RBP-J regulates homeostasis and function of circulating Ly6Clo monocytes
Abstract
Notch-RBP-J signaling plays an essential role in the maintenance of myeloid homeostasis. However, its role in monocyte cell fate decisions is not fully understood. Here, we showed that conditional deletion of transcription factor RBP-J in myeloid cells resulted in marked accumulation of blood Ly6Clo monocytes that highly expressed chemokine receptor CCR2. Bone marrow transplantation and parabiosis experiments revealed a cell-intrinsic requirement of RBP-J for controlling blood Ly6CloCCR2hi monocytes. RBP-J-deficient Ly6Clo monocytes exhibited enhanced capacity competing with wildtype counterparts in blood circulation. In accordance with alterations of circulating monocytes, RBP-J deficiency led to markedly increased population of lung tissues with Ly6Clo monocytes and CD16.2+ interstitial macrophages. Furthermore, RBP-J deficiency-associated phenotypes could be genetically corrected by further deleting Ccr2 in myeloid cells. These results demonstrate that RBP-J functions as a crucial regulator of blood Ly6Clo monocytes and thus derived lung-resident myeloid populations, at least in part through regulation of CCR2.
eLife assessment
This study presents a valuable examination into the role Notch-RBP-J signaling in regulating monocyte subset homeostasis. The data were collected and analyzed using solid and validated methodology and can be used as a starting point for exploring the mechanisms involved in RBP-J signaling in non-classical monocytes. The data presented strongly confirm the authors conclusions. However, this article primarily focuses on providing a description, and additional studies are necessary to fully elucidate the mechanisms through which RBP-J deficiency contributes to the specific increase in Ly6Clo monocyte numbers in both the blood and lungs.
https://doi.org/10.7554/eLife.88135.3.sa0Introduction
Monocytes are integral components of the mononuclear phagocyte system that develop from monocyte precursors in the bone marrow (Guilliams et al., 2018). Several functionally and phenotypically distinct subsets of blood monocytes have been defined on the basis of expression of surface markers (Cros et al., 2010; Geissmann et al., 2003; Passlick et al., 1989; Weber et al., 2000). In mice, Ly6Chi monocytes (also called inflammatory or classical monocytes) characterized by Ly6ChiCCR2hiCX3CR1lo exhibit a short half-life and are recruited to tissue and differentiate into macrophages and dendritic cells (Ginhoux and Jung, 2014). Ly6Chi monocytes are analogous to CD14+ human monocytes based on gene expression profiling (Ingersoll et al., 2010). A second subtype of monocytes is called Ly6Clo monocytes (also termed patrolling monocytes or non-classical monocytes), which express high level of CX3CR1 and low level of CCR2, equivalent to CD14loCD16+ human monocytes. Ly6Clo monocytes are regarded as blood-resident macrophages that patrol blood vessels and scavenge microparticles attached to the endothelium under physiological conditions, and exhibit a long half-life in the steady state (Auffray et al., 2007; Carlin et al., 2013; Yona et al., 2013). Ly6Clo monocytes may exert a protective effect by suppressing atherosclerosis in mice, while high levels of non-classical monocytes have been associated with more advanced vascular dysfunction and oxidative stress in patients with coronary artery disease (Hamers et al., 2012; Hanna et al., 2012; Urbanski et al., 2017). In the inflammatory settings, Ly6Clo monocytes often show anti-inflammatory properties, yet also exhibit a proinflammatory role under certain circumstances (Amano et al., 2005; Brunet et al., 2016; Carlin et al., 2013; Cros et al., 2010; Misharin et al., 2014; Nahrendorf et al., 2007; Puchner et al., 2018). In addition, Ly6Clo monocytes have demonstrated protective functions in tumorigenesis, such as engulfing tumor materials to prevent cancer metastasis, activating and recruiting NK cells to the lungs (Hanna et al., 2015; Thomas et al., 2016). While the functions of Ly6Clo monocytes are complex and sometimes contradictory depending on the animal models and experimental approaches, it is clear that they play a crucial role in both health and disease.
The generation and maintenance of Ly6Clo monocytes are regulated by several transcription factors, including Nr4a1 and CEBPβ (Hanna et al., 2011; Tamura et al., 2017). The colony-stimulating factor 1 receptor (CSF1R) signaling pathway is important for the generation and survival of Ly6Clo monocytes, and neutralizing antibodies against CSF1R can reduce their numbers (MacDonald et al., 2010). Interestingly, recent studies have suggested that different subsets of monocytes may be supported by distinct cellular sources of CSF-1 within bone marrow niches, in which targeted deletion of Csf1 from sinusoidal endothelial cells selectively reduces Ly6Clo monocytes but not Ly6Chi monocytes (Emoto et al., 2022). LAIR1 has been shown to be activated by stromal protein Colec12, and LAIR1 deficiency leads to aberrant proliferation and apoptosis in bone marrow non-classical monocytes (Keerthivasan et al., 2021). These findings further underscore the complexity of the mechanisms that regulate Ly6Clo monocytes and suggest that multiple factors are involved in this process.
Recombinant recognition sequence binding protein at the Jκ site (RBP-J; also named CSL) is commonly known as the master nuclear mediator of canonical Notch signaling (Radtke et al., 2013). In the absence of Notch intracellular domain, RBP-J associates with co-repressor proteins to repress transcription of downstream target genes. In the immune system, the best studied functions of Notch signaling are its roles in regulating the development of lymphocytes, such as T cells and marginal zone B cells (Radtke et al., 2013). Notch signaling also has been reported to regulate the differentiation and function of myeloid cells including granulocyte/monocyte progenitors, osteoclasts, and dendritic cells (Caton et al., 2007; Klinakis et al., 2011; Lewis et al., 2011; Zhao et al., 2012). In osteoclasts, RBP-J represses osteoclastogenesis, particularly under inflammatory conditions (Zhao et al., 2012). In dendritic cells, Notch-RBP-J signaling controls the maintenance of CD8- DC (Caton et al., 2007). In addition, Notch2-RBP-J pathway is essential for the development of CD8-ESAM+ DC in the spleen and CD103+CD11b+ DC in the lamina propria of the intestine (Lewis et al., 2011). However, the role of Notch-RBP-J signaling pathway in regulating monocyte subsets remains elusive. Here, we demonstrated that RBP-J controlled homeostasis of blood Ly6Clo monocytes in a cell-intrinsic manner. RBP-J deficiency led to a drastic increase in blood Ly6Clo monocytes, lung Ly6Clo monocytes, and CD16.2+ IM. Our results suggest that RBP-J plays a critical role in regulating the population and characteristics of Ly6Clo monocytes in the blood and lung.
Results
RBP-J is essential for the maintenance of blood Ly6Clo monocytes
To investigate the role of RBP-J in monocyte subsets, we utilized mice with RBP-J specific deletion in the myeloid cells (Rbpjfl/flLyz2cre/cre mice). Efficient deletion of Rbpj in blood monocytes was confirmed by quantitative real-time PCR (qPCR) (Figure 1—figure supplement 1A). Flow cytometric analysis revealed that RBP-J-deficient mice had a significant increase in the proportion of Ly6Clo monocytes, but not Ly6Chi monocytes, in blood, compared to age-matched control mice with the genotype Rbpj+/+Lyz2cre/cre (Figure 1A). In contrast to circulating monocytes, RBP-J-deficient mice exhibited minimal alterations in the percentages of monocyte subsets in bone marrow (BM) and spleen (Figure 1B, Figure 1—figure supplement 1C). Additionally, RBP-J deficiency in myeloid cells did not appear to have an effect on neutrophils (Figure 1—figure supplement 1B). Therefore, among circulating myeloid populations, RBP-J selectively controlled the subset of Ly6Clo monocytes.

