Epidemiological and viral genome characteristics of the first human H7N9 influenza infection in Guangdong Province, China
Background
On 19 February 2013, the first avian influenza A (H7N9) virus was identified in humans, initiating an influenza outbreak in Eastern and Northern China in domestic birds and humans. As of 27 July, the case-fatality rate in 135 laboratory confirmed human cases was reported to be 32.6% (1). Phylogenetic analysis revealed that the gene segments of H7N9 virus stemmed from three reassortment events: the H7 gene segment are closest to hemagglutinin (HA) of the H7N3 of wild ducks, the six internal genes clustered with those of poultry H9N2 viruses of China (2), and the N9 gene segment shared the highest similarity with wild ducks in South Korea (3) and Baikal teal from Jiangsu Province in China (4). Because the poultry markets are possible locations for the emergence of new influenza viruses by mutations and/or reassortments (5), more monitoring of poultry markets has occurred recently. As a result, many control policies for prevention and response to H7N9 outbreaks have been reinforced, and surveillance has been increased to find people with symptoms early, even in provinces in China that have not had a human H7N9 case before, such as Guangdong Province.
To date, there have been three stages of the H7N9 epidemic in humans. The first stage consisted of sporadic isolations of virus in February and March, followed by an epidemic in April and May in Eastern and Northern China (6). The third stage is currently ongoing, with sporadic H7N9 virus isolations in China. Avian influenza A (H5N1) and seasonal influenza viruses have shown a seasonal pattern whereby human cases are much more common in winter and autumn than in summer. However, two H7N9 human avian influenza cases recently occurred in the summer; these cases have again attracted global concerns of H7N9 influenza. The first of these summer cases was a 61-year-old female from Hebei Province on July 27, 2013. The second case, from Guangdong Province, where had no reported human H7N9 cases prior to this isolation, was a previously healthy 51-year-old female worker with a long history of contact with live poultry.
Here, we analyze the epidemiology and genome features of the first human infected H7N9 virus in Southern China (Guangdong Province), to investigate the H7N9 virus source and transmission in the local community.
Methods
Epidemiological data collection
Collection of data from the H7N9 patient and her close contacts was required by the National Health and Family Planning Commission and conducted by Guangdong Center for Disease Control and Prevention; it was exempt from ethical review by an institutional review board. The information collected included medical records, exposure history and time line of events for the H7N9 patient and close contacts. All close contacts, as well as the environment in which the patient may have been exposed, were sampled to investigate transmission of H7N9 virus.
RNA extraction, genome sequencing, and phylogenetic analysis
The full genome of the virus was amplified by 12 primer sets (designed by our lab basing on the H7N9 sequences published on Genbank) using OneStep RT-PCR Kit (Qiagen, Hilden, Germany) for sequencing. PCR products were gel purified using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sequenced by ABI 3130xl automatic DNA analyzer (Life Technologies, Carlsbad, USA). The full genome of the virus was deposited into the GenBank database on 13 September 2013 (GenBank: KF662943-KF66295). Multiple alignments of HA and neuraminidase (NA) genes were conducted using the ClustalW algorithm. Phylogenetic trees were constructed by the maximum likelihood method with 1,000 bootstrap replications in ClustalW. Accession numbers for reference sequences used in the phylogenetic analysis are listed in Table S1.

Full table
Genetic signatures of A/Guangdong/1/2013(H7N9) (H7N9 Guangdong strain)
To identify the genetic signatures of virus, the full genome of the H7N9 Guangdong strain was compared with sequences of H7N9 viruses previously isolated from humans, birds, and the environment. In addition, key amino acid sequences that have been previously associated with viral characteristics were identified in the H7N9 Guangdong strain.
