Published online Feb 24, 2021. doi: 10.5306/wjco.v12.i2.95
Peer-review started: November 20, 2020
First decision: December 3, 2020
Revised: December 7, 2020
Accepted: December 22, 2020
Article in press: December 22, 2020
Published online: February 24, 2021
Radiation dose to specific cardiac substructures can have a significant on treatment related morbidity and mortality, yet definition of these structures is labor intensive and not standard. Autosegmentation software may potentially address these issues, however it is unclear whether this approach can be broadly applied across different treatment planning conditions. We investigated the feasibility of autosegmentation of the cardiac substructures in four-dimensional (4D) computed tomography (CT), respiratory-gated, non-contrasted imaging.
To determine whether autosegmentation can be successfully employed on 4DCT respiratory-gated, non-contrasted imaging.
We included patients who underwent stereotactic body radiation therapy for inoperable, early-stage non-small cell lung cancer from 2007 to 2019. All patients were simulated via 4DCT imaging with respiratory gating without intravenous contrast. Generated structure quality was evaluated by degree of required manual edits and volume discrepancy between the autocontoured structures and its edited sister structure.
Initial 17-structure cardiac atlas was generated with 20 patients followed by three successive iterations of 10 patients using MIM software. The great vessels and heart chambers were reliably autosegmented with most edits considered minor. In contrast, coronary arteries either failed to be autosegmented or the generated structures required major alterations necessitating deletion and manual definition. Similarly, the generated mitral and tricuspid valves were poor whereas the aortic and pulmonary valves required at least minor and moderate changes respectively. For the majority of subsites, the additional samples did not appear to substantially impact the quality of generated structures. Volumetric analysis between autosegmented and its manually edited sister structure yielded comparable findings to the physician-based assessment of structure quality.
The use of MIM software with 30-sample subject library was found to be useful in delineating many of the heart substructures with acceptable clinical accuracy on respiratory-gated 4DCT imaging. Small volume structures, such as the coronary arteries were poorly autosegmented and require manual definition.
Core Tip: Autosegmentation is an attractive tool to reduce the labor involved with manual delineation of anatomy. However, it is unclear whether this approach is viable for all treatment conditions. Stereotactic body radiation therapy frequently utilizes respiratory gated, non-contrasted computed tomography imaging for radiation planning and involuntary heart motion as well as lack of intravenous contrast may impact the quality of generated structures. In our study, MIM software successfully contoured the great vessels and heart chambers yet failed in generating coronary arteries. We provide evidence that MIM software can reliably autocontour the larger cardiac substructures, but not coronary arteries or heart valves.
- Citation: Farrugia M, Yu H, Singh AK, Malhotra H. Autosegmentation of cardiac substructures in respiratory-gated, non-contrasted computed tomography images. World J Clin Oncol 2021; 12(2): 95-102
- URL: https://www.wjgnet.com/2218-4333/full/v12/i2/95.htm
- DOI: https://dx.doi.org/10.5306/wjco.v12.i2.95
Cardiotoxicity is a significant concern in modern radiotherapy (RT). RTOG 0617 was a large, phase III trial investigating the utility of dose escalation in locally advanced non-small cell lung cancer (NSCLC), where the experimental dose escalated arm reduced patient survival, in part due to excessive cardiotoxicity[1]. In a follow up study, Thor et al[2] revealed that dose to specific cardiac substructures such as the atria, ventricles, and pericardium were highly correlated with overall survival. These findings have been supported by several other groups, who also demonstrate a dose-dependent relationship between cardiac substructures and outcome[3,4]. Despite these findings, definition of such structures is not standard in radiation planning at this time.
Conventionally, the heart is contoured as a single structure which encompasses the atria, ventricles, valve, pericardium, and coronary arteries. Individual definition and subsequent dose calculation to the cardiac substructures is not routinely performed. Delineation of cardiac subsites can be challenging due to heart and respiratory motion, lack of intravenous contrast on treatment planning computed tomography (CT) imaging, and anatomic variances[5-10]. Furthermore, contouring these structures can be time consuming and not necessarily practical in typical care[10].