RBP-J-deficient mice display more blood Ly6Clo monocytes.
(A) Blood Ly6Chi and Ly6Clo monocytes in Rbpj+/+Lyz2cre/cre control and Rbpjfl/flLyz2cre/cre mice were determined by flow cytometry analyses (FACS). Representative FACS plots (left) and cumulative data of cell ratio (right) are shown. (B, C) Representative FACS plots and cumulative data quantitating percentages of bone marrow (BM) monocyte subsets (B) and myeloid progenitor cells (C) (CD45+CD11b+Ly6G-CD115+ Ly6Chi monocyte; CD45+CD11b+Ly6G-CD115+Ly6Clo monocyte; MDP, CD45+Lin-CD117+CD115+CD135+ Ly6C-; cMoP, CD45+Lin-CD11b-CD117+CD115+ CD135-Ly6C+). Lin: CD3, B220, Ter119, Gr-1 and CD11b. (D) Experimental outline for panel (E). (E) Cumulative data quantitating percentages of sinusoidal monocytes (CD45+) within total BM Ly6Clo monocytes. Data are pooled from at least two independent experiments; n ≥ 4 in each group. Data are shown as mean ± SEM; n.s., not significant; **p<0.01 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse.
-
Figure 1—source data 1
Data for Figure 1.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig1-data1-v1.xlsx
BM progenitors that give rise to circulating monocytes are monocyte-dendritic cell progenitors (MDPs) and common monocyte progenitors (cMoPs) (Hanna et al., 2011; Hettinger et al., 2013; Liu et al., 2019; Varol et al., 2007). Next, we analyzed BM progenitors and found that the percentages of MDPs and cMoPs were equivalent in control and RBP-J-deficient mice (Figure 1C). These results in conjunction with the observations of normal BM monocyte populations, suggest that alterations of peripheral blood Ly6Clo monocytes may not originate from BM. Next, we examined whether RBP-J may influence the egress of Ly6Clo monocytes from BM and measured BM exit rate of Ly6Clo monocytes using in vivo labeling of sinusoidal cells as previously described (Figure 1D; Debien et al., 2013). The results showed that RBP-J deficiency did not affect egress of Ly6Clo monocytes from BM (Figure 1E). Taken together, these data revealed RBP-J as a critical regulator controlling homeostasis of peripheral blood Ly6Clo monocytes.
RBP-J deficiency does not affect Ly6Chi monocyte conversion or Ly6Clo monocyte survival and proliferation
Next, we aimed to determine whether the increase in blood Ly6Clo monocytes in RBP-J-deficient mice was due to decreased cell death or enhanced proliferation. We first stained monocytes with Annexin V and 7-amino-actinomycin D (7-AAD) to identify apoptotic cells and observed comparable percentages of apoptotic blood Ly6Clo monocytes in control and RBP-J-deficient mice (Figure 2A). Given the crucial role of Nr4a1 in the survival of Ly6Clo monocytes (Hanna et al., 2011), we detected the expression of Nr4a1, which was similar in Ly6Clo monocytes from control and RBP-J-deficient mice (Figure 2—figure supplement 1A). We then assessed the proliferative capacity by analyzing Ki-67 expression as well as in vivo EdU incorporation. Ki-67 levels in blood monocytes displayed no differences between control and RBP-J-deficient mice (Figure 2B). The percentage of EdU+ monocytes did not significantly differ between control and RBP-J-deficient mice at the indicated time points, implying that the turnover of Ly6Clo monocytes was normal in RBP-J-deficient mice (Figure 2C). Fluorescent latex beads as particulate tracers could be phagocytosed by monocytes after intravenous injection and stably label Ly6Clo monocytes (Tacke et al., 2006). Thus, we intravenously injected fluorescent latex beads into control and RBP-J-deficient mice to track circulating monocytes (Figure 2D). By day 7 post injection, only Ly6Clo monocytes were latex+ as previously reported (Tacke et al., 2006), whereas control and RBP-J-deficient mice groups presented a similar frequency of latex+ monocytes (Figure 2E). Together, these data indicated that RBP-J did not influence monocyte survival and proliferation.
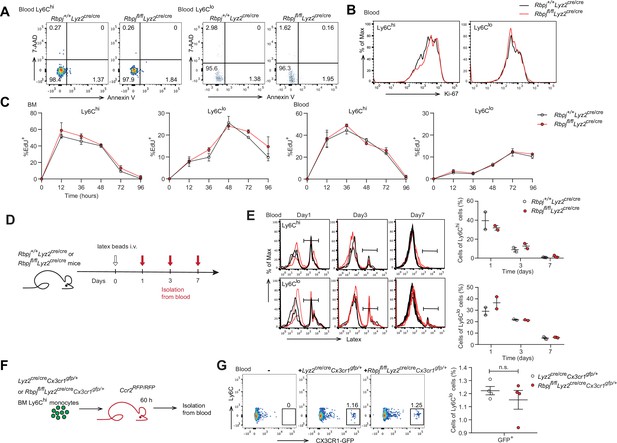
Monocyte subsets in RBP-J-deficient mice display normal cell death.
(A) Representative FACS plots of monocyte subsets in blood stained with 7-amino-actinomycin D (7-AAD) and Annexin V. (B) FACS analysis of Ki-67 expression in Rbpj+/+Lyz2cre/cre control and Rbpjfl/flLyz2cre/cre blood monocyte subsets. Black lines represent control mice, and red lines represent RBP-J-deficient mice. (C) Analysis of time course of EdU incorporation of monocyte subsets in bone marrow (BM) and blood after a single 1 mg EdU pulsing. The percentages of EdU+ cells among the indicated monocyte subsets are shown. (D) Experimental outline for panel (E). (E) Analysis of time course of latex beads incorporation of monocyte subsets in blood after latex beads injection. The percentages of latex+ cells among the indicated monocyte subsets are shown. (F) Cartoon depicting the adoptive transfer. BM GFP+Ly6Chi monocytes were sorted from Lyz2cre/creCx3cr1gfp/+ or Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice and transferred into Ccr2RFP/RFP recipient mice. Sixty hours after transfer, cell fate was analyzed. (G) Representative FACS plots are shown in the left panel, and the frequencies of GFP+Ly6Clo monocytes within total Ly6Clo monocytes are shown in the right panel. Data are pooled from two independent experiments (G); n ≥ 2 in each group (C, E, G). Data are shown as mean ± SEM; n.s., not significant; (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse (E, G).
-
Figure 2—source data 1
Data of Figure 2.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig2-data1-v1.xlsx
Previous studies have shown that Ly6Clo monocytes are observed in recipient mice following the adoptive transfer of Ly6Chi monocytes, indicating that Ly6Chi monocytes can convert into Ly6Clo monocytes (Varol et al., 2007; Yona et al., 2013). We next wished to evaluate whether conversion of Ly6Chi monocyte was regulated by RBP-J by isolating BM GFP+Ly6Chi monocytes from Lyz2cre/creCx3cr1gfp/+ control or Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice and adoptively transferring them into Ccr2RFP/RFP recipients (Figure 2F). Sixty hours after transfer, a subset of cells from donors were converted into Ly6Clo monocytes (Figure 2—figure supplement 1B), and equal percentages of Ly6Clo monocytes were derived from control and Rbpjfl/flLyz2cre/creCx3cr1gfp/+ donors (Figure 2G). Thus, the conversion of Ly6Chi monocyte into Ly6Clo monocyte was not affected by RBP-J deficiency.
