Published online Dec 28, 2017. doi: 10.3748/wjg.v23.i48.8500
Peer-review started: July 30, 2017
First decision: August 30, 2017
Revised: October 15, 2017
Accepted: November 8, 2017
Article in press: November 8, 2017
Published online: December 28, 2017
To investigated the mechanism of the association between the TBX21 T-1993C promoter polymorphism and autoimmune hepatitis type 1 (AIH-1) development.
In vivo, In vivo, and reporter analyses were performed to determine the function of transcription factors binding to the T-1993C element of the TBX21 promoter in human CD4+ T and B cell lines. Flow cytometry and quantitative real-time PCR were used to analyze T-box transcription factor (T-bet) and interferon-γ (IFN-γ) expressions in CD4+ T cells, B cells and monocytes from the peripheral blood of AIH-1 patients including 5-1993TC and 15-1993TT genotype carriers, and healthy controls including 10-1993TC and 25-1993TT genotype carriers. Furthermore, a range of biochemical indices was measured simultaneously in the blood of AIH-1 patients.
TBX21-1993C allele created a strong Yin-Yang 1 (YY1)-binding site and decreased transcriptional activity of TBX21 promoter in human CD4+ T and B cells. Higher levels of T-bet and IFN-γ were detected in the circulating CD4+ T cells and B cells of AIH-1 patients carrying the TBX21-1993 TT genotype compared with the patients carrying the -1993 TC genotype and controls with the -1993 TC genotype. T-bet expression levels of circulating T cells and B cells were positively correlated with AIH-1 disease activity. Knockdown of YY1 with siRNA caused increased expression of T-bet and IFN-γ in peripheral blood mononuclear cells in AIH-1 patients.
The repression of TBX21 expression by high-affinity binding of YY1 to the -1993C allele may contribute to a decreased development of AIH-1 via suppression of type 1 immunity.
Core tip: The -1993C allele in the TBX21 gene (encoding T-bet) promoter has been shown to associated with protection against autoimmune hepatitis type 1 (AIH-1), but the underlying mechanisms are unknown. We found that the TBX21-1993C allele created a strong Yin-Yang 1 (YY1)-binding site and decreased T-bet expression. Reduced T-bet and IFN-γ expression of circulating CD4+ T cells and B cells existed in the individuals carrying the -1993C allele compared with those without the -1993C allele and played a protective role in AIH-1 development. The repression of T-bet expression by high-affinity binding of YY1 to the -1993C allele may contribute to a decreased development of AIH-1 via the suppression of type 1 immunity.
- Citation: Sun W, Wu HY, Chen S. Influence of TBX21 T-1993C variant on autoimmune hepatitis development by Yin-Yang 1 binding. World J Gastroenterol 2017; 23(48): 8500-8511
- URL: https://www.wjgnet.com/1007-9327/full/v23/i48/8500.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i48.8500
Autoimmune hepatitis (AIH) is a progressive inflammatory liver disorder characterized by hypergammaglobulinemia, circulating autoantibodies, and histological evidence of a florid mononuclear cell infiltration, which is referred to as interface hepatitis[1,2]. AIH type 1 (AIH-1) is distinguished by the presence of circulating antinuclear antibodies (ANAs) and/or smooth muscle antibodies (SMAs), and is the predominant form of AIH in adults. Although genetic factors, such as human leucocyte antigen HLA-DR3 and HLA-DR4 are associated with the development of AIH, Other factors contributing to the aetiology and the pathogenic process of AIH remains unclear. While most patients with AIH respond well to immunosuppressive therapy, about 10%-20% of the cases still progress to cirrhosis and require the liver transplantation[3,4]. Therefore, It is important to understand the pathogenesis of AIH in the management of patients with AIH.
