Published online Jun 28, 2020. doi: 10.3748/wjg.v26.i24.3401
Peer-review started: December 29, 2019
First decision: April 1, 2020
Revised: April 24, 2020
Accepted: May 20, 2020
Article in press: May 20, 2020
Published online: June 28, 2020
Long noncoding RNAs (lncRNAs) are important regulators of cell processes that are usually dysregulated in gastric cancer (GC). Based on their high specificity and ease of detection in tissues and body fluids, increasing attention has spurred the study of the roles of lncRNAs in GC patients. Thus, it is necessary to elucidate the molecular mechanisms and further explore the clinical applications of lncRNAs in GC. In this review, we summarize current knowledge to examine dysregulated lncRNAs in GC and their underlying molecular mechanisms and activities in GC, which involve microRNA sponging, mRNA stability, genetic variants, alternative splicing, transcription factor binding, and epigenetic modification. More significantly, the potential of lncRNAs as prognostic, circulating, and drug-resistant biomarkers for GC is also described. This review highlights the method of dissecting molecular mechanisms to explore the clinical application of lncRNAs in GC. Overall, this review offers assistance in using lncRNAs as novel candidates for molecular mechanisms and for the identification of revolutionary biomarkers for GC.
Core tip: The noncoding genome exhibits an extensive landscape of cancer hallmarks. With the emergence of the promising effects of long noncoding RNAs (lncRNAs) in the treatment and diagnosis of cancer, advancements in the understanding of the molecular mechanisms of lncRNAs reveal a new era of therapeutic methods against gastric cancer and biomarkers. Although significant data imply the great translational application potential of lncRNAs in gastric cancer, these approaches still require further validation and a large cohort of patients to assess the long-term clinical outcomes.
- Citation: Gao Y, Wang JW, Ren JY, Guo M, Guo CW, Ning SW, Yu S. Long noncoding RNAs in gastric cancer: From molecular dissection to clinical application. World J Gastroenterol 2020; 26(24): 3401-3412
- URL: https://www.wjgnet.com/1007-9327/full/v26/i24/3401.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i24.3401
Gastric cancer (GC) is a type of malignant tumor with a high recurrence rate and is the third most dangerous malignant tumor after lung cancer and liver cancer worldwide[1,2]. GC accounted for over 1 million new diagnoses and nearly 800000 patient deaths in 2018[3]. Statistics indicate that the incidence and mortality of GC are increasing annually in low- and middle-income countries, especially in Eastern Asia, Eastern Europe, and South America[3,4]. Although the overall incidence of the disease showed a downward trend, gastroesophageal junction cancers and cases in selected special populations, such as young adults, and in developed countries are on the rise[5].
The carcinogenesis of GC is a multistage process with a slowly progressive and multifactorial pathology. The risk factors for GC include Helicobacter pylori infection, obesity, excessive ingestion of salt and nitrates, and blood group A[6]. In addition, the risk factors involved at molecular levels, such as genomic variation, abnormal regulation, and epigenetic alterations, also participate in the development of GC[7]. Thus, identification of specific and effective diagnostics, treatments, and prognostic biomarkers for GC is pivotal.
Accumulating evidence indicates that noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), act in the regulation of the carcinogenesis process of GC[8-12]. lncRNAs are RNA transcripts of more than 200 nucleotides that are not translated into proteins[13] and are less conserved than mRNAs in defining cell ontogeny[14]. lncRNAs are a highly versatile class of transcripts that have sparked new lines of research in nearly all fields of life sciences[15]. More recently, emerging studies have identified lncRNAs as major players in many types of cancer processes[16]. Increasing evidence has suggested that lncRNAs might be able to be used as a potential biomarker for diagnosis, medical treatment, and prognostics in human cancer[17-19].
Evidence suggests that aberrant expression of lncRNAs is associated with GC[11,20], and certain lncRNAs have been implicated in diagnosis and prognostication[21]. The molecular mechanisms of lncRNAs in GC, including genetic mutations, miRNA sponging, transcription factor (TF) interactions, and epigenomic modifications, have also been investigated over a wide range. Remarkably, certain new research directions have emerged in the field of lncRNAs related to GC. For example, studies have investigated the application of circulating lncRNAs in body fluids in GC patients. A number of research studies have shown that abnormal expression of lncRNAs is one of the major causes of drug resistance (drug-resistant lncRNAs). An increasing number of lncRNAs have been suggested to have prognostic value in predicting survival in patients with GC (prognostic lncRNAs). The role of lncRNAs in cancer has been gradually amplified, and the potential mechanism between lncRNAs and GC must be further elucidated. Therefore, this review aims to clarify the molecular regulatory mechanism of lncRNAs and their clinical applications in GC.
Numerous studies have shown that thousands of lncRNAs have the potential to act as “miRNA sponges” and are also recognized as competing endogenous RNAs (ceRNAs) that can decrease the number of miRNAs available to target mRNAs[22]. These results suggest that mRNAs could share the same group of miRNA binding sites and thus the function of indirectly regulating lncRNA cellular abundance based on competition for miRNA binding[23]. There are four major features of ceRNAs: (1) The effectiveness of their interactions can be appreciably impacted by the relative concentration of the lncRNAs and the corresponding miRNAs in a ceRNA motif; (2) The function of miRNA suppression of ceRNAs can be effective when the expression of ceRNAs is changed to a certain extent under diverse conditions; (3) Subcellular localization and tissue or cell specificity are key factors for the range of miRNAs in a ceRNA; and (4) miRNA response elements (MREs) are unequal in ceRNAs. For example, certain differences exist in the nucleotide composition of MREs for two diverse MREs of the same miRNA, resulting in the diverse affinity of each binding MRE. The ability of ceRNAs to identify carcinogenesis, cancer development, and cancer progression has been continually verified.
