Abstract
Background:
The number of cases of HBV infection reported by the WHO for each district and country is positively correlated with the number of HBV sequences in the database isolated from the corresponding district and country.Objectives:
This study determined distribution characteristics of HBV genotypes and subtypes in 14 countries neighboring China. The progress made in genomic research involving HBV was also reviewed.Materials and Methods:
Nine hundred fifty-one complete genome sequences of HBV from 14 countries neighboring China were selected from NCBI. The sequence-related information was analyzed and recorded. One hundred seventy-two sequences of HBV genotype B were screened for alignment using DNA star and MEGA 5.1.Results:
Dominant HBV genotypes in the countries neighboring China were genotypes B, C and D and dominant subtypes were adw2 and adrq+. The association between genotype and serotype of HBV in these countries was shown to differ from previous research results. As shown by sequence alignment, the sequence divergence between five subgenotypes (B3, B5, B7, B8 and B9) was below 4%. The B subgenotypes shared six common specific amino acid sites in the S region.Conclusions:
The B3, B5, B7, B8 and B9 subgenotypes can be clustered into quasi-sub-genotype B3 and the open reading frame of HBV has a start codon preference; however, whether a mutation in the start codon in the pre-S2 region has an impact on survival and replication of HBV remains to be determined.Keywords
1. Background
Hepatitis B virus (HBV) infection poses an enormous threat to public health. According to the report of the World Health Organization (WHO), More than 240 million people have chronic (long-term) liver infections. More than 780000 people die annually due to acute or chronic consequences of hepatitis B (1). There is about 55% cancer risk caused by HBV (2).
Complete genome sequence of HBV was first determined by Galibert et al. (3). in 1979. Since then, more and more research has been devoted to sequencing the genomes of different HBV. Japanese scholars cloned three HBV belonging to subtype adw isolate from the sera of chronic and asymptomatic HBV carriers (AsC) and a 3.9% - 5.6% divergence of DNA sequences was demonstrated between the three clones. Compared with the two adw serotypes isolated in the US, the divergence was 8.3% - 9.3%. According to the sequences of 18 clones already obtained at that time, HBV clones were divided into four genotypes (A, B, C, and D), defined by the divergence of ≥ 8% in complete genome sequence (4). As more complete genome sequences of various clones of HBV were described, genotypes E, F, G and H were identified using phylogenetic analysis. Ten genotypes and several subtypes of HBV have been recognized to date (5). Each genotype was broken down into several subgenotypes. Thus far, several recombinant genotypes have been discovered. HBV infection prevalence differs worldwide with a significant regional variation (6). By 1 August 2013, there were 75373 sequences related to HBV in the NCBI, 5066 of which were complete genome sequences. These sequences were from wild-type clones, natural mutants, drug-induced dominant mutants with immune tolerance and drug resistance, as well as cloned sequences from quasi-species in patients with different diseases worldwide. These sequences are highly valuable to investigate polymorphisms, genotype characteristics, epidemiologic trends and phylogenetic analysis of HBV (7). HBV infection was highly prevalent in Asia and moderately prevalent in India and Russia (8). The largest number of complete genome sequences of HBV in the nucleic acid database is from mainland China, accounting for about one-fourth of the total. Hong Kong and Japan were in the second and third places, followed by Africa, Europe and the US.
2. Objectives
The aim of this study was to assess distribution and epidemiologic trends of HBV genotypes and subtypes in countries neighboring China based on genome analysis to promote clinical practice results.
3. Materials and Methods
3.1. Data Collection
A total of 5066 complete genome sequences of HBV were screened from the NCBI. Of these sequences, 952 were from the clones of HBV isolated in 14 countries neighboring China (Japan, Vietnam, India, Burma, Thailand, Laos, Cambodia, Indonesia, the Philippines, North Korea, South Korea, Mongolia, Malaysia and Russia). Genotyping was performed for these sequences online using the NCBI analysis tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi). The sequences were divided into different serotypes by the amino acids at positions 122, 160, 127, 134, 159, 177 and 178 in the S region of HBV (9). Preliminary analysis of the sequences in this district suggested that the distance among B subgenotyes (B3,B5,B7,B8,B9) was less than 4%, so we selected genotype B for further analysis. S, C, X and P open reading frames of 172 complete genotype B sequences (colonial sequences and incomplete sequences were excluded) from Japan (20), Indonesia (58), Thailand (15), Vietnam (16), Philippine (6) Cambodia (2) and Malaysia (55) were chosen for nucleotide sequence analysis, respectively (10). (Genotype B was only seen in the seven countries).