Although the pathogenic initiating events are unknown, AIH-1 is believed to be mediated by CD4 T cells that recognize one or more liver-specific self antigenic peptides, and histological evidences show the presence of interferon-γ (IFN-γ)-producing T helper type 1 (Th1) cells in liver[5,6]. These imply the importance of Th1-like immune response in the pathogenesis of AIH. A recent study suggests that B cells also contribute to the development and progression of AIH[7]. Activated B cells can differentiate into CD38+ plasma cells that secrete antibodies, such as ANA and SMA[8,9]. B effector 1 cells (Be1) can function as antigen presenting cells to present antigen determinants to trigger T-cell activation and secrete IFN-γ to regulate autoimmunity[10,11]. The T-box transcription factor, T-bet, is required for IFN-γ production and the generation of type 1 immunity[12]. Initial studies demonstrated that T-bet is expressed in developing the Th1 cells and mediates IFN-γ production, Th1 differentiation, and repression of the alternative Th2 program[13]. Recent studies have highlighted the importance of T-bet in other cellular subtypes implicated in the type 1 immune response, such as dendritic cells, natural killer (NK) cells, natural killer T (NKT) cells, and CD8+ T cells. T-bet has also been shown to regulate IFN-γ production by Be1 cells[14,15]. The high level of T-bet production which is associated with increased numbers of IFN-γ-producing B cells have been identified in the blood and lymphoid tissues of mice and humans with autoimmune disorders, such as systemic lupus erythematosus (SLE) and rheumatoid arthritis[10,11]. Furthermore, high T-bet production is associated with the pathogenesis of T-cell-mediated liver injury in conA murine model[16]. T-bet-deficient mice immunized with myelin oligodendrocyte glycoprotein were resistant to the development of experimental autoimmune encephalomyelitis[17]. In a murine lupus model, the absence of T-bet strikingly reduces B cell-dependent manifestations, including autoantibody production, hypergammaglobulinemia, and immune-complex renal disease, and abrogates IFN-γ-mediated IgG2a production[18]. Although the role of T-bet has been studied in a variety of autoimmune diseases in mice, its relative importance in human AIH is not characterized.
Genetic variations in the nucleotide sequences of promoters can contribute to the altered expression of genes in complex inherited diseases[19,20]. In this regard, a genetically induced increase in T-bet production may result in the expansion of either polarized Th1 cells to promote the type 1 immune response or the Be1 cell compartment to gain a greatly enhanced T cell activation capability. Three single nucleotide polymorphisms (SNPs) at -1993(C/T), -1514(C/T), and -999(G/A) sites of the TBX21 gene (encoding T-bet) promoter have been identified[21]. We have previously found that the T-1993C variant, which increased the transcription factor YY1 binding to the TBX21 promoter in Jurkat T cell lines, subsequently caused lower transactivation of the promoter[22]. Furthermore, our case-control study (84 cases and 318 control subjects) demonstrated that TBX21 T-1993C SNP was associated with susceptibility to AIH-1[23]. Individuals carrying the -1993C allele had a decreased risk to AIH-1 compared with those without the -1993C allele (OR = 0.22; 95%CI: 0.09-0.56; P = 0.0016). Recently, the TBX21 T-1993C variant has been shown to be associated with a significant decrease in T-bet expression and IFN-γ production by stimulated peripheral blood CD4+ T cells from healthy participants[24].
In the present study, we further determined the characterization of transcription factor binding to the T-1993C SNP site of the TBX21 promoter in CD4+ T and B cell lines. In addition, we measured the expression of T-bet and IFN-γ in peripheral blood CD4+ T cells and B cells from active AIH-1 patients carrying -1993TC and -1993TT genotypes, compared with healthy controls carrying -1993TC and -1993TT genotypes, in an attempt to explore the mechanism of the association between the TBX21 T-1993C promoter polymorphism and AIH-1 development.
A total of 20 patients with new onset were enrolled in this study at Southwest Hospital (Chongqing, China). Of these 20 patients, 5 were carriers of the -1993TC genotype and 15 were carriers of the -1993TT genotype. All the patients fulfilled the criteria for AIH according to the 1999 revised scoring system of the International Autoimmune Hepatitis Group (IAIHG)[25] and were therefore found to be eligible for the study. Only pretreatment scores were analyzed. A definite diagnosis of AIH based on the IAIHG criteria requires a pretreatment score exceeding 15. Patients were excluded from the study if there was histological evidence of cholangitis or non-alcoholic steatohepatitis. In addition, patients who were positive for hepatitis B virus (HBV)-surface antigen (HBsAg) or hepatitis C virus (HCV)-RNA were also excluded. Patients with other causes of liver disease, such as excess alcohol or drug use, or those who had received immune-suppressive therapies or glucocorticoid therapies within the past 6 mo were excluded based on reviews of their appropriate history and investigations. The control population comprised 35 healthy Chinese Han adults, who were volunteer blood donors from Chongqing, China. They were matched with type 1 AIH patients for age, gender, and TBX21 genotyping (10-1993TC and 25-1993TT genotype carriers), and had no history of any chronic inflammatory disease. All subjects provided written consent to participate in the present study. This study was approved by the Ethics Committee of Southwest Hospital, Chongqing. The demographic and clinical characteristics of these subjects are shown in Table 1.