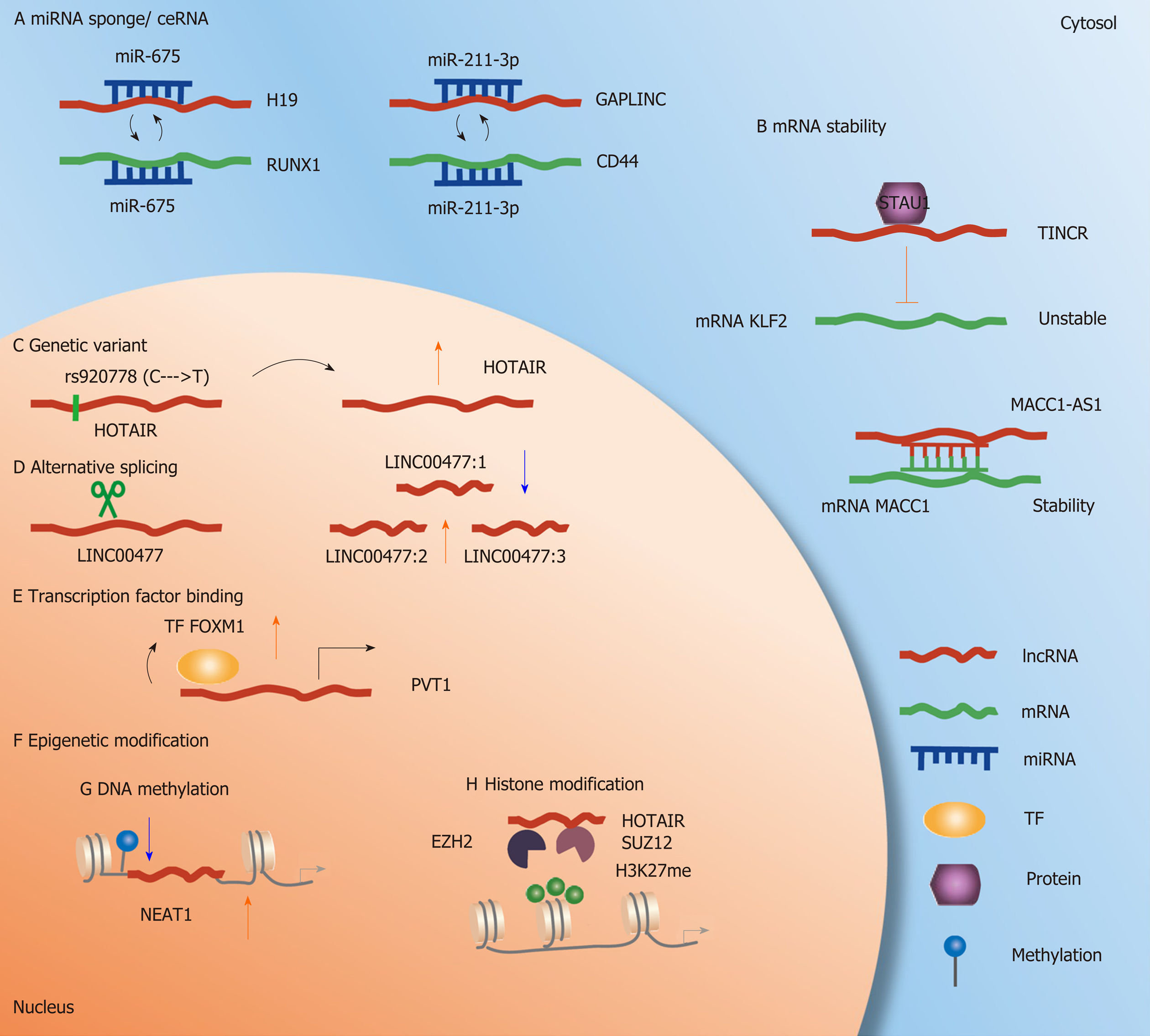
In GC, a large number of lncRNA-mediated ceRNAs have been identified to contribute to the exploration of the molecular mechanisms of carcinogenesis and to develop novel treatment methods in GC (Table 1). For example, studies have reported that some lncRNAs are abnormally upregulated[24-26] and associated with the proliferation of GC cells. The expression of miR-675 and lncRNA H19 shows a strong positive correlation and can act as a potential mediator for H19-related GC cells (Figure 1A). Comprehensive analysis shows that RUNX1-mediated GC cell phenotypic changes are induced by H19/miR-675. Interestingly, this ceRNA has been validated in many studies[27-29]. Other studies have reported that GAPLINC, an lncRNA located at chromosome 18, is upregulated in GC compared with normal matched tissues[30]. GAPLINC is necessary for the efficient invasion and proliferation of GC cells. GAPLINC inhibition and gene expression profiling in GC cells show changes in the cell migration pathway, with a high correlation with CD44 expression. Additionally, the lncRNA GAPLINC and CD44 can both target miR-211-3p according to predictive computational methods. GAPLINC can regulate CD44 expression by competing with miR211-3p in vitro and in vivo[30] (Figure 1A). Collectively, the novel association between gene expression disorders and lncRNA-mediated ceRNAs offers novel directions for understanding GC from a global perspective.