Researchers have attempted to overcome these obstacles utilizing a number of approaches. For example, several groups have provided cardiac atlases for these subsites, including the recently closed clinical trial RTOG 1106 (NCT01507428) to provide guidance on how to define these structures[5,10,11]. Additionally, there is evidence that autosegmentation is a feasible option to mitigate the added labor associated with including these structures[6-10,12]. While this is a provocative strategy, this method may not be viable in all treatment conditions. For example, four-dimensional (4D) CT imaging with respiratory gating is a treatment delivery technique for stereotactic body radiation therapy (SBRT) utilized by approximately 31%-54% of providers in the United States[13,14]. As this requires multiple scans to construct each phase of respiration, timed intravenous contrast is not possible and cardiac motion may be amplified. Therefore, it is unclear how well autosegmentation would perform in this setting. We investigated whether autosegmentation for cardiac substructures could be accurately employed on non-contrasted, respiratory gated imaging.
We included patients who underwent SBRT for inoperable, early-stage NSCLC from 2007 to 2019. Data was collected under approval from the institutional review board at Roswell Park Comprehensive Cancer Center. CT simulation was previously described[15]. All patients were simulated in the supine position with a Body Fix immobilizer (Elekta, Stockholm, Sweden) with respiratory gating. Real-Time Position Management by Varian Medical System (Palo Alto, CA, United States) was utilized to assess respiratory motion. After evaluating tumor motion, treatment planning was performed on CT average of combined respiratory phases where tumor motion was ≤ 5 mm. With the exception of one patient who was treated using a breath-hold technique due to an irregular breathing pattern, all patients were treated using a respiratory phase-based 4D approach.
Cardiac atlas development was approached in two phases. First, 20 patients who were planned for lung SBRT using 4DCT imaging were chosen for initial atlas construction. Cardiac substructures were manually contoured by a single senior Radiation Oncology resident in the Eclipse treatment planning system (Varian Medical System, Palo Alto, CA, United States) on a CT average scan. The 17 cardiac substructures were the four heart chambers (left/right atria, left/right ventricles), four coronary arteries (left main common, left circumflex, left anterior descending, right coronary artery), four heart valves (aortic, pulmonary, mitral, tricuspid), pulmonary artery, ascending aorta, descending aorta, whole heart, and superior vena cava. These structures were well defined by a previously described cardiac atlas and an example is provided in Supplementary Figure 1[5].