3.2. Phylogenetic Analysis
DNA star was used in the sequence alignment. The alignment results were imported into MEGA 5.1 (Kimura 2-parameter matrix, neighbor-joining method, and Bootstrap for 1000 times). The phylogenetic tree was established based on the complete genome sequences of HBV clones and the genetic distance was calculated (11).
3.3. Statistical Analysis
Data was analyzed using SPSS10.0 (Chicago, IL). Data were compared by chi-square and t-tests. A P < 0.05 was considered statistically significant.
4. Results
4.1. Genotype Distribution
Genotype C accounted for the highest proportion (40.65%387/951) of HBV amongst the 14 countries neighboring China, thus genotype C was considered the dominant genotype. The less dominant genotypes were B, D and A, accounting for 19.87% (189/951), 18.51% (176/951) and 11.46% (109/951), respectively. The number of sequences belonging to genotypes F, G, H, I and J were 5, 2, 6, 19 and 3, respectively (Figure 1).
HBV Genotype Distribution Histogram
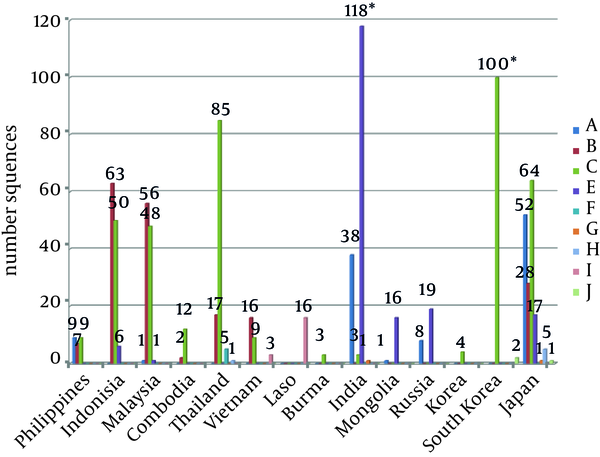
Pearson chi-square test (χ2 = 1765.2, P < 0.0001) indicated that the 14 countries differed considerably regarding HBV genotypes. A t-test was performed for 10 genotypes. Genotype C showed the largest difference in distribution among the countries (t = 2.97, P < 0.05), followed by genotype B (t = 2.364, P < 0.05). The difference in the distribution of other genotypes was relatively small (P > 0.05).
4.2. Recombinant Genotype
Based on the literature, there were 45 sequences belonging to recombinant genotype, accounting for 5.94% (12-19). Malaysia had the greatest diversity of recombinant genotypes and the largest number of recombinant genotype sequences (51.11%; Table 1).
4.3. Serotype Distribution
All 10 genotypes of HBV existed in 14 countries neighboring China, with adw2 and adrq+ dominant and accounting for 29.38% (260/885) and 24.75% (219/885), respectively. The serotype adw4 was only found in 12 sequences from the Philippines and adw3 was found in one sequence from Malaysia and Japan, respectively (Figure 2).
Serotype Distribution in Different Countries
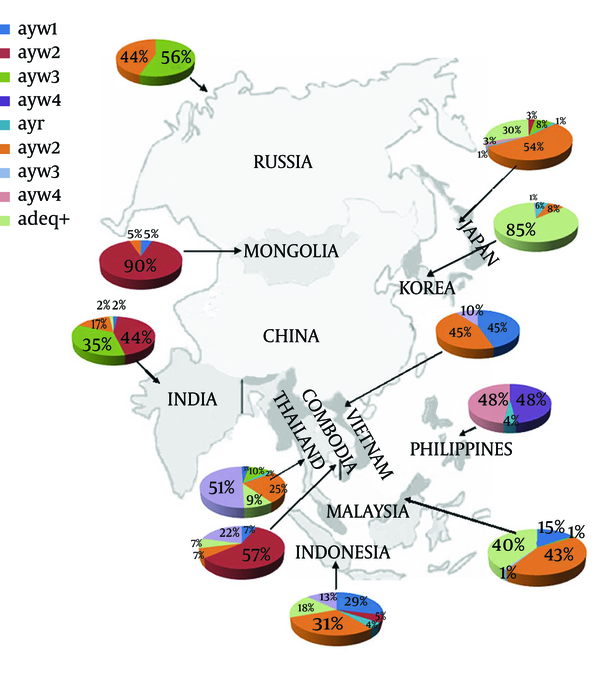
4.4. Correspondence Between Serotype and Genotype
The association between HBV serotype and genotype in 14 countries neighboring China is shown in Table 2.