| lncRNA | Synonym | Pattern | miRNA | Function | Ref. |
| H19 | ENSG00000130600 | Up | miR-675 | Proliferation and invasion | [27-29] |
| GAPLINC | ENSG00000266835 | Up | miR-211-3p | Invasion and survival | [30] |
| ANRIL | ENSG00000240498 | Up | miR-99a/449a | Tumor growth and invasion | [80] |
| BANCR | ENSG00000278910 | Up | miR-532 | Angiogenesis and metastasis | [81] |
| GAS5 | ENSG00000234741 | Down | miR-23a | Critical role in tumorigenesis | [82] |
| HOTAIR | ENSG00000228630 | Up | miR-331-3p | Unknown | [83] |
| MALAT1 | ENSG00000251562 | Up | miR-23b-3p | Related to chemoresistance | [84] |
| MEG3 | ENSG00000214548 | Down | miR-21 | Suppression of epithelial-mesenchymal transition | [85] |
| NEAT1 | ENSG00000245532 | Up | miR-506 | Critical role in tumorigenesis | [86] |
| SNHG12 | ENSG00000197989 | Up | miR-199a/b-5p | Proliferation | [87] |
mRNA can stabilize or decay target transcripts by binding lncRNAs. The expression of the lncRNA TINCR is higher in GC tissues than in control normal tissues. Patients with high TINCR expression in GC cells are associated with a poorer prognosis. Results indicate that the lncRNA TINCR can affect the stability and expression of KLF2 mRNA by binding to the STAU1 protein. KLF2 regulates the transcription and expression of the cyclin-dependent kinase genes CDKN1A/p21 and cdkn2b/p15, thus influencing the proliferation and apoptosis of GC cells[31] (Figure 1B). The lncRNA MACC1-AS1 is the antisense lncRNA of MACC1, and its expression in GC tissues is significantly higher than that in normal control tissues, indicating a poor prognosis in GC patients. MACC1-AS1 is increased under metabolic stress, and MACC1 expression is promoted by enhancing the stability of MACC1 mRNA, thus promoting metabolic plasticity[32] (Figure 1B). A similar mechanism is used by the lncRNA EGFR-AS1. The lncRNA EGFR-AS1 is significantly upregulated in GC tissues, and the expression of EGFR-AS1 and EGFR is positively correlated in GC tissues. Furthermore, in vitro and in vivo data indicate that the proliferation of GC cells can be inhibited via knockdown of EGFR-AS1. According to its mechanism, EGFR mRNA stability is decreased and leads to the downregulation of EGFR with the depletion of EGFR-AS1[33]. In summary, alteration of mRNA stability is crucial for lncRNAs to play their roles in GC.
Single nucleotide polymorphisms (SNPs) are a common type of genetic variant and are considered to be genomic variation phenomena that directly or indirectly alter gene expression, protein activities, and signaling pathways[34]. A number of genetic variants have been identified in human lncRNA regions and are reported to be related to various diseases, including cancers. Compared with CC carriers in Jinan and Huainan populations, HOTAIR rs920778 TT carriers have an increased risk of GC. In detecting the functional correlation of the rs920778 SNP, researchers observed that rs920778 regulated the allele of HOTAIR expression in GC cell lines and tissue samples, and the expression level of HOTAIR was higher in T allele carriers[35] (Figure 1C). SNPs influence gene expression and dysregulation and further contribute to cancer development or prognosis. Wang et al[36] indicated that patients with the GA genotype of the rs2839698 SNP in a group treated with surgery alone had significantly lower risks of recurrence and death and significantly increased survival. Chen et al[37] indicated that GC patients with the AG or GG genotype of rs3737589 had lower overall survival rates than those with the AA genotype. These results indicated that the risk of GC increases if the lncRNA TP73-AS1 rs3737589 polymorphism is present. Moreover, the rs3737589 polymorphism might become a potential specific biomarker for predicting the survival of GC patients[37]. Other types of genetic variants, such as copy number variants (CNVs), lead to the overexpression of oncogenes or the inactivation of tumor suppressor genes and have important effects on cellular functions in cancers[38,39]. CNV is a type of genomic structural variation that leads to an abnormal copy number of genes, e.g., gene amplification, gain, loss, and deletion. Hu et al[30] indicated that one-third of dysregulated lncRNAs are related to CNVs in the genome of GC patients. Because lncRNAs might play a role in tumorigenesis, whether CNV-related lncRNA has a tumorigenic effect and its diagnostic or prognostic significance in GC deserve further study. The studies on associations between genetic variants and lncRNAs in GC are insufficient in both quantity and depth. Thus, further use of GWAS (genome-wide association study) and public databases, such as LincSNP 2.0[40], and the combination of these methods can be highly useful in the investigation of lncRNAs associated with various pathological conditions.
Alternative splicing is a type of post-transcriptional regulation that increases transcriptome diversity by producing multiple tissue- and cell-specific splicing subtypes[41]. The approach based on high-throughput sequencing and its application in transcriptome studies have changed our understanding of alternative splicing[42]. Alternative splicing of lncRNAs increases the diversity and functional capacity of lncRNAs at the post-transcriptional level in carcinogenesis and cancer progression. Conesa et al[43] reported that LINC00477 was downregulated in GC compared with normal gastric tissues at an overall level. Distinguished from normal coding RNAs, low abundance transcripts present a number of lncRNAs because of the large number of isoforms due to alternative splicing in RNA-Seq[43]. Conesa et al[43] suggested that the real expression level of the multiple isoforms of LINC00477 could not be reflected by RNA-Seq and reported that isoform 1 of LINC00477 was downregulated whereas isoform 2 was upregulated in GC cells both in vitro and in vivo (Figure 1D). Modulation of the phosphorylation of the SR splicing factor for MALAT1 can regulate alternative splicing in interphase cells. However, this role of MALAT1 has not been reported in GC, and a large gap still exists in the field of alternative splicing of lncRNAs in GC.