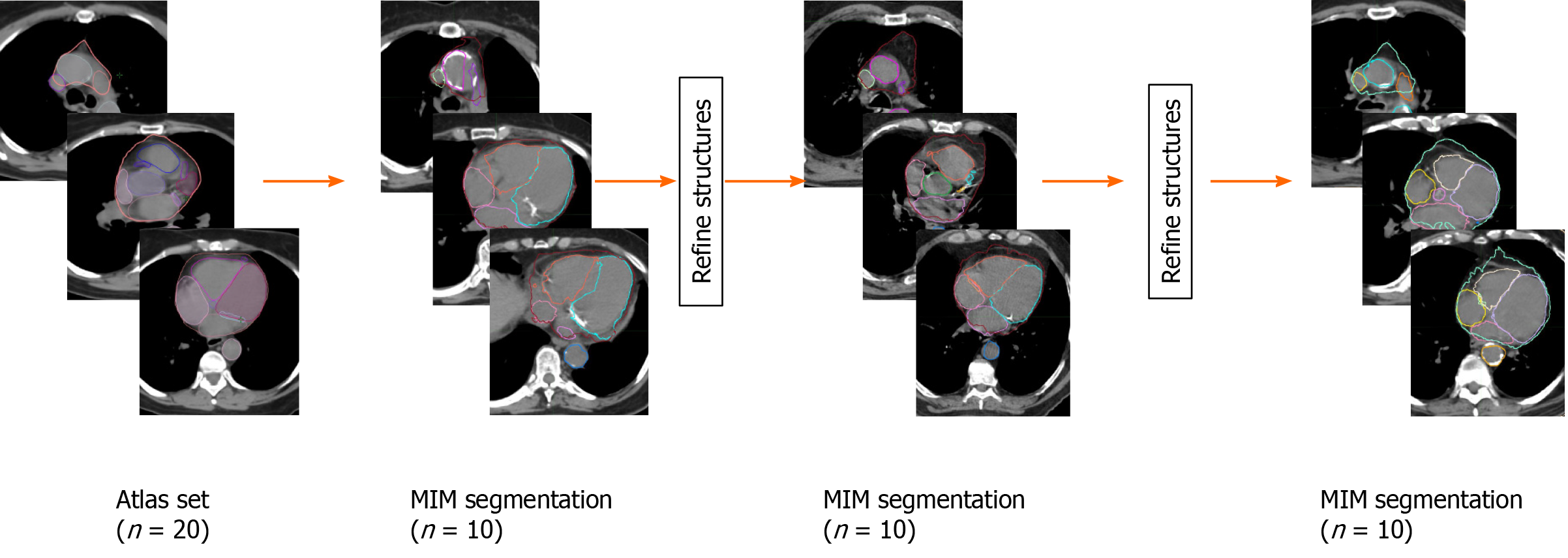
Secondly, these 20 patients were subsequently exported from Eclipse to MIM software (version 6.9.6, Beachwood, OH, United States) and a multi-patient atlas library pretraining to heart was generated for our lung SBRT patients. Using this atlas, 10 additional patients underwent autosegmentation in MIM. These generated structures were then reviewed and refined by the same Radiation Oncology resident, where the initial MIM’s generated structure were saved, copied, and edited to improved accuracy. These edited structures were then added back to the MIM multi-patient atlas library. This process was repeated two additional times adding additional 10 patients every time to the library. These steps were taken to study if incrementally adding patients to the library of patients improves auto contouring accuracy as the MIM software does not recommend minimum number of subjects in the atlas. In two instances, patients were replaced after substantial problems were noticed in the autosegmentation caused by anatomic abnormalities due to a prior pneumonectomy in one patient and a significant hiatal hernia in another. The final number was 10 patients for each iteration in MIM. The entire process is detailed in Figure 1.
Autosegmentated structures by MIM were evaluated by two separate methods. First, autogenerated contours were evaluated manually by the Radiation Oncology resident. A score was given for each structure: 0 = No changes made; 1 = Minor changes made; 2 = Moderate changes required by structure was salvageable; 3 = Structure required deletion and replacement; 4 = Structure failed autosegmentation. Minor changes would describe cleaning up a border of an otherwise accurate structure, where in moderate changes the general shape of the contour may be accurate, but a larger number of edits were necessary. Second, the volumes of autocontoured structures and its edited sister structure were compared. In the setting where a structure failed autosegmentation, the volume measurement was left blank. Data were presented using GraphPad Prism version 8.4.3 (GraphPad Software, San Diego, CA, United States).