The Corresponding Relation of Serotype and Genotype
| Serotype | Genotype |
|---|---|
| ayw1 | BCID |
| ayw2 | ACD |
| ayw3 | BCD |
| ayw4 | ABC |
| ayr | CD |
| adw2 | ABCEIH |
| adw3 | BH |
| adw4 | ABCHF |
| adrq+ | BCDEJ |
| adrq- | C |
4.5. Sequence Alignment
4.5.1. Phylogenetic Tree of Genotype B and the Difference Between the Sub-Genotypes
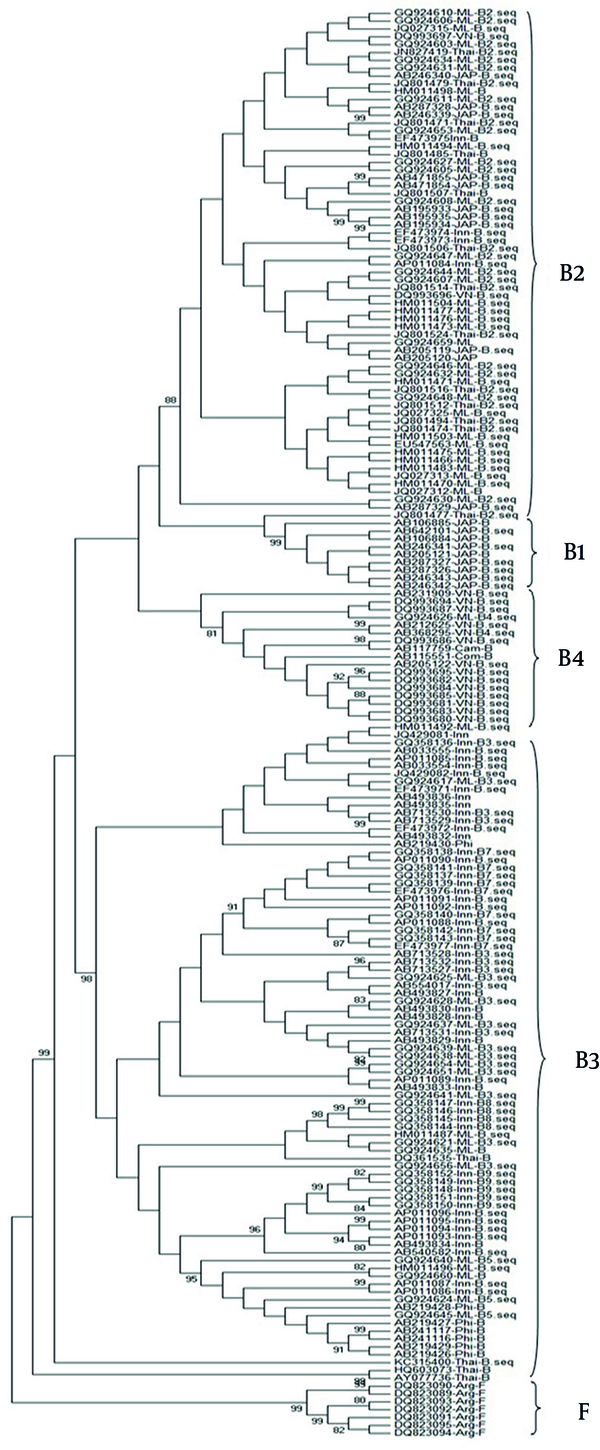
According to the reference sequences reported in the literature, genotype B could be further divided into eight subgenotypes (B1, B2, B3, B4, B5, B7, B8 and B9) (Figure 3, Table 3).
The branch of B2 was far away from other subgenotypes and the B3 was located closer to the trunk. B2 and B3 do not belong to one monophyletic group, but on different branches.