The majority of studies that address lncRNAs focus on defining their regulatory functions, whereas few investigations are directed toward assessing how lncRNAs themselves are transcriptionally regulated. Many studies have suggested that dysregulated lncRNAs are associated with many types of human cancers and other types of complex diseases[44,45] and that dysregulation is common because of the abnormal expression of TFs. TFs perform their functions by binding to the promoters of downstream lncRNAs and activating their transcriptional activities. Many key and essential TFs, such as p53[46], E2F1[47], and EZH2[48], can lead to abnormal expression of lncRNAs in cancers. In GC tissues, the TF SP1 regulates the lncRNA AGAP2-AS1 and leads to its higher expression in GC tissues. AGAP2-AS1 could inhibit P21 and E-cadherin to play an essential role in GC development[49]. Zhang et al[50] revealed that LINC00668 was a direct target of E2F1 and that upregulation of E2F1 might partially contribute to the overexpression of LINC00668 in GC. In addition, there are other types of regulation of lncRNAs mediated by TFs in GC.
In a previous study, the authors demonstrated a positive feedback loop between lncRNA PVT1 and TF FOXM1 in GC patients. The stability of the FOXM1 protein was enhanced with PVT1 binding. The TF FOXM1 also binds directly to and constitutively transactivates the PVT1 promoter. Transcription-induced lncRNAs promote the function of mRNA by stabilizing the transcription of the coding protein, which can be considered a novel mechanism[51] (Figure 1E). Additional regulatory mechanisms of lncRNAs mediated by TFs in GC should be investigated and explored.
Epigenetics is the study of the heritable changes in DNA that can influence the packaging of chromatin. These changes are not caused by any modification in the primary DNA sequence[52,53]. lncRNAs affect the expression of central genes through various epigenetic regulatory mechanisms during GC development, such as DNA methylation, histone modification, and chromatin remodeling[54].
One of the major and common forms of epigenetic modification is DNA methylation[55,56]. DNA methylation usually occurs in CpG islands (cytosine-guanine dinucleotides) or CG-rich regions, which are typically located upstream of the promoter region. DNA methylation of gene promoters is essential for gene silencing and is frequently found in various cancers. Similar to coding genes, DNA methylation of lncRNAs can also regulate the expression of lncRNAs. For example, NEAT1 is a potential binding lncRNA of ALKBH5. In GC cells and tissues, NEAT1 is upregulated and associated with invasion and metastasis. ALKBH5 affects the methylation level of NEAT1 and further accelerates the invasion and metastasis of GC[57] (Figure 1F and G). In vivo and in vitro data found the lncRNA AK058003 is overexpressed under hypoxia in GC patients and accelerates GC migration and invasion. AK058003 can also mediate the metastasis of hypoxia-induced GC cells. The reduction of CpG island methylation in SNCG by the lncRNA AK058003 increases the expression of SNGG under hypoxic conditions. In addition, DNA methylation changes by interacting with DNA methyl transferees (DNMTs) are also found in many lncRNAs in cancers, including GC. In GC tissues and cell lines, upregulation of the lncRNA ecCEBPA inhibits CEBPA promoter methylation because of the interaction of ecCEBPA with DNMT1[58].
The expression of the lncRNA MLK7-AS1 is higher in GC tissues than in matched normal tissues, and GC patients with higher MLK7-AS1 expression are associated with a poorer survival. These findings also suggest that cell proliferation and induced apoptosis can be inhibited via knockdown of MLK7-AS1 in GC cells.
MLK7-AS1 interacts with DNMT1 and recruits it to the miR-375 promotor, hyper-methylating the miR-375 promotor and repressing miR-375 expression in GC[59].
Histones are highly alkaline proteins that wind DNA to form nucleosomes. Histone modifications are considered to be structural changes in histones, and H1/H5, H2A, H2B, H3, and H4 are five families of histones[60,61]. Histone methylation and acetylation are two major types of histone epigenetic processes. Certain lncRNAs can alter the histone methylation status in GC by interacting with chromatin regulatory enzymes, particularly EZH2 (a subunit of PRC2). For example, the binding of EZH2 and H3K27 is reduced with EZH2 and SUZ12 knockdown, which verifies the association between the lncRNA HOTAIR and PRC2[62]. This result indicates that target genes can be silenced by HOTAIR, which recruits the PRC2 complex following H3K27me3 modification in the development of GC (Figure 1H). Histone acetylation is another type of histone modification. Data on GC tissues and cell lines show that the lncRNA MALAT1 is upregulated. The expression of MALAT1 is related to cell invasion and migration by enhanced EGFL7 expression via alteration of the level of H3 histone acetylation in the EGFL7 promoter region[63].
A biomarker is a type of biochemical index that can mark the alteration of a structure or function in subcellular structures, cells, tissues, organs, and systems and has broad application prospects. For the target population in cancers, biomarkers can be applied to many fields, such as diagnosis, staging, and evaluation of therapeutic effectiveness of novel drugs and therapies. In recent years, increasing numbers of studies have suggested that lncRNAs can become effective biomarkers for cancer prognosis and treatment.