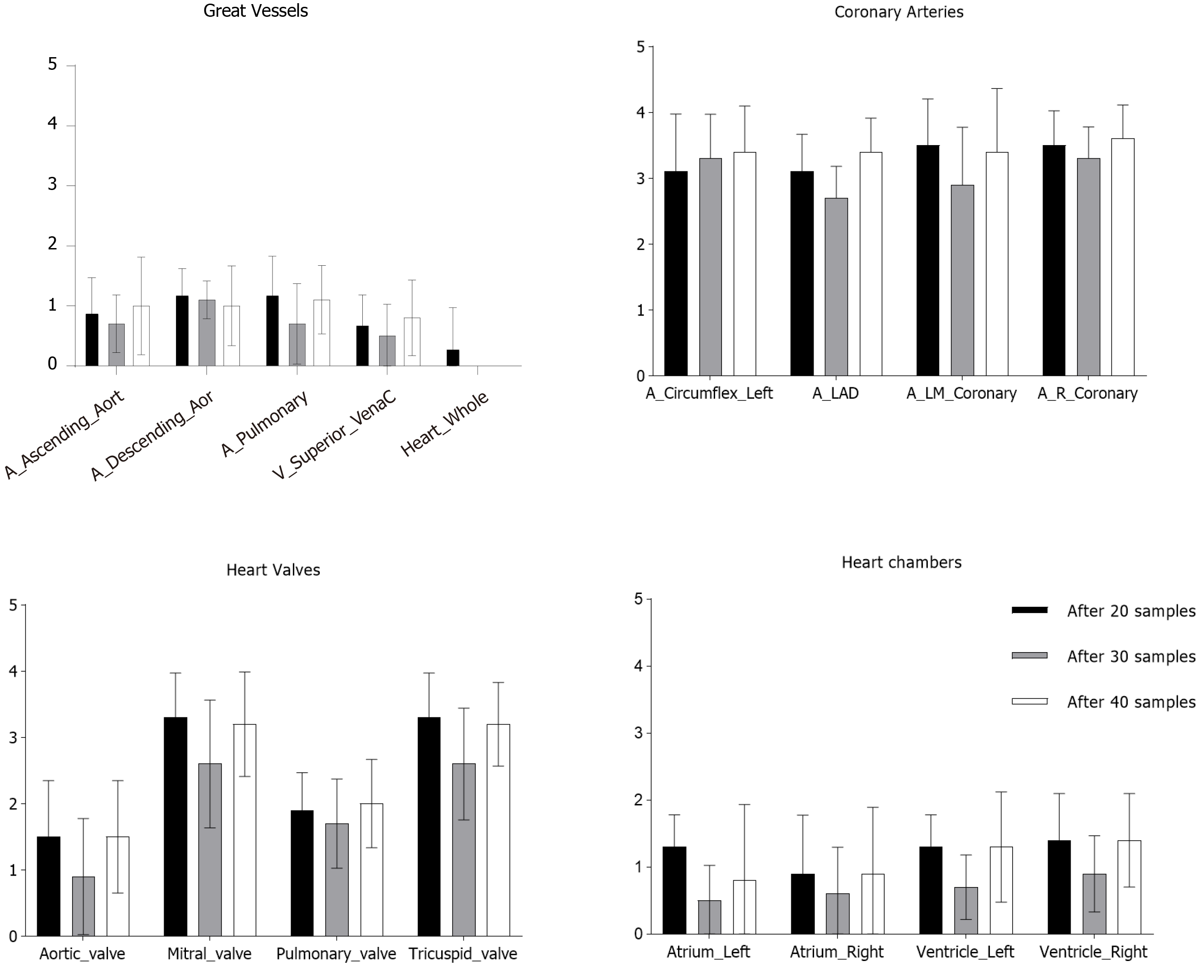
Utilizing MIM software, cardiac substructures were autosegmented in 10 patients and these structures were then manually refined (Figure 1). This process then repeated two subsequent times and representative autosegmented structures following each iteration can be found in Supplementary Figures 2-4. The great vessels including the whole heart were reliably autosegmented with the majority requiring minor changes without a substantial improvement from additional samples (Figure 2). In contrast, coronary arteries either failed to be autosegmented or the generated structures required major alterations necessitating deletion and manual definition. Similarly, the generated mitral and tricuspid valves were poor whereas the aortic and pulmonary valves required at least minor and moderate changes respectively (Figure 2). The heart chambers, particularly the atria, were autocontoured well (Figure 2). As compared to structures generated with 20 samples, there was a general improvement in structure quality after 30 samples, however, this trend did not continue in the 40-sample group.
