Mechanobiology: A brighter force gauge for cells
Under a microscope, cells seem static, but in reality they are constantly pulling and pushing with miniscule forces. Despite being very small (about one millionth the weight of a grain of rice), these forces are important for a number of processes, including wound healing and the immune response. They can also regulate the cell’s fate during development (Mammoto et al., 2013).
To better understand the crosstalk between force and biochemistry in cells, the research community has developed smaller and smaller probes to measure forces (Helenius et al., 2008; Liu et al., 2017). A major breakthrough, reported just under a decade ago, was the development of a genetically encoded force gauge (Grashoff et al., 2010). This biosensor worked like a macroscopic tension gauge in that it contained a ‘spring’ (that stretched when pulled) and a ‘ruler’ (to measure how much the spring extended).
The spring element in the biosensor was adopted from a segment of spider silk and included a 40-amino acid polypeptide chain that formed a random coil. To measure how much it extended under force, fluorescent proteins were engineered at each end of the polypeptide. This pair of proteins was carefully chosen such that energy released after exciting one (the ‘donor’) with a light source was transferred to the other (the ‘acceptor’), causing it to emit light of a different wavelength. This phenomenon, named Förster resonance energy transfer (FRET), only occurs if the proteins are close enough, and it decreases when they move apart. As such, the FRET signal essentially represents the ruler that measures the length of the polypeptide coil.
This tension sensor module, or TSMod for short, was transformative and opened the door to mapping the forces experienced by a number of different mechanosensitive proteins, both in vitro and in vivo (Cost et al., 2015). Yet it was challenging to use, mostly because it lacked sensitivity (Eder et al., 2017). Part of the problem was that the FRET signal was weak, even when the proteins were close to each other. It was also made even weaker because it was concealed by the background glow from other parts of the cell that naturally fluoresce over similar wavelengths (e.g. mitochondria and lysosomes).
Now, in eLife, Andrew LaCroix, Andrew Lynch, Matthew Berginski and Brenton Hoffman of Duke University report how they completely re-engineered the probe to improve its performance (LaCroix et al., 2018). LaCroix et al. first systematically tested different pairs of fluorescent proteins, and whittled away the ‘linker’ region between the fluorescent proteins and the spring element (Austen et al., 2013). They also identified a ‘softer’ and less structured polypeptide spring (Evers et al., 2006), which further maximized the FRET signal (Figure 1).
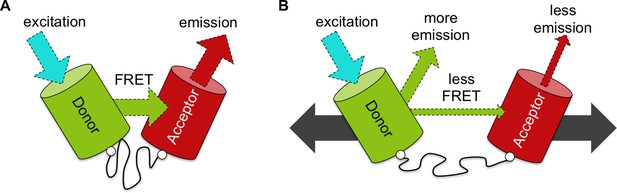
An improved biosensor to visualize tension in mechanosensitive proteins.
(A) Like the original, the optimized tension sensor module contains a spring element (wavy line) attached to two fluorescent proteins (colored cylinders) via linker regions (white circles). However, while the original used cyan and yellow fluorescent proteins, the new version uses a green-red pair. Specifically, a green fluorescent protein called Clover acts as the ‘donor’ (green cylinder), and a red fluorescent protein called mRuby2 acts as the ‘acceptor’ (red cylinder). Excitation of the donor with cyan light causes it to give off a bright green light. If the donor is close enough to the acceptor – for example, because the spring is at rest – the fluorescence from the donor can be transferred to the acceptor via a process called FRET (see main text): the acceptor then emits red light. Dashed arrows of different colors indicate light of different wavelengths. (B) Via genetic engineering, this sensor module can be inserted within proteins of interest. If that protein is put under tension (gray solid arrows), the donor and acceptor proteins are pulled apart. This causes the amount of energy transferred by FRET to decrease, increasing emission of green light from the donor and reducing emission of red light from the acceptor. As such the ratio of emission at these two wavelengths provides a measure of how much the spring is extended, which gives an indication of the forces experienced within the protein of interest.
The optimized TSMod outperforms the original when tested in buffer. However, the gains in performance were lost when the new TSMod was tested in cells. This is an important warning to all researchers developing probes that are dedicated to measuring the forces acting on real cells but are calibrated away from real cells. Nonetheless, based on the data, LaCroix et al. developed a computational model that predicts how the spring element would behave inside cells. Using this model, they then identified the optimal peptide length to measure forces in vinculin, an important force-sensitive protein that is often used to evaluate the performance of this kind of biosensor (Schwartz, 2010).