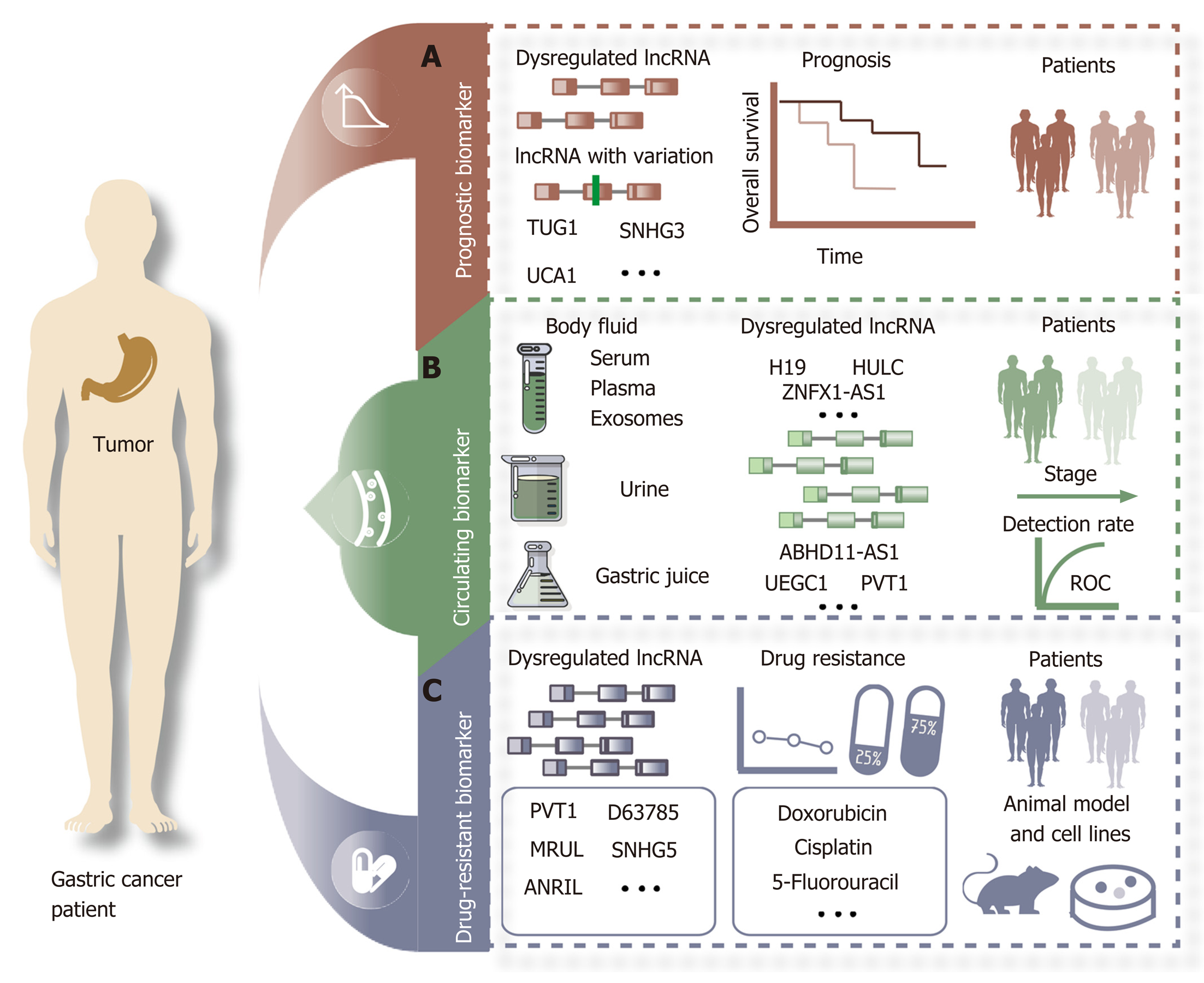
Prognosis is the assessment or prediction of the likely course of a disease and the chance of recovery or survival from diseases, including cancer. Predicting survival is a major challenge that requires a better understanding of the mechanism and treatment for GC. One of the major reasons for the poor prognosis of GC is the lack of early effective prognostic indicators. In recent years, increasing evidence has revealed that lncRNAs may become potential prognostic biomarkers for GC (Figure 2A). The expression of the lncRNA TUG1 is higher in GC tissues than in adjacent control tissues and shows a positive correlation with the invasion and stage of cancer. Higher TUG1 expression is associated with a poorer prognosis and may serve as a specific predictor of prognosis of GC[64]. Expression of the lncRNA SNHG3 is higher in many GC cell lines. In vitro and in vivo results from GC cells have revealed that SNHG3 can promote cell proliferation and metastasis. Overexpression of TUG1 is associated with unfavorable clinical outcomes[11]. The lncRNA UCA1 is upregulated in GC patients and promotes the proliferation and migration of GC cells. A higher UCA1 expression level is associated with a poor overall survival[65]. All of these studies indicate that dysregulated lncRNAs are associated with the survival of GC patients. In addition, variations of certain lncRNAs can also predict the survival of GC patients. For example, the SNP rs145204276 can regulate the expression level of the lncRNA GAS5 in GC samples. The SNP rs145204276 of GAS5 is associated with susceptibility to GC. Kaplan-Meier analysis shows that the genotype del/del of rs145204276 in the lncRNA GAS5 is significantly associated with a higher survival rate.
Based on their advantages of ease and lower invasiveness, circulating biomarkers are ideal in cancers compared with other types of biomarkers. For cancer detection, the advantages of lncRNAs, including good stability, high abundance, tissue specificity, and detection ability in body fluids, such as circulating exosomes, serum, plasma, and urine, make them ideal noninvasive biomarkers. lncRNAs are also detected in the gastric juice of GC patients (Figure 2B, Table 2). Xian et al[66] suggested that the expression of the lncRNAs ZNFX1-AS1 and HULC are differential in the plasma of GC patients compared with healthy control samples. The expression of HULC and ZNFX1-AS1 is higher in the peripheral blood of GC patients because they can be released into circulation via exosomes[66]. A study reported that expression of the lncRNA H19 was higher in the plasma of GC patients than in normal control samples. The expression level of H19 decreased significantly after surgical removal of gastric tumors in selected GC patients[67]. Exosomes are vesicles (30-150 nm) containing complex RNA and proteins. Body fluids such as blood, saliva, urine, cerebrospinal fluid, and milk are all found to have exosomes. Many studies have suggested that cancer cells release exosomes into peripheral circulation and that exosomal lncRNAs may more accurately show changes in GC[68]. The expression of the lncRNAs UEGC1 and UEGC2 was confirmed to be upregulated in exosomes and tumors in GC, especially in early GC patients. These results suggest that the tumor-originated exosomal lncRNA UEGC1 can be used as a circulating biomarker for early-stage GC[69]. Gastric juice is a body fluid that has many advantages, such as ease of sampling, and therefore could be applied in the diagnosis of GC. The expression level of PVT1 is confirmed to be higher in gastric juice from GC samples than in that from normal controls[70]. The expression of the lncRNA ABHD11-AS1 is higher in GC tissues and gastric juice than in control tissues and gastric juice from patients with other gastric diseases, such as mucosa or minimal gastritis, atrophic gastritis, and gastric ulcers. The expression level of the lncRNA ABHD11-AS1 in gastric juice is also correlated with tumor size and stage. The authors also indicated that the positive detection rate of early GC reached 71.4%[71]. Although lncRNAs might become superior biomarkers for GC, additional research based on large samples is required to confirm the sensitivity and specificity of circulating lncRNA biomarkers in GC.
In addition to surgical resection, chemotherapy is one of the main treatment methods for GC. One of the major reasons for chemotherapy failure in GC patients is multidrug resistance during the treatment process. Many studies have reported that lncRNAs can participate in drug resistance in cancer[72]. Abnormal expression of lncRNAs has been found in various stages, including drug resistance of GC cells (Figure 2C). The lncRNA D63785 is overexpressed in GC and promotes proliferation, migration, and invasion in GC. miR-422 expression is increased via knockdown of lnc-D63785, which makes GC cells more sensitive to doxorubicin[73]. The lncRNA PVT1 is dysregulated and promotes the progression of GC. PVT1 can enhance 5-fluorouracil resistance of GC cells by activating BCL2[74]. MRUL is an lncRNA located 400 kb downstream of ABCB1. In multidrug-resistant GC cell lines, including SGC7901/ADR and SGC7901/VCR, MRUL is upregulated. Knockdown of MRUL can reduce the expression level of ABCB1. All of these results suggest that the expression of ABCB1 is increased by MRUL, which is a drug target that can reverse the resistant phenotype in GC patients[75]. The expression of the lncRNA SNHG5 is higher in cisplatin-resistant GC patients than in cisplatin-sensitive GC patients[76].
Targeted therapy based on RNA is a novel treatment strategy for GC that can realize the goal of target specificity. lncRNAs could become a drug target based on the application of small interfering RNAs and the CRISPR-associated protein 9 (CRISPR/Cas9) system in GC. The expression of the lncRNA HOTAIR is higher in GC tissues and cell lines than in other tissues. The proliferation, migration, invasion, and metastasis of GC cells can be repressed by knockdown of HOTAIR[77,78]. The lncRNA TINCR is overexpressed in GC. The silencing of TINCR by siRNA can reduce proliferation, colony formation, and tumorigenicity[31]. Moreover, the lncRNA HULC is overexpressed in GC tissues compared with control normal tissues. Knockdown of HULC can enhance cisplatin-induced apoptosis in GC cells[79]. All of the data support the conclusion that lncRNAs could become direct therapeutic targets for certain drugs in GC. However, a novel therapeutic method based on lncRNAs in GC has not been translated into clinical treatment. More detailed and in-depth studies should be conducted on the roles of lncRNAs in GC drug resistance.
Initially considered to be transcriptional noise, lncRNAs have made significant contributions to various fields of cancer (including GC) study, including mechanistic and functional studies and translational treatments. As outlined in this review, strong evidence currently exists for a major role of lncRNAs in GC. Although the molecular mechanisms of lncRNAs are complex and many questions remain, a large number of lncRNAs have been revealed to perform key and essential functions in carcinogenesis, metastasis, prognosis, and drug resistance in GC. There is little doubt that lncRNAs play an essential role in GC and can become effective clinical biomarkers for diagnosis, prognosis, and treatment. However, certain key obstacles exist to the application of lncRNAs in clinical treatment. First, a systematic and comprehensive understanding of the mechanism of lncRNAs in GC is needed. Second, unknown physiological responses might occur when certain lncRNAs are silenced because of the complex regulatory interactions between lncRNAs and other genes or other types of ncRNAs. Third, accurate measurement of lncRNAs in body fluid and tissues is challenging because of their low abundance, ease of degradation, and instability. Fourth, although lncRNAs are considered a type of tissue-specific RNA, selected lncRNAs are still expressed in many types of tissues, which increases the difficulty of their clinical application for GC. Thus, substantial work remains on the road from molecular dissection to clinical application in GC compared with coding genes.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Chiba T, Ebrahimifar M, Youness RA, Ziogas DE S-Editor: Dou Y L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Smyth EC, Moehler M. Late-line treatment in metastatic gastric cancer: today and tomorrow. Ther Adv Med Oncol. 2019;11:1758835919867522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 2. | Russo AE, Strong VE. Gastric Cancer Etiology and Management in Asia and the West. Annu Rev Med. 2019;70:353-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 3. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50871] [Article Influence: 8478.5] [Reference Citation Analysis (44)] |
| 4. | Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2004] [Cited by in F6Publishing: 2263] [Article Influence: 251.4] [Reference Citation Analysis (0)] |