Phylogenetic of HBV Genotype B
Mean Nucleotide Sequence Divergences Over Complete Genome Sequences of HBV Between Sub Genotypes
| Sub Genotype | B1 | B2 | B3 | B4 | B5 | B7 | B8 |
|---|---|---|---|---|---|---|---|
| B2 | 0.032 ± 0.003 | ||||||
| B3 | 0.048 ± 0.004 | 0.037 ± 0.003 | |||||
| B4 | 0.056 ± 0.003 | 0.046 ± 0.003 | 0.056 ± 0.003 | ||||
| B5 | 0.050 ± 0.004 | 0.038 ± 0.003 | 0.027 ± 0.002 | 0.057 ± 0.003 | |||
| B7 | 0.049 ± 0.004 | 0.038 ± 0.003 | 0.020 ± 0.001 | 0.055 ± 0.003 | 0.027 ± 0.002 | ||
| B8 | 0.050 ± 0.004 | 0.041 ± 0.003 | 0.028 ± 0.002 | 0.058 ± 0.003 | 0.031 ± 0.002 | 0.028 ± 0.002 | |
| B9 | 0.046 ± 0.004 | 0.036 ± 0.003 | 0.025 ± 0.002 | 0.054 ± 0.003 | 0.025 ± 0.002 | 0.025 ± 0.002 | 0.028 ± 0.003 |
4.5.2. Alignment of Complete Genomes of Genotype B
Four open reading, frames (S, P, X, C) of HBV genotype B were analyzed, there were no specificity in P, X and C regions besides S region. Alignment shows that the amino acid sequences translated by the S open reading frame reflects the heterogeneity of HBV from each country (Figure 4).
Specific Amino Acid of HBV Genotype B

The subgenotype B found in Indonesia and Malaysia (B3, B5, B7, B8 and B9) shared six common specific amino acid sites in S-ORF as follows: AA39 D; AA55 S; AA152 N; AA154 V; AA158 A and AA160 S. For most subgenotypes, I and N were found at AA56 and AA84, respectively. However, B7 has a T at position AA56; B9 has an M at position AA84; B1 from Japan has H and I at these two positions, respectively, and for Vietnam, 87.5% of AA84 was I.
In B4, all AA158 was A, with T at AA164, F at AA165 and Y at AA374. Subgenotype B4 from other four countries has L, V, I, L and F at these positions, respectively. For Indonesia, B8 has specific T at AA164 in four sequences.
Generally, there would be no more than four types of amino acids appearing at one position in the complete genome; however, in P-ORF, seven types of amino acids (C, N, S, G, V, I, and D) were found at AA69. In some sequences from Malaysia and Indonesia, five types of amino acids were detected at this position. Moreover, at AA73 there were five types of amino acids (K, D, N, Q, and E). The sequences from Malaysia had all five types of amino acids.
4.6. Stop and Start Codons
The stop codon of S-ORF was TAA in 141 sequences of HBV, belonging to genotype B. The stop codon of P-ORF was TGA. The stop codon was TAG in 95.74% of the sequences in C-ORF. Thus, there were only six sequences with TAA as the stop codon. The stop codon was TAA in 92.9% of the sequences in X-ORF, with the exceptions of TGA in one case and TAG in eight cases (figures omitted).
There was a start codon mutation in the pre-S2 region, with ATG mutated into GTG (DQ993682, AB205122, GQ924641) or ATA (JQ801474, GQ924646, GQ924625, JQ027325) or ACG (JQ027315).
The number of patients with HBV infection reported by the WHO for each district and country of the world was positively correlated with the number of HBV sequences in the database isolated from the corresponding district and country.
5. Discussion
As shown in Figure 1, genotype compositions of HBV from adjacent countries tend to be similar, which was related to a geographical factor. The two southeastern Asian countries, Indonesia and Malaysia, are located near the equator, between the Indian and Pacific Oceans. Both countries have many islands and share a border extending for 874 km. The dominant genotypes of HBV were B and C in the two countries. For Mongolia and Russia (north of China), D was the dominant genotype. Furthermore, the diversity of genotypes differs for each country. Japan has a high diversity of HBV genotypes, probably because Japan was frequent mobility with foreign countries due to its special historical background and developed economy (22). During the exchanges, different genotypes of HBV were introduced. In contrast, for developing Asian countries, such as Laos, Burma and North Korea, fewer genotypes were detected. Harboring a relatively small population on such vast areas of land, Russia and Mongolia reported only two genotypes, as shown by our study. Although India ranks the second in the world regarding population size, India attracts much fewer immigrants because of its underdevelopment (7). Therefore, only four genotypes of HBV have been detected in India. Indeed, land area, population size and level of development are all important factors, which determine the diversity of HBV genotypes and subgenotypes for a country.
An association was found between genotype and serotype. According to our study, such an association in 14 countries neighboring China seems to deviate from the previous results. Thedja MD et al. (20) reported that the serotype ayw1 mainly corresponds to genotype A and very rarely to B on a global scale. The serotype ayw3 corresponds to D in most cases and rarely to C. The serotype ayw4 is expected to correspond to E. The serotype adw2 often corresponds to A, B, G and rarely C. However, based on our results, ayw1 corresponds to B in 57 sequences (79.2%) and no sequence showed correspondence association to A. Although ayw3 corresponds to D in 90 sequences, a correspondence to C was found in a few sequences. The serotype ayw4 does not correspond to E as indicated by our study, but only corresponds to A, B and C. In 72 and 96 sequences, adw2 was found to correspond to genotypes A and B, respectively; however, a correspondence was found between adw2 and C (41, 15.7%). This result may demonstrate a certain regional characteristic or a sampling bias can be suspected.
In the phylogenetic tree, the branch of subgenotype B2 was far away from other subgenotypes. B2 was more phylogenetically ancient. According to a previous study (23), B9 was more phylogenetically ancient than B3 and B7, but in the phylogenetic tree constructed in our study, B3 was located closer to the trunk. Such divergence may be attributed to the difference in the selection of sequences belonging to each subgenotype.
Phylogenetic analysis showed that B2 and B3 do not belong to one monophyletic group, but on different branches. This result contradicts the principle that the genotype and the corresponding subgenotypes of HBV should belong to one monophyletic group (21). Although B7, B5, B9, B4 and B1 have high bootstrap values (> 87%), only B4 and B1 have a sequence divergence more than 4% with other sequences. Thus, only B4 and B1 can be accurately differentiated, which was consistent with a previous study (24). Because B3, B5, B7, B8 and B9 were the recombinant subgenotypes between B and C (25-28), sequence divergence was below 4% for these subgenotypes and can be clustered into quasi-subgenotype B3 (29).
The pre-S2 region of HBV isolated from Vietnam contains three specific amino acid positions (AA158 A, AA164 T, and AA165 F) and the corresponding sequences all belong to subgenotype B4; however, whether this feature could be used as the criterion for differentiating B4 should be verified with a large sample size. For the sequences from Indonesia and Malaysia, six specific amino acid positions simultaneously appear in subgenotypes B3, B5, B7, B8 and B9. It was further confirmed that B3, B5, B7, B8 and B9 can be clustered into quasi-subgenotype B3. More evidence need to be gathered before we can take these six positions as the criterion for differentiating quasi-subgenotype B3. The preS1 and preS2 proteins have very strong antigenicity. It was the reason why they were conserved in that genotype.
Codon bias was a phenomenon that the synonymous codon disequilibrium of organism encoded in the same kinds of amino acids. This phenomenon related with the carrier of genetic information of DNA molecules and biological function of proteins, so it has an important biological significance (30). Similar with amino acids encoded codon bias, this study found that the termination codon of the genotype B has a partiality in different regains.
The stop codon of HBsAg was TAA and TGA, according to the existing literature (31). This study showed that C-ORF and P-ORF have a TAG preference. X-ORF occasionally contains TAG.
Three aspects might explain this feature. First, research showed that synonymous codon usage frequency of one gene and abundance of identify their identify them have a positive correlation, codon bias increases along the length of genes (32). Second, various tRNA on each specific ability to recognize different termination codon. High recognition ability of RNA was preferred to use (33). Last but not least, natural condition, when the terminal codon of HBsAg were TAA,TGA or TGA, the corresponding amino acids of polymerase on overlap region were AA236N,AA236N or AA236D, which means that different terminal codon of HBsAg might change the polarity of aa236. It was known that amino acid 236 located on the catalytic center of polymerase, so the polarity change might have a significant impact on catalytic activity.
The start codon mutation of the pre-S2 region reported herein was consistent with the reports from Japan, Korean and Thailand (34, 35). Occurring in a variety of species, this mutation might be considered to be a new start codon for HBV. Several studies reported the presence of GTG or TTG in a few bacterial species as the start codon (36-38). Kadowaki et al. showed that the mitochondrial ACG codon encodes protein (39). There were also mitochondria and chloroplasts with ATT and ATA as the start codons (40-42). Some studies indicated that this mutation may induce changes in proteins encoded by the pre-S2 region or interfere with the binding of HBV to hepatic cells, thus causing a high risk of hepatic cancer (25, 42). These start codons were still translated as Met when at the start of a protein, even if the codon encodes a different amino acid, because a separate transfer RNA (tRNA) was used for initiation (39, 40). Therefore, this start codon mutation of the pre-S2 region of HBV was probably a same-sense mutation, without affecting the survival and replication of HBV in the host cells. Currently, there has been no experimental report on the possible influence of this mutation.
YMDD mutation occurs in polymerase gene of HBV. Until recently, most research about YMDD mutation focused on the occurrence of lamivudine-related YMDD mutation and its impact on antiviral treatment (43). The YMDD mutation, also known as the M204V/I mutation, was the substitution of methionine by valine or isoleucine and designated as the YVDD or YIDD variant (44, 45). Of 150 sequences in our experiments, YVDD was found in one sequence from Thailand and Indonesia and YIDD was found in two sequences from Japan. In addition, the SMDD mutation (AP011093) was found in one sequence from Indonesia and the YMND mutation (DQ993695) was found in one sequence from Vietnam. Similar SMDD mutations have not been previously reported. The causes for the mutation may be drug-induced tolerance or natural variation.
The largest number of complete genome sequences of HBV in the nucleic acid database was from mainland China, accounting for about one-fourth of the total. Hong Kong and Japan were the second and third, followed by Africa, Europe and the US. The number of cases of HBV infection reported by the WHO for each district and country of the world was positively correlated with the number of HBV sequences in the database isolated from the corresponding district and country.
China is a country with a high incidence of hepatitis. The high prevalence of chronic hepatitis B has already threatened the public health. In NCBI, nearly 1000 sequences of HBV were submitted from 14 countries neighboring China, accounting for one-fifth of the total. It is an integral part of the basic research of HBV to use data of genome sequences from China and the neighboring countries and to promote clinical practice based on genome analysis results.
Acknowledgements
References
-
1.
Yuen MF, Lai CL. Natural history of chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2000;15(s2):E20-4. https://doi.org/10.1046/j.1440-1746.2000.02123.x.
-
2.
Kew MC. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol (Paris). 2010;58(4):273-7. [PubMed ID: 20378277]. https://doi.org/10.1016/j.patbio.2010.01.005.
-
3.
Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281(5733):646-50. [PubMed ID: 399327].
-
4.
Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69 ( Pt 10):2575-83. [PubMed ID: 3171552].
-
5.
Shi W, Zhang Z, Ling C, Zheng W, Zhu C, Carr MJ, et al. Hepatitis B virus subgenotyping: history, effects of recombination, misclassifications, and corrections. Infect Genet Evol. 2013;16:355-61. [PubMed ID: 23538336]. https://doi.org/10.1016/j.meegid.2013.03.021.
-
6.
Xia GL, Liu CB, Cao HL, Bi SL, Zhan MY, Su CA, et al. Prevalence of hepatitis B and C virus infections in the general Chinese population. Results from a nationwide cross-sectional seroepidemiologic study of hepatitis A, B, C, D, and E virus infections in China, 1992. Int Hepatol Commun. 1996;5(1):62-73. https://doi.org/10.1016/S0928-4346(96)82012-3.
-
7.
Margeridon S, Lachaux A, Trepo C, Zoulim F, Kay A. A quasi-monoclonal anti-HBs response can lead to immune escape of 'wild-type' hepatitis B virus. J Gen Virol. 2005;86(Pt 6):1687-93. [PubMed ID: 15914846]. https://doi.org/10.1099/vir.0.80810-0.
-
8.
Wu GH, Ding HG, Zeng CQ. Overview of hepatitis b virus complete genome sequence of the public nucleic acid database and the establishment of the Chinese reference sequence. Prog Nat. 2008;18(2):121-9.
-
9.
Huang Y, Zhang Z, Liu J, Xu QG. Adaptive evolution analysis of hepatitis B virus epidemic strains in China. J Jiangsu Univ. 2012;22(4):312-23.
-
10.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-9. [PubMed ID: 21546353]. https://doi.org/10.1093/molbev/msr121.
-
11.
Phung TB, Alestig E, Nguyen TL, Hannoun C, Lindh M. Genotype X/C recombinant (putative genotype I) of hepatitis B virus is rare in Hanoi, Vietnam--genotypes B4 and C1 predominate. J Med Virol. 2010;82(8):1327-33. [PubMed ID: 20572069]. https://doi.org/10.1002/jmv.21775.
-
12.
Arankalle VA, Gandhe SS, Borkakoty BJ, Walimbe AM, Biswas D, Mahanta J. A novel HBV recombinant (genotype I) similar to Vietnam/Laos in a primitive tribe in eastern India. J Viral Hepat. 2010;17(7):501-10. [PubMed ID: 20059669]. https://doi.org/10.1111/j.1365-2893.2009.01206.x.
-
13.
Chauhan R, Kazim SN, Kumar M, Bhattacharjee J, Krishnamoorthy N, Sarin SK. Identification and characterization of genotype A and D recombinant hepatitis B virus from Indian chronic HBV isolates. World J Gastroenterol. 2008;14(40):6228-36. [PubMed ID: 18985816].
-
14.
Suwannakarn K, Tangkijvanich P, Theamboonlers A, Abe K, Poovorawan Y. A novel recombinant of Hepatitis B virus genotypes G and C isolated from a Thai patient with hepatocellular carcinoma. J Gen Virol. 2005;86(Pt 11):3027-30. [PubMed ID: 16227224]. https://doi.org/10.1099/vir.0.81241-0.
-
15.
Tangkijvanich P, Sa-Nguanmoo P, Avihingsanon A, Ruxrungtham K, Poovorawan K, Poovorawan Y. Characterization of hepatitis B virus mutations in untreated patients co-infected with HIV and HBV based on complete genome sequencing. J Med Virol. 2013;85(1):16-25. [PubMed ID: 23024025]. https://doi.org/10.1002/jmv.23430.
-
16.
Pancawardani P, Depamede SN, Wahyono A, Jirintai S, Nagashima S, et al. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012;163(1):129-40. [PubMed ID: 21925554]. https://doi.org/10.1016/j.virusres.2011.09.002.
-
17.
Elkady A, Tanaka Y, Kurbanov F, Oynsuren T, Mizokami M. Virological and clinical implication of core promoter C1752/V1753 and T1764/G1766 mutations in hepatitis B virus genotype D infection in Mongolia. J Gastroenterol Hepatol. 2008;23(3):474-81. [PubMed ID: 18318825]. https://doi.org/10.1111/j.1440-1746.2008.05321.x.
-
18.
Chook JB, Ngeow YF, Yap SF, Tan TC, Mohamed R. Combined use of wild-type HBV precore and high serum iron marker as a potential tool for the prediction of cirrhosis in chronic hepatitis B infection. J Med Virol. 2011;83(4):594-601. [PubMed ID: 21328372]. https://doi.org/10.1002/jmv.22016.
-
19.
Pecoud A, Guchteneire P. Immigrants without borders (M). United States: Berghahn Books; 2007.
-
20.
Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9. Arch Virol. 2011;156(5):855-68. [PubMed ID: 21318309]. https://doi.org/10.1007/s00705-011-0926-y.
-
21.
Shi W, Zhu C, Zheng W, Carr MJ, Higgins DG, Zhang Z. Subgenotype reclassification of genotype B hepatitis B virus. BMC Gastroenterol. 2012;12:116. [PubMed ID: 22925657]. https://doi.org/10.1186/1471-230X-12-116.
-
22.
Li YP, Jiang W. India's 2011 census and prospected. Popolation J. 2013;5(35):36-46.
-
23.
Pourkarim MR, Amini-Bavil-Olyaee S, Lemey P, Maes P, Van Ranst M. Are hepatitis B virus "subgenotypes" defined accurately? J Clin Virol. 2010;47(4):356-60. [PubMed ID: 20153973]. https://doi.org/10.1016/j.jcv.2010.01.015.
-
24.
Sakamoto T, Tanaka Y, Simonetti J, Osiowy C, Borresen ML, Koch A, et al. Classification of hepatitis B virus genotype B into 2 major types based on characterization of a novel subgenotype in Arctic indigenous populations. J Infect Dis. 2007;196(10):1487-92. [PubMed ID: 18008228]. https://doi.org/10.1086/523111.
-
25.
Nurainy N, Muljono DH, Sudoyo H, Marzuki S. Genetic study of hepatitis B virus in Indonesia reveals a new subgenotype of genotype B in east Nusa Tenggara. Arch Virol. 2008;153(6):1057-65. [PubMed ID: 18463783]. https://doi.org/10.1007/s00705-008-0092-z.
-
26.
Depamede SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, et al. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154(7):1047-59. [PubMed ID: 19499283]. https://doi.org/10.1007/s00705-009-0406-9.
-
27.
Shi W, Carr MJ, Dunford L, Zhu C, Hall WW, Higgins DG. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology. 2012;427(1):51-9. [PubMed ID: 22374235]. https://doi.org/10.1016/j.virol.2012.01.030.
-
28.
Wu XM, Wu FS, Reng DM. Codon bias analysis methods and research progress. Genet. 2007;29:420-6.
-
29.
Li JQ, Tian JH, Liu F, Du SC. [Hepatitis B surface antigen terminates codon bias selection]. Beijing Da Xue Xue Bao. 2008;40(3):270-2. [PubMed ID: 18560454].
-
30.
Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24(2):374-81. [PubMed ID: 17101719]. https://doi.org/10.1093/molbev/msl166.
-
31.
Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346-53. [PubMed ID: 15245907]. https://doi.org/10.1016/j.tibtech.2004.04.006.
-
32.
Huy TT, Ushijima H, Win KM, Luengrojanakul P, Shrestha PK, Zhong ZH, et al. High prevalence of hepatitis B virus pre-s mutant in countries where it is endemic and its relationship with genotype and chronicity. J Clin Microbiol. 2003;41(12):5449-55. [PubMed ID: 14662924].
-
33.
Choi MS, Kim DY, Lee DH, Lee JH, Koh KC, Paik SW, et al. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007;14(3):161-8. [PubMed ID: 17305881]. https://doi.org/10.1111/j.1365-2893.2006.00784.x.
-
34.
Suwannakarn K, Tangkijvanich P, Thawornsuk N, Theamboonlers A, Tharmaphornpilas P, Yoocharoen P, et al. Molecular epidemiological study of hepatitis B virus in Thailand based on the analysis of pre-S and S genes. Hepatol Res. 2008;38(3):244-51. [PubMed ID: 17711443]. https://doi.org/10.1111/j.1872-034X.2007.00254.x.
-
35.
Blattner FR. The Complete Genome Sequence of Escherichia coli K-12. Science. 1997;277(5331):1453-62. https://doi.org/10.1126/science.277.5331.1453.
-
36.
Gilbert W, Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973;70(12):3581-4. [PubMed ID: 4587255].
-
37.
Farabaugh PJ. Sequence of the lacI gene. Nature. 1978;274(5673):765-9. [PubMed ID: 355891].
-
38.
Sakamoto W, Tan SH, Murata M, Motoyoshi F. An unusual mitochondrial atp9-rpl16 cotranscript found in the maternal distorted leaf mutant of Arabidopsis thaliana: implication of GUG as an initiation codon in plant mitochondria. Plant Cell Physiol. 1997;38(8):975-9. [PubMed ID: 9327595].
-
39.
Kadowaki K, Ozawa K, Kazama S, Kubo N, Akihama T. Creation of an initiation codon by RNA editing in the coxI transcript from tomato mitochondria. Curr Genet. 1995;28(5):415-22. [PubMed ID: 8575013].
-
40.
Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37(5):1616-27. [PubMed ID: 19151083]. https://doi.org/10.1093/nar/gkp001.
-
41.
Fearnley IM, Walker JE. Initiation codons in mammalian mitochondria: differences in genetic code in the organelle. Biochemistry. 1987;26(25):8247-51. [PubMed ID: 2964865].
-
42.
Utama A, Siburian MD, Fanany I, Intan MD, Dhenni R, Kurniasih TS, et al. Hepatitis B virus pre-S2 start codon mutations in Indonesian liver disease patients. World J Gastroenterol. 2012;18(38):5418-26. [PubMed ID: 23082059]. https://doi.org/10.3748/wjg.v18.i38.5418.
-
43.
Yang JH, Zhang H, Chen XB, Chen G, Wang X. Relationship between hepatocellular carcinoma and hepatitis B virus genotype with spontaneous YMDD mutations. World J Gastroenterol. 2013;19(24):3861-5. [PubMed ID: 23840126]. https://doi.org/10.3748/wjg.v19.i24.3861.
-
44.
Moskovitz DN, Osiowy C, Giles E, Tomlinson G, Heathcote EJ. Response to long-term lamivudine treatment (up to 5 years) in patients with severe chronic hepatitis B, role of genotype and drug resistance. J Viral Hepat. 2005;12(4):398-404. [PubMed ID: 15985011]. https://doi.org/10.1111/j.1365-2893.2005.00613.x.
-
45.
Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36(6):687-96. [PubMed ID: 12627352]. https://doi.org/10.1086/368083.