With the aid of the computational modeling, the optimized TSMod mapped vinculin tension within cells much better than its predecessors. As a result of this improved performance, LaCroix et al. found a tension gradient across vinculin molecules within focal adhesions – the microstructures that anchor cells to their external environment. This asymmetry, which suggests that tension can be transmitted unevenly across the focal adhesion, was not detectable using with the original TSMod. Furthermore, using three optimized TSMods with springs of different lengths, LaCroix et al. showed that level of tension experienced by each sensor was different, but that all three sensors extended to approximately the same length. This result is intriguing because it suggests that focal adhesion formation and cell spreading may be governed by the physical extension of adaptor proteins (i.e., by how long they are), rather than the absolute magnitude of the forces they transmit (i.e. the level of tension they experience).
Another important result comes from systematic simulations that provide a ‘cheat-sheet’ to guide cell biologists in using the most optimal tension probe for a desired force range and range of light wavelengths. Those familiar with the original TSMod know that it often required extensive trial and error, and this road map will make the process more rational and predictable.
Moving forward, it remains to be seen whether other adaptor proteins within the focal adhesion complex, or other mechanosensitive molecules in general, get extended in the way that vinculin does. Nonetheless, the work by LaCroix et al. should motivate other researchers to look at other systems and processes that involve the transmission of forces (such as T cell receptors and the Notch-Delta signaling pathway; Liu et al., 2016; Luca et al., 2017). This finding could be the proverbial tip-of-the-iceberg for new discoveries in mechanobiology research.
References
-
Generation and analysis of biosensors to measure mechanical forces within cellsMethods in Molecular Biology 1066:169–184.https://doi.org/10.1007/978-1-62703-604-7_15
-
How to measure molecular forces in cells: a guide to evaluating genetically-encoded FRET-based tension sensorsCellular and Molecular Bioengineering 8:96–105.https://doi.org/10.1007/s12195-014-0368-1
-
Single-cell force spectroscopyJournal of Cell Science 121:1785–1791.https://doi.org/10.1242/jcs.030999
-
Molecular tension probes for imaging forces at the cell surfaceAccounts of Chemical Research 50:2915–2924.https://doi.org/10.1021/acs.accounts.7b00305
-
Mechanobiology and developmental controlAnnual Review of Cell and Developmental Biology 29:27–61.https://doi.org/10.1146/annurev-cellbio-101512-122340
-
Integrins and extracellular matrix in mechanotransductionCold Spring Harbor Perspectives in Biology 2:a005066.https://doi.org/10.1101/cshperspect.a005066
Article and author information
Author details
Publication history
- Version of Record published: July 19, 2018 (version 1)
Copyright
© 2018, Ma et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,495
- views
-
- 203
- downloads
-
- 3
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Structural Biology and Molecular Biophysics
Mutations in the human PURA gene cause the neurodevelopmental PURA syndrome. In contrast to several other monogenetic disorders, almost all reported mutations in this nucleic acid-binding protein result in the full disease penetrance. In this study, we observed that patient mutations across PURA impair its previously reported co-localization with processing bodies. These mutations either destroyed the folding integrity, RNA binding, or dimerization of PURA. We also solved the crystal structures of the N- and C-terminal PUR domains of human PURA and combined them with molecular dynamics simulations and nuclear magnetic resonance measurements. The observed unusually high dynamics and structural promiscuity of PURA indicated that this protein is particularly susceptible to mutations impairing its structural integrity. It offers an explanation why even conservative mutations across PURA result in the full penetrance of symptoms in patients with PURA syndrome.
-
- Cell Biology
Endogenous tags have become invaluable tools to visualize and study native proteins in live cells. However, generating human cell lines carrying endogenous tags is difficult due to the low efficiency of homology-directed repair. Recently, an engineered split mNeonGreen protein was used to generate a large-scale endogenous tag library in HEK293 cells. Using split mNeonGreen for large-scale endogenous tagging in human iPSCs would open the door to studying protein function in healthy cells and across differentiated cell types. We engineered an iPS cell line to express the large fragment of the split mNeonGreen protein (mNG21-10) and showed that it enables fast and efficient endogenous tagging of proteins with the short fragment (mNG211). We also demonstrate that neural network-based image restoration enables live imaging studies of highly dynamic cellular processes such as cytokinesis in iPSCs. This work represents the first step towards a genome-wide endogenous tag library in human stem cells.