RBP-J regulates blood Ly6Clo monocytes in a cell-intrinsic manner
We next wondered whether the increase in Ly6Clo monocytes in RBP-J-deficient mice was BM-derived and cell-intrinsic. We performed BM transplantation by engrafting lethally irradiated mice with a 1:4 mixture of Rbpj+/+Lyz2cre/cre control and Rbpjfl/flLyz2cre/cre BM cells (CD45.2) and Cx3cr1gfp/+ BM cells (CD45.1) (Figure 3A). Eight weeks after transplantation, we analyzed the frequencies of CD45.2+ donor cells in the BM and blood of recipient mice and found that the frequencies of CD45.2+ cells within total cells were similar to the mixture ratio of BM cells for Ly6Chi monocytes and neutrophils in both BM and blood (Figure 3B, Figure 3—figure supplement 1A and B). Specifically, more blood Ly6Clo monocytes were derived from RBP-J-deficient donors than control donors (Figure 3B), reflecting that the contribution of RBP-J-deficient cells in blood Ly6Clo cells was significantly higher than that of control cells. These results implied that RBP-J-deficient BM cells were highly efficient in generating blood Ly6Clo monocytes.

The role of RBP-J in blood Ly6Clo monocytes is cell-intrinsic.
(A) Cartoon depicting the bone marrow (BM) transplantation. BM recipient 6-week-old male C57BL/6 mice (CD45.2) were lethally irradiated. BM cells from donor mice (Rbpj+/+Lyz2cre/cre or Rbpjfl/flLyz2cre/cre and CD45.1, Cx3cr1gfp/+) were collected and transferred into recipient mice. Mice were used after 8 wk of BM reconstitution. (B) Representative FACS plots (left) and cumulative data (right) quantitating the frequency of Rbpj+/+Lyz2cre/cre and Rbpjfl/flLyz2cre/cre donor cells among Ly6Chi and Ly6Clo monocytes in the blood of recipient mice. (C) Cartoon depicting the generation of Rbpj+/+Lyz2cre/cre control or Rbpjfl/flLyz2cre/cre and Cx3cr1gfp/+ parabiotic pairs. (D) Representative FACS plots (left) and cumulative data (right) quantitating percentages of monocyte subsets derived from control or RBP-J-deficient mice in control or RBP-J-deficient mice. (E) Representative FACS plots (left) and cumulative data (right) quantitating percentages of monocyte subsets derived from control or RBP-J-deficient mice in Cx3cr1gfp/+ mice. Data are pooled from at least two independent experiments; n ≥ 6 in each group. Data are shown as mean ± SEM; n.s., not significant; ***p<0.001; ****p<0.0001 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse. SSC-A, side scatter area.
-
Figure 3—source data 1
Data of Figure 3.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig3-data1-v1.xlsx
Given that RBP-J-deficient mice had more Ly6Clo monocytes in blood than did control mice, we performed parabiosis experiments, a surgical union of two organisms, which allowed parabiotic mice to share their blood circulation. The contribution of circulating cells from one animal to another can be estimated by measuring the percentage of blood cells that originated from each animal (Liu et al., 2007). We joined a CD45.1, Cx3cr1gfp/+ mouse with an age- and sex-matched Rbpj+/+Lyz2cre/cre control or Rbpjfl/flLyz2cre/cre mouse (CD45.2) (Figure 3C). Four weeks after the procedure, about 50% of B cells and T cells in parabiotic mice displayed efficient exchange of their circulation (Figure 3—figure supplement 1C). As expected, RBP-J-deficient mice exhibited higher percentages of Ly6Clo monocytes than control animals (Figure 3D). Intriguingly, in the parabiotic Cx3cr1gfp/+ mice, RBP-J-deficient cells still constituted significantly higher proportion of circulating Ly6Clo, but not Ly6Chi monocytes, than RBP-J sufficient counterparts as indicated by the percentages of the GFP-negative population (Figure 3E). These results implicated that the enhanced ability of Ly6Clo monocytes to circulate in the peripheral blood as a result of RBP-J deficiency was cell-intrinsic.
RBP-J regulates phenotypical marker genes in blood Ly6Clo monocytes
To further study the consequences of RBP-J loss of function in blood Ly6Clo monocytes, we performed gene expression profiling by RNA-seq, which revealed that Ly6Clo monocytes in RBP-J-deficient mice exhibited low expression of Itgax but high expression of Ccr2, in comparison to those in control mice (Figure 4A). These differential expression patterns of phenotypic marker genes were also confirmed by qPCR and flow cytometry analysis (Figure 4B). However, blood Ly6Chi monocytes and BM monocytes displayed normal levels of CD11c and CCR2 (Figure 4B, Figure 4—figure supplement 1A). Moreover, principal component analysis showed distinct expression pattern of monocytes between control and RBP-J-deficient mice (Figure 4C). To determine whether the regulation of phenotypic markers by RBP-J in blood Ly6Clo monocytes was cell-intrinsic, we performed BM transplantation as described above (Figure 3A). The findings indicated that Ly6Clo monocytes derived from RBP-J-deficient BM cells expressed high levels of CCR2 but low levels of CD11c, in comparison to those derived from control BM cells, whereas no significant changes were observed in blood Ly6Chi and BM monocytes, as expected (Figure 4D, Figure 4—figure supplement 1B). These results collectively suggested that RBP-J regulated the expression of CCR2/CD11c in a cell-intrinsic manner.

Phenotypic markers in Ly6Clo monocytes are changed in RBP-J-deficient mice.
(A) Heatmap of RNA-seq dataset showing the top 20 downregulated and upregulated genes in blood Ly6Clo monocytes from Rbpjfl/flLyz2cre/cre versus Rbpj+/+Lyz2cre/cre control mice. Blue and red font indicates downregulated and upregulated genes in RBP-J-deficient Ly6Clo monocytes respectively. (B) Representative FACS plots, cumulative mean fluorescence intensity (MFI), and quantitative real-time PCR (qPCR) analysis of CCR2/CD11c expression in control and RBP-J-deficient blood monocyte subsets. Shaded curves represent isotype control, black lines represent control mice, and red lines represent RBP-J-deficient mice. (C) Principal component analysis (PCA) of indicated cell types. (D) Representative FACS plots and cumulative MFI of CCR2/CD11c expression in blood monocyte subsets derived from control or RBP-J-deficient mice. Black lines represent control mice, and red lines represent RBP-J-deficient mice. Data are pooled from two independent experiments (B, D); n ≥ 6 in each group. Data are shown as mean ± SEM; n.s., not significant; **p<0.01; ****p<0.0001 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse.
-
Figure 4—source data 1
Data of Figure 4.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig4-data1-v1.xlsx
RBP-J-mediated control of blood Ly6Clo monocytes is CCR2 dependent
Given that RBP-J deficiency led to enhanced CCR2 expression in Ly6Clo monocytes and that CCR2 is essential for monocyte functionality under various inflammatory and non-inflammatory conditions (Shi and Pamer, 2011), we wished to genetically investigate the role of CCR2 in the RBP-J-deficient background. We generated RBP-J/CCR2 double-deficient (DKO) mice with the genotype Rbpjfl/flLyz2cre/creCcr2RFP/RFP with Lyz2cre/creCcr2RFP/+ and Rbpjfl/flLyz2cre/creCcr2RFP/+ mice serving as controls. As expected, the expression of CCR2 in Ly6Clo monocytes was reduced in DKO mice compared to RBP-J-deficient mice (Figure 5A), confirming the successful deletion of the Ccr2 gene. DKO mice showed a lower percentage of both Ly6Chi and Ly6Clo monocytes than RBP-J-deficient mice (Figure 5B and C). Notably, the percentage of Ly6Clo monocytes in DKO mice was comparable to that observed in the control mice (Figure 5B and C), implicating that deletion of CCR2 corrected RBP-J deficiency-associated phenotype of increased Ly6Clo monocytes. Whereas, both RBP-J-deficient and DKO mice exhibited lower expression levels of CD11c in their Ly6Clo monocyte than control mice, suggesting that the regulation of CD11c expression was independent of CCR2 (Figure 5D). These results suggested that RBP-J regulated Ly6Clo monocytes, at least in part, through CCR2.