| Parameters | AIH | HC |
| No | 20 | 35 |
| Age (yr) | 47.2 ± 11.8 | 46.9 ± 9.9 |
| Patient age at diagnosis(yr) | 44.5 ± 11.8 | - |
| Gender: female/male | 16/4 | 27/8 |
| IAIHG score | 17.15 ± 2.23 | - |
| ALT (< 40 IU/L) | 301.7 ± 284.7b | 21.9± 9.5 |
| AST (< 40 IU/L) | 306 ± 224.4b | 21.8 ± 4.9 |
| ALP (< 114 IU/L) | 190 ± 130.4b | 69 ± 21 |
| Total Bilirubin (6-21 μmol/L) | 112.5 ± 116.1b | 13.6 ± 3.7 |
| Albumin (38-51 g/L) | 38.2 ± 6.9a | 45.2 ± 8.4 |
| IgG (7-16 g/L) | 24.1 ± 11.8b | 8.4 ± 2.8 |
| Ant-ANA (+) (≥ 1:100) | 15 (75) | - |
| Ant-SMA (+) (≥ 1:100) | 3 (15) | - |
| Ant-ANA (+) + Ant-SMA(+) | 2 (10) | - |
| Ant-LKM-1 (+) (≥ 1:40) | 0 | - |
| Cirrhosis at presentation | 7 (35) | - |
The clinical data of each subject were collected from the hospital records. These data included age, sex and age at diagnosis, time of onset of symptoms or other evidence of liver disease, markers of infection with hepatitis viruses HBV and HCV, alcohol intake, coexisting autoimmune diseases, serum levels of alanine transaminase (ALT), aspartate aminotransferase (AST), alkaline phosphatase and bilirubin, and full blood cell counts. Autoantibodies were determined by indirect immunofluorescence on murine tissue sections (EuroImmun Medizinische Labordiagnostika AG, Lübeck, Germany), and a serum titer of 1:100 or greater was considered positive for all antibodies. All patients underwent liver biopsies to evaluate disease activity at the time of entry. The histological diagnosis of cirrhosis required a loss of the normal lobular architecture, reconstruction of hepatic nodules and presence of regenerative nodules.
Jurkat cells (CD4+ human T lymphoblasts) and Raji cells (human B lymphocytes) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mmol/L glutamine, 100 U/mL penicillin and 50 mg/mL streptomycin as described previously[23]. Human peripheral blood mononuclear cell (PBMC) were obtained by Ficoll-Paque density centrifugation (Amerhsam Pharmacia, Uppsala, Sweden) of whole blood from AIH-1 patients.
The electrophoretic mobility shift assay (EMSA) was performed by the method described previously[22]. The synthesized oligonucleotides (5’-TACGGAGAAATGG(T)GGGTAAGGTGTTG-3’ corresponding to the -2006/-1980 region of the human TBX21 gene with -1993T) and 5’-TACGGAGAAATGG(C)GGGTAAGGTGTTG-3’, -1993C), labeled with biotin at the 5’-end, were annealed to the complementary oligonucleotide labeled with biotin at the 5’-end. Nuclear extracts were prepared from Jurkat and Raji cells as described previously. For the competition assay, non-labeled double stranded DNA (dsDNA) fragments consisting of the following sequences were used: YY1 5’-CGCTCCCCGGCCATCTTGGCGGCTGGT-3’, and the sequences containing --1993T or -1993C. Supershifts were performed with antibodies to YY1, C/EBPβ (N-10) and c-Jun (N) that were purchased from Santa Cruz Biotechnology. All EMSA experiments were repeated more than three times, and a representative result for each experiment is shown.
The chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT Express kit (Active Motif, Carlsbad, CA, United States) as described previously[22]. The following antibodies were used: a rabbit anti-YY1 (C-20; Santa Cruz Biotechnology), anti-C-Jun (N; Santa Cruz Biotechnology), ant-C/EBPβ (Santa Cruz Biotechnology) and a nonspecific rabbit IgG (Santa Cruz Biotechnology). PCR was performed with the following primer, which were designed to amplify the -2048 to -1912 region of the TBX21 promoter: 5’-CCCACTTTGAACATCAGGCAGA-3’ and 5’-CAGCACATCTTTTGAGGTTGGAG-3’.
A reporter plasmid carrying luciferase gene under the control of a 2.2-kb TBX21 promoter fragment carrying the -1993T or -1993C allele was used as described previously[22]. The pCMV-YY1 vector expressing full-size YY1 and its control pCMV6-XL5 were obtained from OriGene.
Harvested cells were suspended in each culture medium supplemented with additional 10% fetal calf serum (FCS). Jurkat cells and Raji cells (2 × 105 cells in 0.5 mL) were cotransfected with 5 μg of the test construct and 25 ng of the pRL-TK Renilla reporter plasmid (Promega) using Nucleofector Kit V(LONZA) according to the manufacturer’s protocol. On the coexpression analysis, 3 μg of pCMV-YY1 or control pCMV6-XL5 was added into the cell suspension. The luciferase activity was measured, as described previously[22].
The isolated PBMCs were plated at a density of 106 cells/ml as described above. siRNA targeted against YY1 (sc-36864) and siRNA consisting of a scrambled sequence that would not lead to specific degradation of any cellular message (control) were purchased from Santa Cruz Biotechnology. The cells in 1 mL of medium were incubated in 5 mmol/L YY1-siRNA in the Nucleofector transfection reagent at room temperature with gentle agitation. Two hours after transfection, fresh medium was added to provide the cells with sufficient nutrients, and the transfected cells were harvested after 48 h for RNA extraction.
The Trizol-purified (Sangon, China) total RNA (2 μg) from PBMCs was used for cDNA synthesis with the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Canada) following the manufacturer’s instructions. The mRNA expression levels of IFN-γ and of nuclear factors such as YY1 and T-bet were quantified by real-time PCR, and the level of β-actin mRNA was also detected as an internal control for each sample. The relative mRNA expression levels of genes were determined using the comparative (2-ΔΔCt) method. The following primer sequences were used for this analysis: IFN-γ 5’-TCAGATGTAGCGGATAATGGAAC-3’ and 5’-TTCCTTGATGGTCTCCACACTC-3’; T-bet 5’-CCCACTTTGAACATCAGGCAGA-3’ and 5’-CAGCACATCTTTTGAGGTTGGAG-3’; 5’-YY1 GGATAACTCGGCCATGAGAA-3’ and 5’-ATAGGGCCTGTCTCCGGTAT-3’.
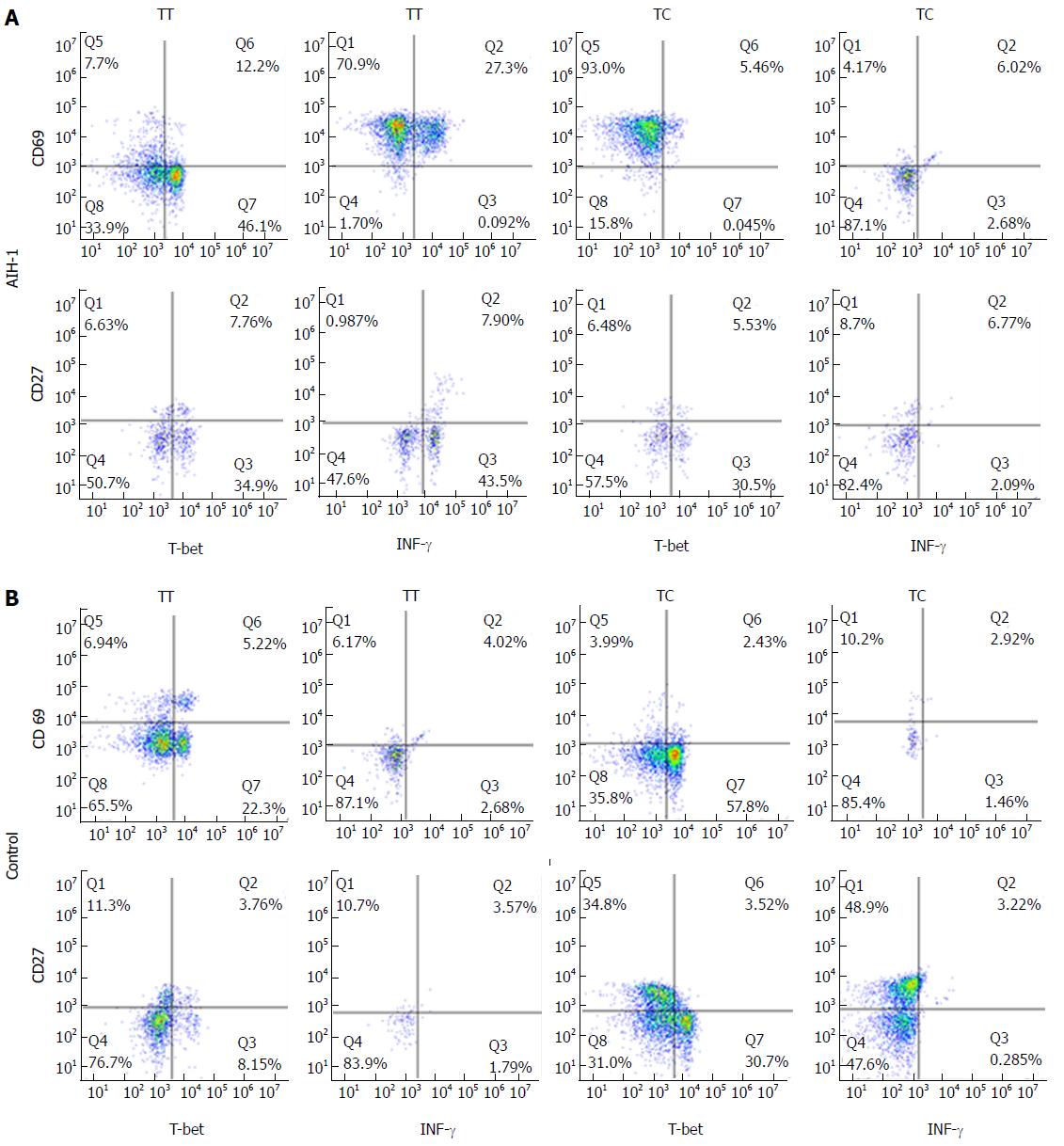