Results
Epidemiological features
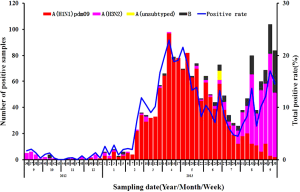
The seasonal distribution of influenza viruses in Guangdong province in 2012-2013 is shown in Figure 1. Before epidemic week (EW) 8 the influenza activity was low. However, after EW8, A (H1N1) pdm09 dramatically increased and predominated until EW30. Prior to August 2013, influenza B and H3N2 viruses were rarely detected, but both became more common in August, with H3N2 predominating. It is particularly intriguing that at EW17 and EW20 two samples from live poultry were found positive for avian influenza A (H7N9) virus in Dongguan and Zengcheng cities both of which located in Guangdong Province (7,8). Despite the fact that human H7N9 cases were distributed across most regions of Eastern or Northern China before July, the most recent animal and human cases detected were concentrated in the cities of the Pearl River delta in Southern China (Figure 2).
The H7N9 patient from Guangdong province did not have any recent travel history and lived in a two-story building, of which the ground floor for was used for selling poultry and the second floor for living. This building was located in a wet poultry market of Boluo (a county in Huizhou city). Because of H7N9 outbreaks reported in other regions in China, Guangdong performed emergency surveillance for avian influenza A (H7N9) in wet poultry markets throughout all 21 prefectures from 15 April to 31 May 2013. A total of 3,235 samples were collected from wet market environments, including 120 samples from Huizhou. No sample was found to be H7N9 positive in this surveillance. We tested 65 throat swabs and 125 blood samples from the case’s close contacts, including her husbands, son, and daughter, but none tested positive for the A (H7N9) virus, suggesting that this patient did not spread the virus to other humans.
At the onset of disease, the patient developed fever, headache and chills. After receiving medical care in a private clinic for three days without improvement, she visited an outpatient department of a county hospital on day 4, and had watery stools and fatigue on day 6. She was hospitalized at Huizhou central hospital on day 7, her condition became more severe, and she was transferred to the intensive care unit (ICU) because of an infection in the right lung on chest radiograph on day 9. Although an influenza test was negative on day 9, the patient became critically ill and was started on a ventilator. A sample from the patient on day 10 was positive for avian influenza A (H7N9) virus, detected by Huizhou Centers for Disease Control and Prevention (CDC) and confirmed by China CDC. On day 13, the patient was transferred to the Guangzhou Institute of Respiratory Diseases for further treatment. The patient recovered and was discharged uneventfully on day 53 (Figure 3).

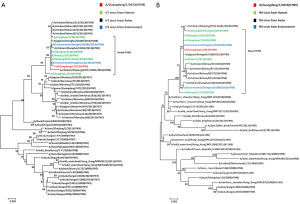
Phylogenetic analysis of A/Guangdong/1/2013 (H7N9)
An H7N9 virus, named A/Guangdong/1/2013 (H7N9), was isolated from day 14 sample (BALF) of the patient using 9-11-day-old embrocated chicken eggs in a BSL-3 laboratory. Multiple sequence alignment showed that the Guangdong strain was 96-100% identical to the first three reported avian influenza A (H7N9) viruses in all eight genes (Table 1), including 99% identical in the HA and NA genes. Blast analysis of the viral genome showed that each gene was from avian origin, and no evidence of genetic reassortment with human viruses was found (Table 1). Phylogenetic analysis suggested that A/Guangdong/1/2013 (H7N9) has similar antigen city to previous avian influenza A (H7N9) viruses reported in China (Figure 4). The HA gene shared the highest identity with A/environment/Hangzhou/34/2013 (H7N9), while the NA gene was most closely related to A/environment/Hangzhou/34/2013 (H7N9) and A/Hangzhou/2/2013 (H7N9) (Table 1). The HA and NA genes were 99.6% and 99.9% identical, respectively, with genes of H7N9 viruses detected in the local environment.

Full table
Genetic signatures of A/Guangdong/1/2013 (H7N9)
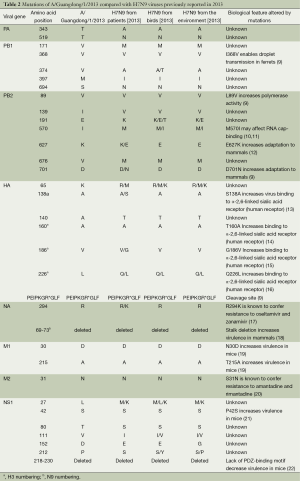
The genome signatures of Guangdong strain were identified, including T160A, V186G, and Q266L in HA, N30D in M1, and E627K in PB2 (Table 2). These mutations were observed in most of the avian influenza A (H7N9) viruses isolated from humans in the epidemic of 2013, and appear to have allowed the virus to adapt to humans by increasing binding to the human receptor as well as increasing virus virulence. Although the genome of A/Guangdong/1/2013 (H7N9) was similar to previously isolated H7N9 viruses in China, some unique mutations existed in the PA, PB1, PB2, HA and NS1 genes when compared with other H7N9 viral genes. However, there is a lack of published evidence that these mutations affect the biological properties of H7N9 viruses. Interestingly, 191E in PB2, 65K in HA and 27L, 111V, and 212P in NS1 were observed in A/Guangdong/1/2013 (H7N9) and the viruses from birds and the environment, but were not seen in other H7N9 viruses isolated from humans. This finding implies that the Guangdong H7N9 patient may have been infected from birds or the environment, due to her working in a poultry market.
Full table
Discussion
In the 20th century, three of four influenza pandemics were thought to have originated in Southern China (23,24). Guangdong Province has often been the focus of attention in influenza epidemiology. In this study, we analyzed the epidemiology and genetic signatures of the H7N9 virus from the first human H7N9 case in Guangdong Province, and compared the virus to those isolated from birds, the environment, and humans in Eastern and Northern China.
Live poultry markets bring together bird and poultry species from different sources in a concentrated environment, providing an environment for reassortment among avian influenza viruses of different subtypes (25,26). Most human H7N9 infections were diagnosed in poultry workers or visitors to poultry markets, implying that the market environment plays a critical role in the epidemic (27). In this study, the patient who is a poultry worker has been involved in this field for over 10 years. That supports the notion that avian influenza A (H7N9) virus-infected poultry are a transmission source.
Many studies based on retrospective investigations have stated that poultry market exposure is a key risk factor for H7N9 infection. Notably, before the isolation of A/Guangdong/1/2013 (H7N9), avian influenza A (H7N9) viruses were isolated from chickens in live poultry markets of the nearby cities of Dongguan and Zengcheng in Guangdong Province. This finding provides chronological evidence for poultry markets as a source of H7N9 infection. Therefore, it is thought that those who have close contact to live poultry in Southern China have a high risk of infection with avian influenza A (H7N9) virus. Vigilance is needed to continue surveillance of live poultry markets and those who frequent them.
Guangdong Province, located in a subtropical area, has multiple peaks of seasonal influenza activity annually, and the highest peak tends to occur in warmer months (June and July) (28). The activity of avian influenza A (H5N1) is similar to that of seasonal influenza, in that in tropical areas of Asia, avian influenza A (H5N1) has been more common during the warmer months (29). Therefore, this human H7N9 case in Guangdong province occurring in summer season followed the expected seasonality. However, no further positive detection of avian influenza A (H7N9) virus in local live poultry markets and no additional human case has been identified within one month after this isolation in Guangdong or adjacent southern provinces in China. Hence, it does not appear that H7N9 virus is currently moving south.
The avian influenza A (H7N9) virus we identified from the first human case in Guangdong was closely related to A/environment/Hangzhou/34/2013(H7N9) in its surface glycoprotein’s (HA and NA), and also has internal genes similar to other N7N9 viruses detected from birds and the environment. We compared the sequences of A/Guangdong/1/2013 (H7N9) with those of previously reported avian influenza A (H7N9) viruses. In the HA protein, A/Guangdong/1/2013 (H7N9) had a Q226L substitution, which increases virus binding to the human receptor; other previous avian influenza A (H7N9) viruses isolated from humans had the same mutation (12,30). In the PB2 protein, A/Guangdong/1/2013 (H7N9) had an E627K mutation; this change is thought to increase the virulence of avian viruses in mice. Avian influenza A (H7N9) viruses from birds and the environment typically have 627E. Interestingly, four previously reported human isolates of H7N9 viruses also had 627E in PB2, while two of them had D701N, which is thought to play a key role in viral adaption to mammals and take the place of the E627K mutation (31). This implies that there is more than one possible PB2 mutation for H7N9 viruses to adapt to mammalian hosts. However, it is worth noting that 191E in PB1, 570I in PB2, 65K in HA, and 27L, 111V, and 212P in NS1 were only observed in A/Guangdong/1/2013 (H7N9) and in H7N9 virus isolates from birds and the environment. Among these mutations, M570I is located in the binding region of PB2 and is reported to possibly alter the secondary structure from a strand to a helix, and may affect RNA cap-binding (10,11). Several unique substitutions were observed in A/Guangdong/1/2013 (H7N9) compared with other avian influenza A (H7N9) viruses, including T343A in PA, which also has been found in A (H1N1) pdm09 viruses (32,33). Whereas effects of them on the fitness of viruses need to be studied further due to unknown biological function. Based on the findings above, to study the virus’s virulence more accurately and objectively, it is important that the sequence and virological analyses are considered in combination with the epidemiological findings.
Conclusions
Overall, the epidemiology and genome characteristics of A/Guangdong/1/2013 (H7N9) seemed to be similar with previously reported H7N9 viruses isolated from humans. However, several unique substitutions never reported before need to be investigated, and may already exist in the environment and live poultry locally. Although their impact on the pathogenesis of viruses is unclear, it is necessary to emphasize that agriculture and forestry departments should continue monitoring and sharing animal surveillance information, which can facilitate early warning and intervention.
Acknowledgements
This study was financially supported by Municipal Science and Technology Bureau Foundation of Guangzhou, the Science and Technology Planning Project of Guangdong Province, China (Guangdong–Macau Joint Research Centre for New Drug Discovery Against Respiratory Pathogens, Grant No. 2010B091000018), Emergency Response Project of Ministry of Science and Technology of China (Grant No. KJYJ-2013-01-05), National Science and Technology Major Project of the Ministry of Science and Technology of China (Grant No. 2014ZX10004006), and the National Key Technology R&D Program of the 12th National Five-year Development Plan of China (Grant No. 2012BAI05B01).
Authors’ contributions: Nan-shan Zhong and Yong-hui Zhang designed the study. Shi-guan Wu, Xiao-bo Li, Guo-yun Ding and Ji-cheng Huang conducted H7N9 nucleic acid detection and virus isolation in P3 lab. Wen-da Guan and Si-hua Pan sequenced the full genome of virus, did phylogenetic analysis. Run-feng Li, Min Kang, Jie Wu and Yong-ping Li did analysis of genetic features. Wei-qi Pan, Rong Zhou, Ling Chen, Rong-chang Chen, Yi-min Li collected clinical data. Jianfeng He, Chang-wen Ke, Jin-yan Lin and Wen-long Xiao collected and analyzed epidemiological data. Zi-feng Yang wrote the first draft, and all authors contributed to review and revision and have seen and approved the final version.
Disclosure: The authors declare no conflict of interest.
References
- World Health Organization. Human infection with avian influenza A(H7N9) virus – update. Available online: http://www.who.int/csr/don/2013_08_11/en/index.html
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013;368:1888-97. [PubMed]
- Liu D, Shi W, Shi Y, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 2013;381:1926-32. [PubMed]
- Xiong C, Zhang Z, Jiang Q, et al. Evolutionary characteristics of A/Hangzhou/1/2013 and source of avian influenza virus H7N9 subtype in China. Clin Infect Dis 2013;57:622-4. [PubMed]
- Lai KY, Wing YNG, Wong KF, et al. Human H7N9 avian influenza virus infection: a review and pandemic risk assessment. Emerging Microbes & Infections 2013;2:e48.
- Li Q, Zhou L, Zhou M, et al. Epidemiology of Human Infections with Avian Influenza A(H7N9) Virus in China. N Engl J Med 2014;370:520-32. [PubMed]
- Ministry of Agriculture of China. Avian influenza A(H7N9) virus–update 2013-05-05. Available online: http://www.moa.gov.cn/zwllm/zwdt/201305/t20130505_3450918.htm
- Ministry of Agriculture of China. Avian influenza A(H7N9) virus–update 2013-05-20. Available online: http://www.moa.gov.cn/zwllm/zwdt/201305/t20130520_3467279.htm
- To KK, Chan JF, Chen H, et al. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect Dis 2013;13:809-21. [PubMed]
- Honda A, Mizumoto K, Ishihama A. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 1999;4:475-85. [PubMed]
- Danishuddin, Khan AU. Analysis of PB2 protein from H9N2 and H5N1 avian flu virus. Bioinformation 2008;3:41-6. [PubMed]
- Hatta M, Gao P, Halfmann P, et al. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001;293:1840-2. [PubMed]
- Nidom CA, Takano R, Yamada S, et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis 2010;16:1515-23. [PubMed]
- Wang W, Lu B, Zhou H, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol 2010;84:6570-7. [PubMed]
- Yang H, Chen LM, Carney PJ, et al. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 2010;6:e1001081. [PubMed]
- Srinivasan K, Raman R, Jayaraman A, et al. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One 2013;8:e49597. [PubMed]
- McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol 1998;72:2456-62. [PubMed]
- Matsuoka Y, Swayne DE, Thomas C, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol 2009;83:4704-8. [PubMed]
- Fan S, Deng G, Song J, et al. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009;384:28-32. [PubMed]
- Hay AJ, Wolstenholme AJ, Skehel JJ, et al. The molecular basis of the specific anti-influenza action of amantadine. EMBO J 1985;4:3021-4. [PubMed]
- Jiao P, Tian G, Li Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol 2008;82:1146-54. [PubMed]
- Jackson D, Hossain MJ, Hickman D, et al. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A 2008;105:4381-6. [PubMed]
- Shortridge KF, Stuart-Harris CH. An influenza epicentre? Lancet 1982;2:812-3. [PubMed]
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009;459:931-9. [PubMed]
- Correa P, Haenszel W, Cuello C, et al. Gastric precancerous process in a high risk population: cross-sectional studies. Cancer Res 1990;50:4731-6. [PubMed]
- Lam TT, Wang J, Shen Y, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 2013;502:241-4. [PubMed]
- Han J, Jin M, Zhang P, et al. Epidemiological link between exposure to poultry and all influenza A(H7N9) confirmed cases in Huzhou city, China, March to May 2013. Euro Surveill 2013;18:20481. [PubMed]
- Deng A, He J, Kang M. Epidemiologic characteristics of influenza in Guangdong Province, 2005–2007. South China J Prev Med 2008;34:14-7.
- Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med 2006;355:2174-7. [PubMed]
- Li J, Yu X, Pu X, et al. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Sci China Life Sci 2013;56:485-92. [PubMed]
- de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006;12:1203-7. [PubMed]
- Yang JR, Huang YP, Chang FY, et al. New variants and age shift to high fatality groups contribute to severe successive waves in the 2009 influenza pandemic in Taiwan. PLoS One 2011;6:e28288. [PubMed]
- Xu L, Bao L, Zhou J, et al. Genomic polymorphism of the pandemic A (H1N1) influenza viruses correlates with viral replication, virulence, and pathogenicity in vitro and in vivo. PLoS One 2011;6:e20698. [PubMed]