Blood Ly6Clo monocytes are decreased in double-deficient (DKO) mice.
(A) Representative FACS plots and cumulative mean fluorescence intensity (MFI) of CCR2 expression in Lyz2cre/creCcr2RFP/+ control, Rbpjfl/flLyz2cre/creCcr2RFP/+ and Rbpjfl/flLyz2cre/creCcr2RFP/RFP (DKO) blood Ly6Chi and Ly6Clo monocytes are shown. (B, C) Blood monocyte subsets in control, RBP-J-deficient, and DKO mice were determined by FACS. Representative FACS plots (B) and cumulative data of cell ratio (C) are shown. (D) Representative FACS plots and cumulative MFI of CD11c expression in control, RBP-J-deficient and DKO blood Ly6Chi and Ly6Clo monocytes are shown. Data are pooled from at least two independent experiments; n ≥ 5 in each group. Data are shown as mean ± SEM; n.s., not significant; **p<0.01; ***p<0.001; ****p<0.0001 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse.
-
Figure 5—source data 1
Data for Figure 5.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig5-data1-v1.xlsx
Lung Ly6Clo monocytes are accumulated in RBP-J-deficient mice
In mice, alveolar macrophages (AM) are maintained by local self-renewal and arise from fetal liver-derived precursors, whereas interstitial macrophages (IM) probably originate from monocytes (Guilliams et al., 2013; Hashimoto et al., 2013; Sabatel et al., 2017). Schyns et al. identified three subpopulations of IM according to expression of CD206 and CD16.2, and Ly6Clo monocytes are proposed to give rise to CD64+CD16.2+ IM (Schyns et al., 2019). We next compared the populations of lung monocytes and IM between control and RBP-J-deficient mice. The conditional deletion of RBP-J resulted in a significant increase in the absolute and relative numbers of Ly6Clo monocytes and CD16.2+ IM, while the numbers of Ly6Chi monocytes, CD206- IM, and CD206+ IM remained unchanged (Figure 6A and B). To confirm these findings, lung sections from Lyz2cre/creCx3cr1gfp/+ control and Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice were stained with an anti-GFP antibody, which revealed a significant increase in the number of GFP+ cells in RBP-J-deficient mice (Figure 6C). In addition, we evaluated the proliferative capacity of Ly6Clo monocytes and CD16.2+ IM, but did not observe enhanced EdU incorporation in Ly6Clo monocytes and CD16.2+ IM from RBP-J-deficient mice (Figure 6—figure supplement 1A and B). These findings suggested that the augmented population of these cells might not have resulted from increased in situ proliferation. Previous reports have indicated that lipopolysaccharide (LPS) can increase the population of IM in a CCR2-dependent manner (Sabatel et al., 2017). We thus challenged mice with LPS and analyzed monocytes and IM at days 0 and 4. LPS exposure induced an increase in numbers of Ly6Clo monocytes, CD16.2+ IM, and CD206- IM compared to baseline both in control and RBP-J-deficient mice (Figure 6D and E). There was a trend toward higher Ly6Chi monocyte after LPS treatment, although this observation was not statistically significant. Of note, RBP-J-deficient mice exhibited robustly elevated numbers of Ly6Clo monocytes and CD16.2+ IM compared with control mice after LPS treatment (Figure 6D and E), suggesting that RBP-J-deficient Ly6Clo monocytes were recruited to inflamed tissue in large numbers and differentiated into CD16.2+ IM.
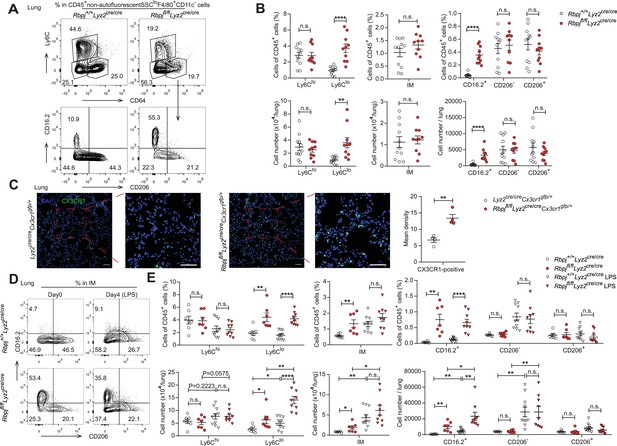
RBP-J-deficient mice exhibit more lung Ly6Clo monocytes and CD16.2+ interstitial macrophages (IM).
(A, B) indicate populations in the lungs of Rbpj+/+Lyz2cre/cre and Rbpjfl/flLyz2cre/cre mice were determined by FACS. Representative FACS plots (A) and cumulative data of cell ratio and absolute numbers (B) are shown. (C) Immunofluorescence staining for GFP+ cells in the lungs from Lyz2cre/creCx3cr1gfp/+ and Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice (CX3CR1 [green]; DAPI [blue]). Scale bars represent 50 µm. (D, E) Rbpj+/+Lyz2cre/cre and Rbpjfl/flLyz2cre/cre mice were instilled intranasally with phosphate buffered saline (PBS) or PBS containing lipopolysaccharide (LPS), and lungs were harvested at the indicated time points. Representative FACS plots (D) and cumulative data of cell ratio and absolute numbers (E) are shown. Data are pooled from at least two independent experiments; n ≥ 4 in each group. Data are shown as mean ± SEM; n.s., not significant; *p<0.05; **p<0.01; ****p<0.0001 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse.
-
Figure 6—source data 1
Data for Figure 6.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Data for Figure 6C.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig6-data2-v1.zip
To investigate whether the elevated levels of lung monocytes and CD16.2+ IM in RBP-J-deficient mice were derived from blood Ly6Clo monocytes, we examined monocytes and IM in Lyz2cre/creCcr2RFP/+ control, Lyz2cre/creCcr2RFP/RFP, Rbpjfl/flLyz2cre/creCcr2RFP/+ and Rbpjfl/flLyz2cre/creCcr2RFP/RFP (DKO) mice. The deletion of CCR2 led to a decrease in Ly6Chi monocytes, and the Ly6Clo monocytes in DKO mice were reduced to a level similar to that of control mice (Figure 7A and B). While the DKO mice had more CD16.2+ IM compared to control mice, these cells were severely reduced compared to RBP-J-deficient mice (Figure 7A and C). The above data supported the notion that increased lung Ly6Clo monocytes and CD16.2+ IM in RBP-J-deficient mice were derived from blood Ly6Clo monocytes. In summary, these results suggested that in RBP-J-deficient mice, recruitment of blood Ly6Clo monocytes to the lung was markedly facilitated by increased cell number as well as heightened expression of CCR2, leading to the increase of lung Ly6Clo monocytes and CD16.2+ IM.