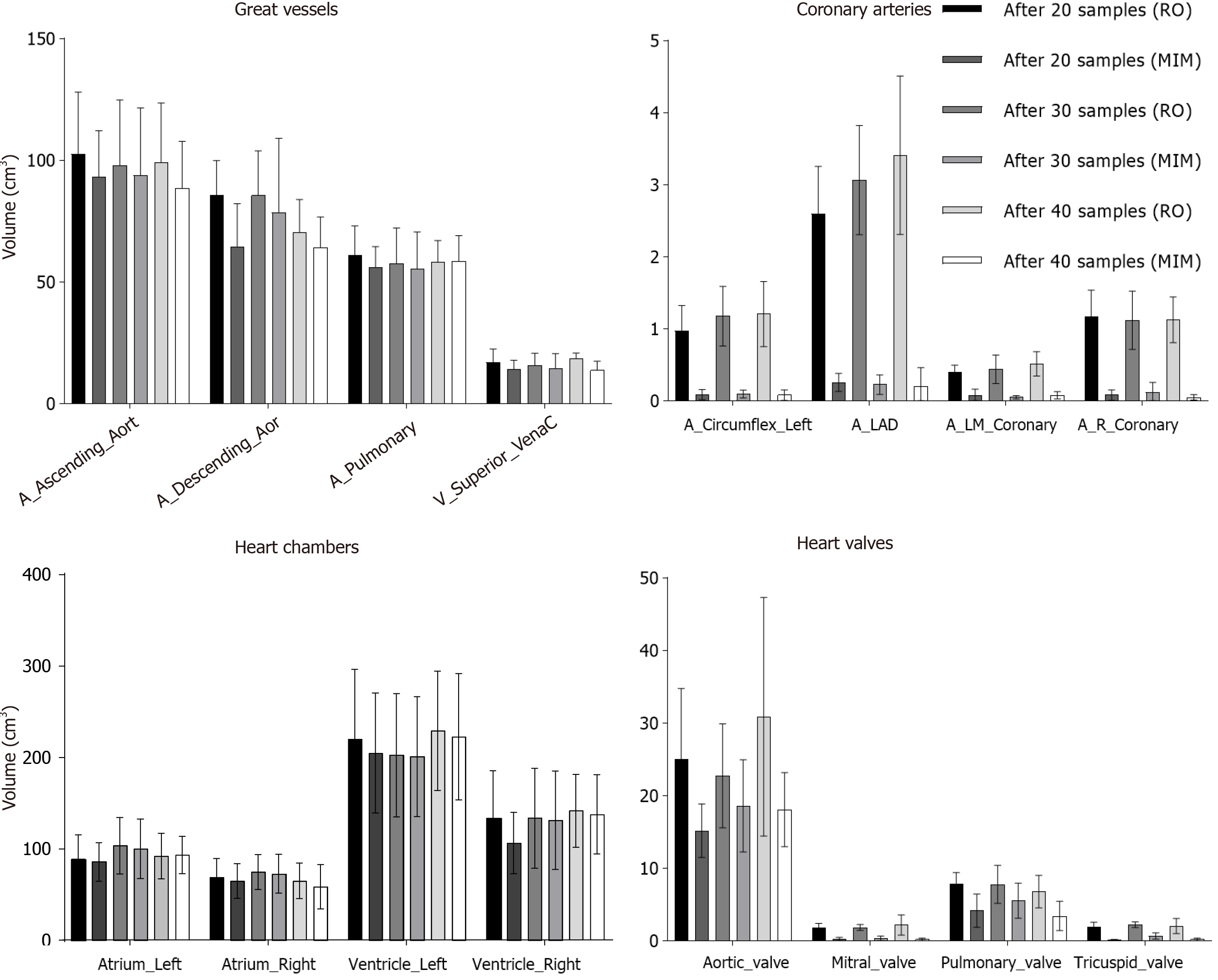
Additionally, volumes of the manually contoured and autosegmented cardiac substructures for the respective groups were compared. In general, MIM undercontoured structures when compared to the manually derived contours (Figure 3). This discrepancy was typically smaller for cardiac substructures with larger volumes such as the great vessels and heart chambers, whereas the difference was more profound in the smaller structures (Figure 3). There was excellent agreement in the volume of autogenerated and manual structures for the whole heart (Supplementary Figure 5). In some structures (right ventricle, descending aorta, pulmonary artery), more samples improved the volume discrepancy between manual and autocontoured structures (Figure 3). Unfortunately, often the autosegmented structures for the coronary arteries, mitral, and tricuspid valves were very small (< 0.1 cm3) and essentially unusable.