| 5. | Anderson WF, Camargo MC, Fraumeni JF Jr, Correa P, Rosenberg PS, Rabkin CS. Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA. 2010;303:1723-1728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 336] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 6. | Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015;16:e60-e70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 246] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 7. | Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 303] [Cited by in F6Publishing: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 8. | Dong L, Zhang Z, Xu J, Wang F, Ma Y, Li F, Shen C, Liu Z, Zhang J, Liu C, Yi P, Yu J. Consistency analysis of microRNA-arm expression reveals microRNA-369-5p/3p as tumor suppressors in gastric cancer. Mol Oncol. 2019;13:1605-1620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B, Zhao J, Xia S, Fan S, Yu X, Du Y, Hou L, Li Z, Ding Z, An S, Huang B, Li L, Tang J, Ju J, Guan H, Song B. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett. 2019;459:122-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 10. | Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol. 2019;11:1758835919874651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10:694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 12. | Zheng J, Zhang H, Ma R, Liu H, Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene. 2019;38:7073-7088. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 13. | Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 587] [Cited by in F6Publishing: 677] [Article Influence: 96.7] [Reference Citation Analysis (0)] |
| 14. | Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-1927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2555] [Cited by in F6Publishing: 2632] [Article Influence: 202.5] [Reference Citation Analysis (0)] |
| 15. | Grote P, Boon RA. LncRNAs Coming of Age. Circ Res. 2018;123:535-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 17. | Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL, Provero P, Church GM, Clohessy JG, Pandolfi PP. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 2018;173:649-664.e20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 210] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 18. | Gao Y, Wang P, Wang Y, Ma X, Zhi H, Zhou D, Li X, Fang Y, Shen W, Xu Y, Shang S, Wang L, Wang L, Ning S, Li X. Lnc2Cancer v2.0: updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019;47:D1028-D1033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 19. | Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018;28:287-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 372] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 20. | Cao C, Xu Y, Du K, Mi C, Yang C, Xiang L, Xie Y, Liu W. LINC01303 functions as a competing endogenous RNA to regulate EZH2 expression by sponging miR-101-3p in gastric cancer. J Cell Mol Med. 2019;23:7342-7348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 21. | Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, Wang Z, Fang JY, Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 22. | Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2305] [Cited by in F6Publishing: 2785] [Article Influence: 278.5] [Reference Citation Analysis (0)] |
| 23. | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 823] [Cited by in F6Publishing: 912] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 24. | Li P, Tong L, Song Y, Sun J, Shi J, Wu Z, Diao Y, Li Y, Wang Z. Long noncoding RNA H19 participates in metformin-mediated inhibition of gastric cancer cell invasion. J Cell Physiol. 2019;234:4515-4527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 25. | Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 26. | Mohamed WA, Schaalan MF, Ramadan B. The expression profiling of circulating miR-204, miR-182, and lncRNA H19 as novel potential biomarkers for the progression of peptic ulcer to gastric cancer. J Cell Biochem. 2019;120:13464-13477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Fijneman RJ, Anderson RA, Richards E, Liu J, Tijssen M, Meijer GA, Anderson J, Rod A, O'Sullivan MG, Scott PM, Cormier RT. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci. 2012;103:593-599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Liu G, Xiang T, Wu QF, Wang WX. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells. Oncol Res. 2016;23:99-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 29. | Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 30. | Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890-6902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 213] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 31. | Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, Huang MD, Shu YQ. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648-5661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 197] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 32. | Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, Dong S, Wen Z, Rao J, Liao W, Shi M. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 169] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 33. | Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y, Li X, Chen X. Long Noncoding RNA EGFR-AS1 Promotes Cell Proliferation by Increasing EGFR mRNA Stability in Gastric Cancer. Cell Physiol Biochem. 2018;49:322-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 34. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5242] [Cited by in F6Publishing: 5196] [Article Influence: 472.4] [Reference Citation Analysis (0)] |
| 35. | Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 117] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | Wang W, Yang Q, Huang Q, Zhang H, Zhang Z, Gao J, Ren W, Hu Y, Lin Y, Dang Y, Zhang F, Wang W, Wang L. The rs2839698 Single Nucleotide Polymorphism of lncRNA H19 is Associated with Post-Operative Prognosis in T3 Gastric Adenocarcinoma. Clin Lab. 2018;64:105-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Chen W, Xiao J, Shi L, Lin L, Jiang M, Ge Y, Li Z, Fan H, Yang L, Xu Z. Association of TP73-AS1 gene polymorphisms with the risk and survival of gastric cancer in a Chinese Han Population. Artif Cells Nanomed Biotechnol. 2019;47:3814-3822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J; Wellcome Trust Case Control Consortium, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1412] [Cited by in F6Publishing: 1371] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 39. | Liu H, Gu X, Wang G, Huang Y, Ju S, Huang J, Wang X. Copy number variations primed lncRNAs deregulation contribute to poor prognosis in colorectal cancer. Aging (Albany NY). 2019;11:6089-6108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Ning S, Yue M, Wang P, Liu Y, Zhi H, Zhang Y, Zhang J, Gao Y, Guo M, Zhou D, Li X, Li X. LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017;45:D74-D78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 41. | Ule J, Blencowe BJ. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol Cell. 2019;76:329-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 342] [Article Influence: 85.5] [Reference Citation Analysis (0)] |
| 42. | Zhao S. Alternative splicing, RNA-seq and drug discovery. Drug Discov Today. 2019;24:1258-1267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 43. | Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1325] [Cited by in F6Publishing: 1317] [Article Influence: 164.6] [Reference Citation Analysis (0)] |