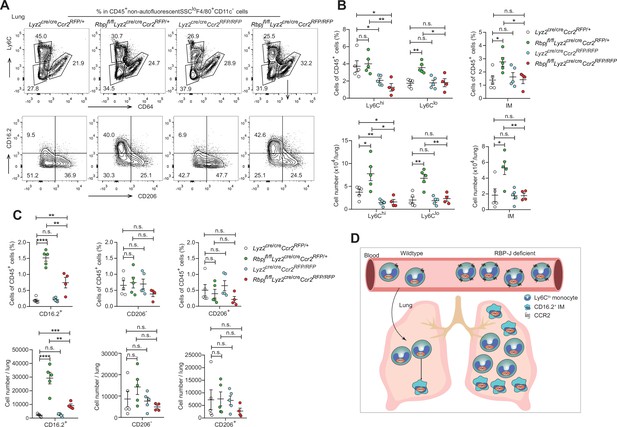
Double-deficient (DKO) mice lack lung Ly6Clo monocytes and CD16.2+ interstitial macrophages (IM).
(A) Representative FACS plots of lung monocyte and IM subsets in Lyz2cre/creCcr2RFP/+, Rbpjfl/flLyz2cre/creCcr2RFP/+, Lyz2cre/creCcr2RFP/RFP and Rbpjfl/flLyz2cre/creCcr2RFP/RFP mice. (B, C) Cumulative data of cell ratio and absolute numbers of monocyte (B) and IM (C) subsets. (D) Proposed model. RBP-J is a crucial regulator of blood Ly6Clo monocytes. Mice with conditional deletion of RBP-J in myeloid cells exhibit a marked increase in blood Ly6Clo monocytes, which highly express CCR2, and subsequently accumulate lung Ly6Clo monocytes and CD16.2+ IM. Data are pooled from two independent experiments; n ≥ 4 in each group. Data are shown as mean ± SEM; n.s., not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 (two-tailed Student’s unpaired t-test). Each symbol represents an individual mouse.
-
Figure 7—source data 1
Data for Figure 7.
- https://cdn.elifesciences.org/articles/88135/elife-88135-fig7-data1-v1.xlsx
Discussion
In this study, we identified RBP-J as a pivotal factor in regulating blood Ly6Clo monocyte cell fate. Our results showed that mice with conditional deletion of RBP-J in myeloid cells exhibited a robust increase in blood Ly6Clo monocytes and subsequently accumulated lung Ly6Clo monocytes and CD16.2+ IM under steady-state conditions (Figure 7D). BM transplantation experiments in which RBP-J-deficient cells were transplanted into recipient mice as well as the parabiosis experiment showed a similar increase in blood Ly6Clo monocytes, demonstrating that RBP-J was an intrinsic factor required for the regulation of Ly6Clo monocytes. Further analysis revealed that RBP-J regulated the expression of CCR2 and CD11c on the surface of Ly6Clo monocytes. Moreover, the phenotype of elevated blood Ly6Clo monocytes and their progeny was further ameliorated by deleting Ccr2 in an RBP-J-deficient background.
Notch-RBP-J signaling has been shown to play a role in regulating functional polarization and activation of macrophages, and regulated formation of Kupffer cells and macrophage differentiation from Ly6Chi monocytes in ischemia (Foldi et al., 2016; Hu et al., 2008; Kang et al., 2020; Krishnasamy et al., 2017; Sakai et al., 2019; Wang et al., 2010; Xu et al., 2012). At steady state, interaction of DLL1 with Notch 2 regulates conversion of Ly6Chi monocytes into Ly6Clo monocytes in special niches of the BM and spleen (Gamrekelashvili et al., 2016). Under inflammatory conditions, Notch2 and TLR7 pathways independently and synergistically promote conversion of Ly6Chi monocytes into Ly6Clo monocytes (Gamrekelashvili et al., 2020). However, in our study, the percentage of blood Ly6Clo monocytes increased in RBP-J-deficient mice. These results somewhat differ from what was observed in mice with conditional deletion of Notch2 (Gamrekelashvili et al., 2016). Actually, four members of the Notch family have been identified in mammals (Radtke et al., 2013). Thus, Notch receptors seem to be non-redundant in regulating monocyte cell fate and distinct Notch receptors are likely to act in a concerted manner to coordinate the monocyte differentiation program.
Colony-stimulating factor 1 receptor (CSF1R) signal, CX3CR1, CEBPβ, and Nr4a1 have been suggested to be involved in the generation and survival of Ly6Clo monocytes. The lifespan of Ly6Clo monocytes is acutely shortened after blockade of CSF1R, and the numbers of these cells are reduced, and the percentage of dead cells increased in mice with endothelial cell-specific depletion of Csf1 (Emoto et al., 2022; MacDonald et al., 2010). CX3CR1 and CEBPβ-knockout mice show accelerated death of Ly6Clo monocyte and display the decreased level of Ly6Clo monocytes (Landsman et al., 2009; Tamura et al., 2017). Nr4a1 knockout mice have significantly fewer monocytes in blood, BM, and spleen due to differentiation deficiency in MDP and accelerated apoptosis of Ly6Clo monocytes in BM (Hanna et al., 2011). RBP-J-deficient mice had normal MDP and Ly6Clo monocytes, expressed normal level of Nr4a1 and Ki-67, and exhibited normal half-life and turnover rate, which suggested that RBP-J may not regulate Ly6Clo monocytes through these factors. Further studies are required to elucidate the precise molecular mechanisms of RBP-J in Ly6Clo monocytes under steady state.
At the steady state, Ly6Clo monocytes patrol endothelium of blood vessel. After R848-induced endothelial injury, Ly6Clo monocytes recruit neutrophils to mediate focal endothelial necrosis and fuel vascular inflammation, and subsequently, the Ly6Clo monocytes remove cellular debris (Carlin et al., 2013; Imhof et al., 2016). In response to Listeria monocytogenes infection, Ly6Clo monocytes can extravasate but do not give rise to macrophages or DCs (Auffray et al., 2007). Additional evidence shows that Ly6Clo monocytes do not merely act as luminal blood macrophages. In lung, exposure to unmethylated CpG DNA expands CD16.2+ IM, which originate from Ly6Clo monocytes and spontaneously produce IL-10, thereby preventing allergic inflammation (Schyns et al., 2019). A recent study shows that Ly6Clo monocytes give rise to CD9+ macrophages, which provide an intracellular replication niche for Salmonella Typhimurium in the spleen, and Ly6Clo-depleted mice are more resistant to Salmonella Typhimurium infection, suggesting that Ly6Clo monocytes exert certain functions in systemic infection (Hoffman et al., 2021). Our data provide evidence for RBP-J in the function of blood Ly6Clo monocytes as RBP-J-deficient Ly6Clo monocytes exhibited enhanced competition in blood circulation, as well as gave rise to increased numbers of lung CD16.2+ IM. In summary, we showed that RBP-J acted as a fundamental regulator of the maintenance of blood Ly6Clo monocytes and their descendent, at least in part through regulation of CCR2. These results provide insights into understanding the mechanisms that regulate monocyte homeostasis and function.
Materials and methods
Mice
Cx3cr1gfp/gfp mice (JAX stock 005582) and Ccr2RFP/RFP mice (JAX stock 017586) were purchased from the Jackson Laboratory. Mice with a myeloid-specific deletion of the Rbpj were generated by crossing Rbpjfl/fl mice to Lyz2-Cre mice as described previously (Hu et al., 2008). Rbpjfl/flLyz2cre/cre were crossed to Cx3cr1gfp/gfp mice to obtain Lyz2cre/creCx3cr1gfp/+and Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice. Cx3cr1gfp/+ mice were obtained by crossing Cx3cr1gfp/gfp with C57/BL6 CD45.1+ mice. Rbpjfl/flLyz2cre/cre mice were crossed with Ccr2RFP/RFP mice to obtain Lyz2cre/creCcr2RFP/+, Lyz2cre/creCcr2RFP/RFP, Rbpjfl/flLyz2cre/creCcr2RFP/+ and Rbpjfl/flLyz2cre/creCcr2RFP/RFP mice. All mice were maintained under specific pathogen-free conditions. All animal experimental protocols were approved by the Institutional Animal Care and Use Committees of Tsinghua University (17-hxy). Gender- and age-matched mice were used at 7–12 weeks old for experiments.