In current study, we explored the feasibility of MIM autocontouring for cardiac substructures on respiratory gated, non-contrasted 4DCT scans. Following autosegmentation, the great vessels and heart chambers required minor to no changes on average, whereas coronary arteries, tricuspid and mitral valves often failed autosegmentation or required complete recontouring. The aortic and pulmonary valves required a greater degree of modification but ultimately were usable structures. Similar observations were seen on volumetric analysis between the autosegmented structures and their manually edited correlates.
Several atlases for cardiac substructures have been described. The current study defined heart subsites per Feng et al[5], who used contrast-enhanced CT images of breast cancer patients to provide a well-defined atlas which is similar to previous studies, including RTOG 1106 (NCT01507428)[5,11,16].
Prior reports have evaluated methods for autocontouring cardiac substructures. Kirisli et al[17] utilized a multi-atlas approach to autosegment the whole heart and cardiac chambers using CT angiography. Shahzad et al[9] relied on contrast enhanced imaging for atlas generation, yet was then able to successfully autosegment structures on non-contrasted imaging via this template.
Others have demonstrated the reliability of non-contrasted images for atlas construction. In a breast cancer population, Finnegan et al[6] used multi-atlas segmentation in non-contrasted, free-breath CT imaging reporting similar success with the great vessels, whole heart, and heart chambers. Moreover, Luo et al[8] utilized non-contrasted, 4DCT images for multi-atlas autosegmentation finding minimal changes needed in generated structures with no dosimetric difference between autosegmented and manually derived structures. Additional groups have successfully used 4DCT imaging in deriving cardiac atlases and subsequent autosegmentation[3,10]. Consistent with our findings, these studies have reported success in autocontouring the great vessels and heart chambers, whereas the coronary arteries and heart valves have too much variability to be reliably used, despite a wide variety of planning techniques[3,6,8,10].
In the current report, the quality and volume agreement of the studied structures improved when using a 30 vs 20 patient atlas, however this trend did not continue when 40 patients were used. The reason for this discrepancy is likely due to anatomic variances. The study cohort consisted of patients planned for intrathoracic SBRT, a group that often has a high proportion of cardiac disease (manuscript in press). As such, it is not uncommon to see cardiac hypertrophy, atherosclerosis, implantable hardware (pacemakers, cardiac stents), and other cardiovascular pathologies within this patient population. These factors can ultimately impact the ability to accurately autosegment heart substructures either due to the relative high hounsfield units of implants/calcium or differences compared to normal anatomy. Indeed, four patients within the 40-sample group who required a high degree of modification of autocontoured structures had such abnormalities (pacemaker and cardiac stents, 1 patient; sternotomy wires and atrial/ventricular hypertrophy, 1 patient; significant atherosclerosis and atrial/ventricular hypertrophy, 2 patients). Therefore, while increasing the sample number in the atlas database can improve the quality of generated structures, individual patient factors can still significantly impact autosegmentation.
There are several limitations in our study. The lack of intravenous contrast can limit the identification of cardiac substructures, particularly the coronary arteries and heart valves. However, given that these are small volume structures significantly impacted by cardiac motion and the difficulty that previous studies had with autosegmenting these subsites, contrast alone may not be sufficient to overcome this issue. Additionally, the conducting pathways of the heart cannot be reliably identified on CT[5]. Future studies incorporating cardiac magnetic resonance imaging could be useful in this setting. Lastly, while increasing the number of patients in the MIM atlas did not consistently improve contour quality in this study, given the limited number patients it is unclear whether this is applicable to much larger datasets. Strengths of this study includes the use of respiratory gated planning CTs, as this technique is commonly used for motion management in intrathoracic SBRT. While respiratory gating can amplify cardiac motion, the current study demonstrates that MIM autosegmentation is viable for the great vessels and heart chambers thus enabling a practical approach to dose calculation to these structures. Future studies will use this approach to evaluate radiation dose to cardiac substructures during SBRT.
Accurate segmentation of various organs in the heart is necessary to understand and correlate cardiac toxicities in the management of NSCLC lung cancer patients being treated using SBRT dose delivery technique. Even though human heart is associated with non-voluntary motion, thereby, exhibiting motion artifacts during imaging, use of MIM software with 30-sample subject library was found to be useful in delineating majority of the heart substructures with acceptable clinical accuracy except some structures having very little volume. Moreover, the autosegmented structures need to be evaluated on case by case basis by the treating physician as presence of calcifications or other sources of imaging artifacts like pacemaker, etc., may inhibit the model to accurately auto segment the heart sub-structures.
Cardiotoxicity from thoracic radiotherapy can significantly contribute to treatment related morbidity and mortality. Despite this, cardiac substructures are not routinely delineated in thoracic radiation planning.
Autosegmentation of cardiac substructures would allow for relative dose calculation to these subsites without the added labor of manual definition.
To determine whether autosegmentation software can be successfully employed for the cardiac substructures in patients planned using respiratory gated, non-contrasted computed tomography (CT) imaging.
This retrospective study included patients who underwent stereotactic body radiation therapy (SBRT) for inoperable, early-stage non-small cell lung cancer from 2007 to 2019. All patients were simulated via CT imaging with respiratory gating without intravenous contrast. A 20-patient atlas of the cardiac substructures was manually constructed and used to facilitate autosegmentation via MIM software. A total of three iterations of autosegmentations were completed, each using 10 patients. Generated structure quality was evaluated by degree of required manual edits and volume discrepancy between the autocontoured structures and its edited sister structure.
The great vessels and heart chambers were reliably autosegmented with most edits considered minor. In contrast, coronary arteries either failed to be autosegmented or the generated structures required major alterations necessitating deletion and manual definition. Similarly, the generated mitral and tricuspid valves were poor whereas the aortic and pulmonary valves required at least minor and moderate changes respectively. For the majority of subsites, the additional samples did not appear to substantially impact the quality of generated structures. Volumetric analysis between autosegmented and its manually edited sister structure yielded comparable findings to the physician-based assessment of structure quality.
Our study indicated that the great vessels and heart chambers can be reliable autocontoured using MIM software. On the other hand, autosegmentation for valves is inconsistent and poor for coronary arteries. Anatomic variances and/or implanted hardware may impact the quality of autosegmentation.
Radiation heart dose is an important dosimetric parameter however dose tolerances for the cardiac substructures in conventional therapy and SBRT are not well-established. Therefore, artificial intelligence based contouring programs allow dose to be calculated to the select cardiac subsites without the added labor of manual definition.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Que J S-Editor: Huang P L-Editor: A P-Editor: Wang LL
| 1. | Bradley JD, Hu C, Komaki RR, Masters GA, Blumenschein GR, Schild SE, Bogart JA, Forster KM, Magliocco AM, Kavadi VS, Narayan S, Iyengar P, Robinson CG, Wynn RB, Koprowski CD, Olson MR, Meng J, Paulus R, Curran WJ Jr, Choy H. Long-Term Results of NRG Oncology RTOG 0617: Standard- Versus High-Dose Chemoradiotherapy With or Without Cetuximab for Unresectable Stage III Non-Small-Cell Lung Cancer. J Clin Oncol. 2020;38:706-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 287] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 2. | Thor M, Deasy JO, Hu C, Gore E, Bar-Ad V, Robinson C, Wheatley M, Oh JH, Bogart J, Garces YI, Kavadi VS, Narayan S, Iyengar P, Witt JS, Welsh JW, Koprowski CD, Larner JM, Xiao Y, Bradley J. Modeling the Impact of Cardiopulmonary Irradiation on Overall Survival in NRG Oncology Trial RTOG 0617. Clin Cancer Res. 2020;26:4643-4650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 3. | McWilliam A, Khalifa J, Vasquez Osorio E, Banfill K, Abravan A, Faivre-Finn C, van Herk M. Novel Methodology to Investigate the Effect of Radiation Dose to Heart Substructures on Overall Survival. Int J Radiat Oncol Biol Phys. 2020;108:1073-1081. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 4. | Reshko LB, Kalman NS, Hugo GD, Weiss E. Cardiac radiation dose distribution, cardiac events and mortality in early-stage lung cancer treated with stereotactic body radiation therapy (SBRT). J Thorac Dis. 2018;10:2346-2356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Feng M, Moran JM, Koelling T, Chughtai A, Chan JL, Freedman L, Hayman JA, Jagsi R, Jolly S, Larouere J, Soriano J, Marsh R, Pierce LJ. Development and validation of a heart atlas to study cardiac exposure to radiation following treatment for breast cancer. Int J Radiat Oncol Biol Phys. 2011;79:10-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 442] [Cited by in F6Publishing: 498] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 6. | Finnegan R, Dowling J, Koh ES, Tang S, Otton J, Delaney G, Batumalai V, Luo C, Atluri P, Satchithanandha A, Thwaites D, Holloway L. Feasibility of multi-atlas cardiac segmentation from thoracic planning CT in a probabilistic framework. Phys Med Biol. 2019;64:085006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Lorenzen EL, Ewertz M, Brink C. Automatic segmentation of the heart in radiotherapy for breast cancer. Acta Oncol. 2014;53:1366-1372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Luo Y, Xu Y, Liao Z, Gomez D, Wang J, Jiang W, Zhou R, Williamson R, Court LE, Yang J. Automatic segmentation of cardiac substructures from noncontrast CT images: accurate enough for dosimetric analysis? Acta Oncol. 2019;58:81-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Shahzad R, Bos D, Budde RP, Pellikaan K, Niessen WJ, van der Lugt A, van Walsum T. Automatic segmentation and quantification of the cardiac structures from non-contrast-enhanced cardiac CT scans. Phys Med Biol. 2017;62:3798-3813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Zhou R, Liao Z, Pan T, Milgrom SA, Pinnix CC, Shi A, Tang L, Yang J, Liu Y, Gomez D, Nguyen QN, Dabaja BS, Court L, Yang J. Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiother Oncol. 2017;122:66-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 11. | Duane F, Aznar MC, Bartlett F, Cutter DJ, Darby SC, Jagsi R, Lorenzen EL, McArdle O, McGale P, Myerson S, Rahimi K, Vivekanandan S, Warren S, Taylor CW. A cardiac contouring atlas for radiotherapy. Radiother Oncol. 2017;122:416-422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 12. | Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, van Ginneken B. Multi-atlas-based segmentation with local decision fusion--application to cardiac and aortic segmentation in CT scans. IEEE Trans Med Imaging. 2009;28:1000-1010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 214] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 13. | Daly ME, Perks JR, Chen AM. Patterns-of-care for thoracic stereotactic body radiotherapy among practicing radiation oncologists in the United States. J Thorac Oncol. 2013;8:202-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Pan H, Simpson DR, Mell LK, Mundt AJ, Lawson JD. A survey of stereotactic body radiotherapy use in the United States. Cancer. 2011;117:4566-4572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 15. | Singh AK, Gomez-Suescun JA, Stephans KL, Bogart JA, Hermann GM, Tian L, Groman A, Videtic GM. One Versus Three Fractions of Stereotactic Body Radiation Therapy for Peripheral Stage I to II Non-Small Cell Lung Cancer: A Randomized, Multi-Institution, Phase 2 Trial. Int J Radiat Oncol Biol Phys. 2019;105:752-759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 16. | Milo MLH, Offersen BV, Bechmann T, Diederichsen ACP, Hansen CR, Holtved E, Josipovic M, Lörincz T, Maraldo MV, Nielsen MH, Nordsmark M, Nyström PW, Pøhl M, Rose HK, Schytte T, Yates ES, Lorenzen EL. Delineation of whole heart and substructures in thoracic radiation therapy: National guidelines and contouring atlas by the Danish Multidisciplinary Cancer Groups. Radiother Oncol. 2020;150:121-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 17. | Kirişli HA, Schaap M, Klein S, Papadopoulou SL, Bonardi M, Chen CH, Weustink AC, Mollet NR, Vonken EJ, van der Geest RJ, van Walsum T, Niessen WJ. Evaluation of a multi-atlas based method for segmentation of cardiac CTA data: a large-scale, multicenter, and multivendor study. Med Phys. 2010;37:6279-6291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |