns showed that the CmPRRs were highly expressed in flower and stem tissues, and the expression levels were increasing across the Chrysanthemum developmental process. In addition, we found that these genes appear to be light regulated/responsive and are differentially expressed under LD and SD conditions. These CmPRRs could be involved in regulating flowering time in C. morifolium.
Supplemental Information
Raw data of cDNA and protein sequences of four CmPRRs.
CmPRR2, CmPRR7, CmPRR37 and CmPRR73 were cloned from Aoyunbaixue using the tender leaves by RT-PCR.
Full length PCR amplification of CmPRRs..
PCR prducts were used to validate the amplificaion of four CmPRRs. Non-reverse transcribed RNA was used as the negative control. M-DNA marker; NC-negative control.
Sequence logos for the conserved motifs of PRR proteins.
Conserved motifs and the sequence logos were generated using the MEME search tool (http://meme-suite.org/tools/meme). Numbers on the horizontal axis represent the sequence positions in the motifs and the vertical axis represents the information content measured in bits.