Quantitative RT-PCR
Request a detailed protocolBlood monocytes were sorted by flow cytometry. The total RNA was extracted using total RNA purification kit (GeneMarkbio) and reversely transcribed to cDNA by M-MLV Reverse Transcriptase (Takara). qPCR was performed on a real-time PCR system (StepOnePlus; Applied Biosystems) using FastSYBR mixture (CWBIO). Gapdh messenger RNA was used as internal control to normalize the expression of target genes. Primer sequences are provided in Table 1.
Primers sequences for regular quantitative real-time PCR (qPCR) used in this study.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Gapdh | ATCAAGAAGGTGGTGAAGCA | AGACAACCTGGTCCTCAGTGT |
| Rbpj | ACCCCTGTGCCTGTCGTAGAA | TCCCGGAATGCAGAAATGTC |
| Nr4a1 | TTGAGCTTGAATACAGGGCA | AGTTGGGGGAGTGTGCTAGA |
| Ccr2 | CCTTGGGAATGAGTAACTGTGTGAT | ATGGAGAGATACCTTCGGAACTTCT |
RNA-seq analysis
Request a detailed protocolLy6Chi and Ly6Clo blood monocytes were isolated from Rbpj+/+Lyz2cre/cre and Rbpjfl/flLyz2cre/cre mice, and total RNA was extracted using total RNA purification kit (GeneMarkbio). RNA was converted into RNA-seq libraries, which were sequenced with the pair-end option using an Illumina-HiSeq2500 platform at Beijing Genomics Institute (BGI), China. The significantly downregulated genes were identified with p-value<0.05 and (FPKM + 1) fold changes ≤ 0.2, and significantly upregulated genes were identified with p-value <0.05 and (FPKM + 1) fold changes ≥ 5.7. The RNA-seq data are deposited in Gene Expression Omnibus under accession number GSE208772.
Annexin V staining
Request a detailed protocolAnnexin V and 7-AAD were used for identification of apoptotic monocytes by flow cytometry. Blood cells were stained with annexin V (eBioscience) and 7-AAD (BioLegend) according to the manufacturer’s protocols.
EdU pulsing and latex beads labeling
Request a detailed protocolMice were injected intravenously with a single 1 mg EdU (Thermo Scientific). BM and blood cells were collected and stained with fluorescence-conjugated mAb against CD45, CD11b, Ly6G, CD115, and Ly6C. Cells were then fixed, permeabilized, and stained with reaction cocktail using EdU Assay Kit. Labeled cells were analyzed by flow cytometry.
Mice were injected intravenously with a single 10 µl latex beads (Polysciences) in 250 µl phosphate buffered saline (PBS). Blood cells were harvested at indicated time. Cells were then stained for CD45, CD11b, Ly6G, CD115, Ly6C, and analyzed by flow cytometry.
In vivo labeling of sinusoidal leukocytes
Request a detailed protocolMice were injected intravenously with 1 μg of PE-conjugated anti-CD45 antibodies. Two minutes after antibody injection, mice were sacrificed, and BM were harvested. BM cells were then stained and analyzed by flow cytometry.
Generation of BM chimera
Request a detailed protocolC57/BL6 CD45.2+ mice were lethally irradiated in two doses of 5.5 Gy 2 hr apart. 0.8 × 105 BM cells from Rbpj+/+Lyz2cre/cre mice (CD45.2+) or Rbpjfl/flLyz2cre/cre mice (CD45.2+) were mixed with 3.2 × 105 BM cells from Cx3cr1gfp/+ mice (CD45.1+), and injected intravenously into recipient mice. Mice were used for experiments 8 wk after irradiation.
Adoptive transfers
Request a detailed protocolBM GFP+Ly6Chi monocytes were sorted from Lyz2cre/creCx3cr1gfp/+ or Rbpjfl/flLyz2cre/creCx3cr1gfp/+ mice, and transferred to Ccr2RFP/RFP mice. Blood cells were collected 60 hr later for flow cytometry analyses.
Parabiosis
Request a detailed protocolCx3cr1gfp/+ CD45.1+ mice were surgically joined with age-matched female Rbpjfl/flLyz2cre/cre or Rbpj+/+Lyz2cre/cre CD45.2+ mice at the age of 6 wk. Bloods were obtained via cardiac puncture, and cell populations were analyzed by flow cytometry at 4 wk after the surgery.
Intranasal instillations of LPS
Request a detailed protocolRbpj+/+Lyz2cre/cre or Rbpjfl/flLyz2cre/cre mice were anesthetized with isoflurane and intranasally instilled with 10 μg LPS in 25 μl of PBS. Lungs were harvested 4 d later for flow cytometry analyses.
Immunofluorescence histology
Request a detailed protocolLungs from Rbpjfl/flLyz2cre/creCx3cr1gfp/+ and Lyz2cre/creCx3cr1gfp/+ mice were fixed in 1% paraformaldehyde and incubated in 30% sucrose separately overnight at 4℃. The samples were then incubated in the mixture of 30% sucrose and OCT compound (Sakura Finetek) overnight at 4℃. The tissues were embedded and frozen in OCT compound and then cut at 10 µm thickness. Tissue sections were dried for 10 min at 50℃ and then fixed in 1% paraformaldehyde at room temperature for 10 min, permeabilized in PBS/0.5% Triton X-100/0.3 M glycine for 10 min, and blocked in PBS/5% goat serum for 1 hr at room temperature. Sections were stained with rabbit anti-GFP antibodies (1:200; Proteintech) overnight at 4℃ and washed with PBS/0.1% Tween-20 for 30 min three times at room temperature. Sections were then incubated with AF488-conjugated goat anti-rabbit antibodies (1:1000; Cell Signaling Technology) for 2 hr at room temperature and washed with PBS/0.1% Tween-20 for 30 min three times at room temperature. Sections were stained with DAPI (Solarbio) for 7 min, washed in PBS for 8 min two times at room temperature, and mounted with SlowFade Diamond Antifade Mountant (Life Technologies).
Cell isolation and flow cytometry
Request a detailed protocolPeripheral blood (PB) was sampled by eyeball extirpating, spleens were mashed through a 70 μm strainer, and BM cells were collected from femurs. Lungs were perfused with 5 ml of HBSS (MACGENE) through the right ventricle, excised, and digested in HBSS containing 5% FBS, 1 mg/ml collagenase type I (Sigma-Aldrich) and 0.05 mg/ml DNase I (Sigma-Aldrich) for 1 hr at 37°C. The digested tissues were homogenized by shaking, passed through a 70 µm cell strainer to create a single-cell suspension. The suspension was enriched in mononuclear cells and harvested from the 1.080:1.038 g/ml interface using a density gradient (Percoll from GE Healthcare). BM, spleen, PB, and lung cells were stained with fluorescently conjugated antibodies. The absolute number of cells was counted by using CountBright Absolute Counting Beads (Invitrogen).
Antibodies against CD45 (30-F11), Ly6C (HK1.4), CD64 (X54-5/7.1), CD206 (C068C2), CD16.2 (9E9), CD45.1 (A20), F4/80 (BM8), Biotin, CD19 (6D5), CCR2 (SA203G11), and CD3ε (145-2C11) were purchased from BioLegend. Antibodies against Ly6G (1A8-Ly6g), CD11b (M1/70), CD115 (AFS98), CD117 (2B8), CD135 (A2F10), CD11c (N418), CD206 (MR6F3), CD45.2 (104), Nur77 (12.14), Ki-67 (SolA15), CD4 (RM4-5), and lineage maker were purchased from eBioscience. Fluorescence-conjugated mAb against Ly6C (AL-21) were purchased from BD Biosciences. Isotype-matched antibodies (eBioscience) were used for control staining. All antibodies were used in 1:400 dilutions, and surface antigens were stained on ice for 30 min.
For intracellular staining, cells were stained with antibodies to surface antigens, fixed and permeabilized with the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences), and stained with antibodies or isotype control diluted in permeabilization buffer separately for 30 min at room temperature.
Cells were analyzed on FACSFortessa or FACSAria III flow cytometer (BD Biosciences) using FlowJo software.
Statistical analysis
Request a detailed protocolStatistical analysis was performed using GraphPad Prism software. All results are shown as mean ± SEM. Statistical significance was determined using Student’s unpaired t-test. Statistical significance was defined as p<0.05.
Appendix 1
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Cx3cr1gfp/gfp | Jackson Laboratory | Strain #: 005582 from Jackson Laboratory | |
| Strain, strain background (M. musculus) | Ccr2RFP/RFP | Jackson Laboratory | Strain #: 017586 from Jackson Laboratory | |
| Strain, strain background (M. musculus) | Rbpjfl/fl | Tasuku Honjo of Kyoto University | ||
| Strain, strain background (M. musculus) | Lyz2-Cre | Jackson Laboratory | Strain #: 004781 from Jackson Laboratory | |
| Strain, strain background (M. musculus) | C57BL6/J | Jackson Laboratory | Strain #: 000664 from Jackson Laboratory | |
| Strain, strain background (M. musculus) | CD45.1 | Jackson Laboratory | Strain #:002014 from Jackson Laboratory | |
| Antibody | APC/Cy7 anti-mouse CD45 antibody | BioLegend | 103116 | 1:400 |
| Antibody | PE anti-mouse CD45 | BioLegend | 103106 | 1:400 |
| Antibody | Brilliant Violet 510 anti-mouse CD45 antibody | BioLegend | 103137 | 1:400 |
| Antibody | Alexa Fluor 700 anti-mouse Ly-6C | BioLegend | 128024 | 1:400 |
| Antibody | PE anti-mouse Ly-6C | BD Biosciences | 560592 | 1:400 |
| Antibody | CD4 monoclonal antibody, PerCP-Cyanine5.5 | eBioscience | 45-0042-82 | 1:400 |
| Antibody | PE anti-mouse CD3ε antibody | BioLegend | 100307 | 1:400 |
| Antibody | BV605 anti-mouse CD19 antibody | BioLegend | 115539 | 1:400 |
| Antibody | BV421 anti-mouse CD16.2 antibody | BioLegend | 149521 | 1:400 |
| Antibody | CD11c monoclonal antibody, PE-Cyanine7 | eBioscience | 25-0114-82 | 1:400 |
| Antibody | CD11c monoclonal antibody, PerCP-Cyanine5.5 | eBioscience | 45-0114-82 | 1:400 |
| Antibody | Ly-6G monoclonal antibody, APC | eBioscience | 17-9668-82 | 1:400 |
| Antibody | Ly-6G monoclonal antibody, PerCP-eFluor 710 | eBioscience | 46-9668-82 | 1:400 |
| Antibody | CD11b monoclonal antibody, PerCP-Cyanine5.5 | eBioscience | 45-0112-82 | 1:400 |
| Antibody | CD11b monoclonal antibody, PE-Cyanine7 | eBioscience | 25-0112-82 | 1:400 |
| Antibody | CD117 (c-Kit) monoclonal antibody, APC | eBioscience | 17-1171-82 | 1:400 |
| Antibody | CD135 (Flt3) monoclonal antibody, APC, | eBioscience | 17-1351-82 | 1:400 |
| Antibody | CD135 (Flt3) monoclonal antibody, PE | eBioscience | 12-1351-82 | 1:400 |
| Antibody | Brilliant Violet 421 anti-mouse CD192 (CCR2) antibody | BioLegend | 150605 | 1:400 |
| Antibody | Mouse Hematopoietic Lineage Antibody Cocktail, eFluor 450 | eBioscience | 88-7772-72 | 1:400 |
| Antibody | Brilliant Violet 421 anti-mouse CD45.1 antibody | BioLegend | 110732 | 1:400 |
| Antibody | CD45.2 monoclonal antibody, PE-Cyanine7 | eBioscience | 25-0454-80 | 1:400 |
| Antibody | FITC anti-mouse F4/80 antibody | BioLegend | 123107 | 1:400 |
| Antibody | APC/Cyanine7 anti-mouse F4/80 antibody | BioLegend | 123117 | 1:400 |
| Antibody | APC anti-mouse CD64 antibody | BioLegend | 139306 | 1:400 |
| Antibody | CD206 monoclonal antibody, PE | eBioscience | 12-2061-82 | 1:400 |
| Antibody | PerCP/Cyanine5.5 anti-mouse CD206 antibody | BioLegend | 141715 | 1:400 |
| Antibody | Nur77 monoclonal antibody, PerCP-eFluor 710 | eBioscience | 46-5965-82 | 1:400 |
| Antibody | Ki-67 monoclonal antibody, APC | eBioscience | 17-5698-82 | 1:400 |
| Antibody | CD115 monoclonal antibody, Biotin | eBioscience | 13-1152-85 | 1:400 |
| Antibody | Brilliant Violet 605 Streptavidin | BioLegend | 405229 | 1:400 |
| Antibody | Rabbit anti-GFP antibody | Proteintech | 50430-2-AP | 1:200 |
| Antibody | Goat anti-rabbit Alexa Fluor 488 | Cell Signaling Technology | 4412S | 1:1000 |
| Chemical compound, drug | Phosphate buffered saline (PBS) | Gibco | C10010500BT | |
| Chemical compound, drug | CountBright Absolute Counting Beads | Invitrogen | C36950 | |
| Chemical compound, drug | DAPI | Solarbio | C0060-1ml | |
| Chemical compound, drug | HBSS | MACGENE | CC016.1 | |
| Chemical compound, drug | FBS | Gibco | 16000-044 | |
| Chemical compound, drug | Collagenase type I | Sigma-Aldrich | C0130-500MG | |
| Chemical compound, drug | DNase I | Sigma-Aldrich | 10104159001 | |
| Chemical compound, drug | Percoll | GE Healthcare | 17-0891-01 | |
| Chemical compound, drug | SlowFade Diamond Antifade Mountant | Life Technologies | S36972 | |
| Chemical compound, drug | Tween-20 | Amresco | 0777-1L | |
| Chemical compound, drug | Paraformaldehyde | Sigma-Aldrich | 158127-500G | |
| Chemical compound, drug | OCT | Sakura Finetek | 4583 | |
| Chemical compound, drug | Triton X-100 | Merck Millipore | 648466 | |
| Chemical compound, drug | Glycine | Amresco | 0167-5kg | |
| Chemical compound, drug | Lipopolysaccharide (LPS) | Sigma-Aldrich | L2630 | |
| Commercial assay or kit | Cytofix/Cytoperm Fixation/Permeabilization Solution Kit | BD Biosciences | 554715 | |
| Commercial assay or kit | Click-iT EdU AF488 Flow Cytometry Assay Kit | Invitrogen | C10425 | |
| Commercial assay or kit | Fluoresbrite Polychromatic Red Microspheres | Polysciences | 19507-5 | |
| Commercial assay or kit | 7-AAD Viability Staining Solution | BioLegend | 420403 | |
| Commercial assay or kit | Total RNA Purification Kit | GeneMarkbio | TR01-150 | |
| Commercial assay or kit | FastSYBR mixture | CWBIO | CW2622M | |
| Commercial assay or kit | Reverse Transcriptase M-MLV | Takara | 2641B | |
| Commercial assay or kit | Annexin V Apoptosis Detection Kit APC | eBioscience | 88-8007-72 | |
| Software, algorithm | FlowJo | FlowJo | RRID:SCR_008520 | |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 |
Data availability
Sequencing data have been deposited in GEO under accession code GSE208772.
-
NCBI Gene Expression OmnibusID GSE208772. RNA-seq for blood Ly6Chi and Ly6Clo monocyte in WT and RBP-Jfl/fl Lyz2-Cre mice.
References
-
NR4A1-dependent Ly6Clow monocytes contribute to reducing joint inflammation in arthritic mice through Treg cellsEuropean Journal of Immunology 46:2789–2800.https://doi.org/10.1002/eji.201646406
-
Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleenThe Journal of Experimental Medicine 204:1653–1664.https://doi.org/10.1084/jem.20062648
-
S1PR5 is pivotal for the homeostasis of patrolling monocytesEuropean Journal of Immunology 43:1667–1675.https://doi.org/10.1002/eji.201343312
-
Regulation of monocyte cell fate by blood vessels mediated by Notch signallingNature Communications 7:12597.https://doi.org/10.1038/ncomms12597
-
Monocytes and macrophages: developmental pathways and tissue homeostasisNature Reviews. Immunology 14:392–404.https://doi.org/10.1038/nri3671
-
Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSFThe Journal of Experimental Medicine 210:1977–1992.https://doi.org/10.1084/jem.20131199
-
Bone marrow-specific deficiency of nuclear receptor Nur77 enhances atherosclerosisCirculation Research 110:428–438.https://doi.org/10.1161/CIRCRESAHA.111.260760
-
Origin of monocytes and macrophages in a committed progenitorNature Immunology 14:821–830.https://doi.org/10.1038/ni.2638
-
The colonic macrophage transcription factor RBP-J orchestrates intestinal immunity against bacterial pathogensThe Journal of Experimental Medicine 217:e20190762.https://doi.org/10.1084/jem.20190762
-
Origin of dendritic cells in peripheral lymphoid organs of miceNature Immunology 8:578–583.https://doi.org/10.1038/ni1462
-
The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functionsThe Journal of Experimental Medicine 204:3037–3047.https://doi.org/10.1084/jem.20070885
-
Identification and characterization of a novel monocyte subpopulation in human peripheral bloodBlood 74:2527–2534.
-
Non-classical monocytes as mediators of tissue destruction in arthritisAnnals of the Rheumatic Diseases 77:1490–1497.https://doi.org/10.1136/annrheumdis-2018-213250
-
Regulation of innate and adaptive immunity by NotchNature Reviews. Immunology 13:427–437.https://doi.org/10.1038/nri3445
-
Monocyte recruitment during infection and inflammationNature Reviews. Immunology 11:762–774.https://doi.org/10.1038/nri3070
-
Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the peripheryThe Journal of Experimental Medicine 203:583–597.https://doi.org/10.1084/jem.20052119
-
Monocytes give rise to mucosal, but not splenic, conventional dendritic cellsThe Journal of Experimental Medicine 204:171–180.https://doi.org/10.1084/jem.20061011
-
Differential chemokine receptor expression and function in human monocyte subpopulationsJournal of Leukocyte Biology 67:699–704.https://doi.org/10.1002/jlb.67.5.699
-
TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-JThe Journal of Experimental Medicine 209:319–334.https://doi.org/10.1084/jem.20111566
Article and author information
Author details
Funding
National Natural Science Foundation of China (31821003)
- Xiaoyu Hu
National Natural Science Foundation of China (31991174)
- Xiaoyu Hu
National Natural Science Foundation of China (32030037)
- Xiaoyu Hu
National Natural Science Foundation of China (82150105)
- Xiaoyu Hu
Ministry of Science and Technology of the People's Republic of China (2020YFA0509100)
- Xiaoyu Hu
Center for Life Sciences
- Xiaoyu Hu
Tsinghua University
- Xiaoyu Hu
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank the core facility at Institute for Immunology, Tsinghua University, for valuable technical assistance.
Ethics
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocols of the Tsinghua University. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Tsinghua University (17-hxy). All surgery was performed under sodium pentobarbital or avertin anesthesia, and every effort was made to minimize suffering.
Version history
- Sent for peer review: April 13, 2023
- Preprint posted: April 14, 2023 (view preprint)
- Preprint posted: June 7, 2023 (view preprint)
- Preprint posted: February 13, 2024 (view preprint)
- Version of Record published: February 26, 2024 (version 1)
Cite all versions
You can cite all versions using the DOI https://doi.org/10.7554/eLife.88135. This DOI represents all versions, and will always resolve to the latest one.
Copyright
© 2023, Kou, Kang et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 477
- views
-
- 67
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Immunology and Inflammation
SARS-CoV-2 vaccines have been used worldwide to combat COVID-19 pandemic. To elucidate the factors that determine the longevity of spike (S)-specific antibodies, we traced the characteristics of S-specific T cell clonotypes together with their epitopes and anti-S antibody titers before and after BNT162b2 vaccination over time. T cell receptor (TCR) αβ sequences and mRNA expression of the S-responded T cells were investigated using single-cell TCR- and RNA-sequencing. Highly expanded 199 TCR clonotypes upon stimulation with S peptide pools were reconstituted into a reporter T cell line for the determination of epitopes and restricting HLAs. Among them, we could determine 78 S epitopes, most of which were conserved in variants of concern (VOCs). After the 2nd vaccination, T cell clonotypes highly responsive to recall S stimulation were polarized to follicular helper T (Tfh)-like cells in donors exhibiting sustained anti-S antibody titers (designated as ‘sustainers’), but not in ‘decliners’. Even before vaccination, S-reactive CD4+ T cell clonotypes did exist, most of which cross-reacted with environmental or symbiotic microbes. However, these clonotypes contracted after vaccination. Conversely, S-reactive clonotypes dominated after vaccination were undetectable in pre-vaccinated T cell pool, suggesting that highly responding S-reactive T cells were established by vaccination from rare clonotypes. These results suggest that de novo acquisition of memory Tfh-like cells upon vaccination may contribute to the longevity of anti-S antibody titers.
-
- Chromosomes and Gene Expression
- Immunology and Inflammation
Ikaros is a transcriptional factor required for conventional T cell development, differentiation, and anergy. While the related factors Helios and Eos have defined roles in regulatory T cells (Treg), a role for Ikaros has not been established. To determine the function of Ikaros in the Treg lineage, we generated mice with Treg-specific deletion of the Ikaros gene (Ikzf1). We find that Ikaros cooperates with Foxp3 to establish a major portion of the Treg epigenome and transcriptome. Ikaros-deficient Treg exhibit Th1-like gene expression with abnormal production of IL-2, IFNg, TNFa, and factors involved in Wnt and Notch signaling. While Ikzf1-Treg-cko mice do not develop spontaneous autoimmunity, Ikaros-deficient Treg are unable to control conventional T cell-mediated immune pathology in response to TCR and inflammatory stimuli in models of IBD and organ transplantation. These studies establish Ikaros as a core factor required in Treg for tolerance and the control of inflammatory immune responses.