| 44. | Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152-R161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 393] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 45. | Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577-4587. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 717] [Cited by in F6Publishing: 816] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 46. | Melo CA, Léveillé N, Rooijers K, Wijchers PJ, Geeven G, Tal A, Melo SA, de Laat W, Agami R. A p53-bound enhancer region controls a long intergenic noncoding RNA required for p53 stress response. Oncogene. 2016;35:4399-4406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 47. | Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086-1095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 48. | Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, Lu BB, Yin R, Zhang EB, Xu L, De W, Wang ZX. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 49. | Qi F, Liu X, Wu H, Yu X, Wei C, Huang X, Ji G, Nie F, Wang K. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10:48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 50. | Zhang E, Yin D, Han L, He X, Si X, Chen W, Xia R, Xu T, Gu D, De W, Guo R, Xu Z, Chen J. E2F1-induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget. 2016;7:23212-23226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 51. | Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q, Tan C, Ni SJ, Dong L, Yang Y, Lin W, Xu Q, Huang D, Huang Z, Ma Y, Zhang W, Sheng W, Du X. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res. 2017;23:2071-2080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 185] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 52. | Ashapkin VV, Linkova NS, Khavinson VKh, Vanyushin BF. Epigenetic mechanisms of peptidergic regulation of gene expression during aging of human cells. Biochemistry (Mosc). 2015;80:310-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 53. | Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20:245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 270] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 54. | Fattahi S, Kosari-Monfared M, Golpour M, Emami Z, Ghasemiyan M, Nouri M, Akhavan-Niaki H. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: A novel approach to personalized medicine. J Cell Physiol. 2020;235:3189-3206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 55. | Alipour S, Nouri M, Sakhinia E, Samadi N, Roshanravan N, Ghavami A, Khabbazi A. Epigenetic alterations in chronic disease focusing on Behçet's disease: Review. Biomed Pharmacother. 2017;91:526-533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 56. | Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1878] [Cited by in F6Publishing: 1851] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 57. | Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 199] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 58. | Nasrollahzadeh-Khakiani M, Emadi-Baygi M, Nikpour P. Augmented expression levels of lncRNAs ecCEBPA and UCA1 in gastric cancer tissues and their clinical significance. Iran J Basic Med Sci. 2017;20:1149-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 12] [Reference Citation Analysis (0)] |
| 59. | Quan Y, Zhang Y, Lin W, Shen Z, Wu S, Zhu C, Wang X. Knockdown of long non-coding RNA MAP3K20 antisense RNA 1 inhibits gastric cancer growth through epigenetically regulating miR-375. Biochem Biophys Res Commun. 2018;497:527-534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 60. | Bhasin M, Reinherz EL, Reche PA. Recognition and classification of histones using support vector machine. J Comput Biol. 2006;13:102-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 61. | Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, Zheng H, Wang H, Jiang Y, Xu L. LncRNA Structural Characteristics in Epigenetic Regulation. Int J Mol Sci. 2017;18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 62. | Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W, Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 63. | Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17:135-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 157] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 64. | Zhang E, He X, Yin D, Han L, Qiu M, Xu T, Xia R, Xu L, Yin R, De W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 128] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 65. | Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, Zhang ZZ. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 66. | Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689-4698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 67. | Yörüker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep. 2018;9:181-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 68. | Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226-1232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1828] [Cited by in F6Publishing: 1940] [Article Influence: 242.5] [Reference Citation Analysis (0)] |
| 69. | Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, Li BA, Cai JC, Cai WY. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 70. | Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63:442-449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 71. | Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, Xu C, Xiao B, Sun Y, Guo J. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37:1183-1188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 72. | Wang Y, Wang Z, Xu J, Li J, Li S, Zhang M, Yang D. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat Commun. 2018;9:3192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 73. | Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M, Ao X, Shan P, Tariq MA, Li P, Wang J. The Long Noncoding RNA D63785 Regulates Chemotherapy Sensitivity in Human Gastric Cancer by Targeting miR-422a. Mol Ther Nucleic Acids. 2018;12:405-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 74. | Du P, Hu C, Qin Y, Zhao J, Patel R, Fu Y, Zhu M, Zhang W, Huang G. LncRNA PVT1 Mediates Antiapoptosis and 5-Fluorouracil Resistance via Increasing Bcl2 Expression in Gastric Cancer. J Oncol. 2019;2019:9325407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 75. | Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182-3193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 76. | Li M, Zhang YY, Shang J, Xu YD. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23:4185-4191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 21] [Reference Citation Analysis (0)] |
| 77. | Emadi-Andani E, Nikpour P, Emadi-Baygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 78. | Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K, Shimosegawa T, Sugamura K, Satoh K. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 79. | Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC Enhances Chemotherapy Induced Apoptosis in Human Gastric Cancer. J Med Biochem. 2016;35:137-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 80. | Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen Jf. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276-2292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 299] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 81. | Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW, Gao Y, Xu Y, Chen C, Zheng ZS, Weng HW, Ye S, Kuang M, Xie D, Peng S. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660-2675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 82. | Liu X, Jiao T, Wang Y, Su W, Tang Z, Han C. Long non-coding RNA GAS5 acts as a molecular sponge to regulate miR-23a in gastric cancer. Minerva Med. 2016;. [PubMed] [Cited in This Article: ] |
| 83. | Elsayed ET, Salem PE, Darwish AM, Fayed HM. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int J Biol Markers. 2018;1724600818760244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 245] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 85. | Xu G, Meng L, Yuan D, Li K, Zhang Y, Dang C, Zhu K. MEG3/miR‑21 axis affects cell mobility by suppressing epithelial‑mesenchymal transition in gastric cancer. Oncol Rep. 2018;40:39-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 86. | Tan HY, Wang C, Liu G, Zhou X. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J Cell Biochem. 2019;120:4827-4836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 87. | Yang BF, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. 2018;22:1297-1306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 19] [Reference Citation Analysis (0)] |
| 88. | Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, Liang W, Hu C, Liu Y, Li J, Wang N, Wei B, Chen L. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7:213-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |