Supporting data and materials for the de novo assembly of Dekkera bruxellensis CBS11270 using multiple technologies.
Dataset type: Genome-Mapping, Genomic, Software
Data released on November 04, 2015
Olsen R; Bunikis I; Tiukova I; Holmberg K; Lotstedt B; Pettersson OV; Passoth V; Kaller M; Vezzi F (2015): Supporting data and materials for the de novo assembly of Dekkera bruxellensis CBS11270 using multiple technologies. GigaScience Database. https://doi.org/10.5524/100179
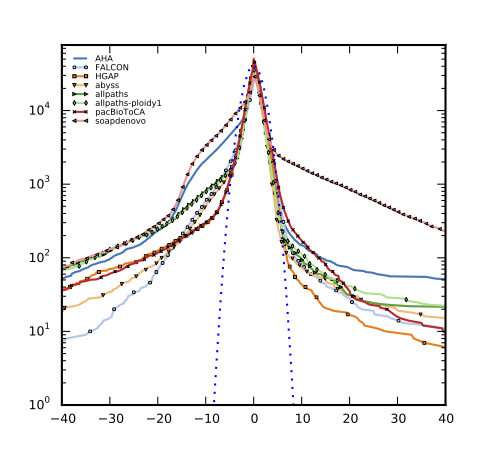
We present a genomic dataset sampled from the yeast Dekkera bruxellensis using three different technologies: Illumina short-read sequencing, PacBio long-read sequencing and optical mapping. The Illumina data consists of four different libraries of differing insert sizes (ie. paired-end fragments and mate-pair libraries), following the ALLPATHS recipe.
The purpose was to generate a draft genome assembly of high quality by combining these three different and somewhat complementary technologies. As a by-product of our work we present a pipeline for de novo assembly, NouGAT. It is a semi-automated pipeline for read pre-processing, de novo assembly with support of a wide range of assemblers and final assembly evaluation.
The version of the pipeline hosted here in GigaDB is the version as published (02-Nov-2015), for the most upto date version users are directed to the GitHub repository.
Additional details
Read the peer-reviewed publication(s):
- Olsen, R.-A., Bunikis, I., Tiukova, I., Holmberg, K., Lötstedt, B., Pettersson, O. V., Passoth, V., Käller, M., & Vezzi, F. (2015). De novo assembly of Dekkera bruxellensis: a multi technology approach using short and long-read sequencing and optical mapping. GigaScience, 4(1). https://doi.org/10.1186/s13742-015-0094-1
Additional information:
https://github.com/SciLifeLab/NouGAT/
Accessions (data included in GigaDB):
ENA: ERP012947
Click on a table column to sort the results.
Table Settings| Sample ID | Common Name | Scientific Name | Sample Attributes | Taxonomic ID | Genbank Name |
|---|---|---|---|---|---|
| CBS11270 | Brettanomyces bruxellensis | Dekkera bruxellensis | Alternative accession-BioSample:SAMEA3639848 Isolate:CBS11270 Alternative names:Dekkera bruxellensis ... |
5007 |
Click on a table column to sort the results.
Table Settings| File Name | Description | Sample ID | Data Type | File Format | Size | Release Date | File Attributes | Download |
|---|---|---|---|---|---|---|---|---|
| Readme | TEXT | 2.47 kB | 2015-11-02 | MD5 checksum: 9f7fd540b0fc9bc6ee0dee3fe99588eb |
||||
| ABySS assembly | Sequence assembly | FASTA | 5.20 MB | 2015-11-02 | MD5 checksum: 59ec48f561d0db5499e7ca2e131580d5 |
|||
| AHA assembly | Sequence assembly | FASTA | 4.04 MB | 2015-11-02 | MD5 checksum: 0259820ecb7b9fcaf7276a85ed5eb1e2 |
|||
| ALLPATHS-LG assembly | Sequence assembly | FASTA | 4.14 MB | 2015-11-02 | MD5 checksum: 87938800da784d3c55cbdeb84f8f4a46 |
|||
| Optical map guided assembly version chr1-5 | Sequence assembly | FASTA | 4.04 MB | 2015-11-02 | MD5 checksum: 454dd822c3903a938114c245b795fc5e |
|||
| Optical map guided assembly version chr1-7 | Sequence assembly | FASTA | 4.42 MB | 2015-11-02 | MD5 checksum: 58fffd51a30bbc874eba617c731b5b5e |
|||
| FALCON assembly | Sequence assembly | FASTA | 3.15 MB | 2015-11-02 | MD5 checksum: 77da463e4965afbebd3175c532caa883 |
|||
| HGAP assembly | Sequence assembly | FASTA | 4.57 MB | 2015-11-02 | MD5 checksum: 137a824c0a7f762123dced204969e0d1 |
|||
| PacBioToCA assembly | Sequence assembly | FASTA | 5.03 MB | 2015-11-02 | MD5 checksum: 7f1138f9c830900a40b9c5b1b5854fb4 |
|||
| SOAPdenovo assembly | Sequence assembly | FASTA | 6.59 MB | 2015-11-02 | MD5 checksum: 68486a3b03da7554c1c0ee495180be60 |
| Date | Action |
|---|---|
| November 4, 2015 | Dataset publish |
| November 27, 2015 | Manuscript Link added : 10.1186/s13742-015-0094-1 |