The experiments were performed on whole blood using the BD FastImmune CD4 Intracellular Cytokine Detection Kit (cat# 340970, BDIS, San Jose, CA, United States) according to the manufacturer’s instructions. In order to identify Th1 and B effector cells, 0.5 mL heparinized whole blood was stimulated with 2.5 μg/mL PMA and either the anti-CD28/ant-CD49d co-stimulatory monoclonal antibodies (final concentration of 1 μg/mL) or IFN-α2b (2000 U/mL, Schering-Plough, United States)[26]. For the five-color analysis of the CD4+ and CD20+ cytokine responses, the staining antibody cocktail consisted of anti-IFNγ-FITC/anti-CD4-PerCP(PE)-Cy5.5/anti-CD69-PE/anti-T-bet-Alexa-Fluor 647 (eBioscience, San Diego, United States) and anti-IFNγ-Cy5.5/anti-CD27-PE/anti-CD20-PE- Cy5.5/anti-T-bet-APC (eBioscience). Isotype-matched control antibodies were included to detect non-specific binding to the cells. Each analysis was performed using at least 50000 cells that were gated in the region of the lymphocyte population, as determined by light-scatter properties (forwardscatter versus side-scatter). To analyze the expression of IFN-γ and T-bet in lymphocytes (CD4+ T cells and CD20+ B cells), cells were gated in both the lymphocyte and CD4+/CD20+ regions. The dot plots display the events as cytokine+ versus CD69+ T or CD27+ B cells. The specific responses of the lymphocytes to stimuli were obtained by subtracting the percentage of positive events in the unstimulated sample from that in the activated samples. The isotype-corrected responses of the unstimulated samples were subtracted from those of the activated samples.
Data are expressed as mean ± SD. All statistical analyses were performed in SPSS 19.0 software. The difference between the groups was analyzed by parametric Student’s t-test or nonparametric Mann-Whitney U test. Correlations were examined by Pearson correlation. P values < 0.05 were considered significant.
To determine the potential role of CD4+ T and B cells in the pathogenesis of AIH, 20 patients with active AIH and 35 gender- and age-matched healthy controls were recruited. There was no significant difference in the distribution of age and gender between the patients and healthy controls (Table 1). As expected, the concentrations of serum ALT and AST and total bilirubin in the patients were significantly higher than that in the healthy controls. Furthermore, 15 of 20 patients with active AIH were positive for anti-ANA antibodies, three were positive for anti-SMA antibodies, and two were positive for both. In addition, abnormally higher levels of serum IgG were detected in the patients, demonstrating the liver damage and hypergammaglobulinaemia in the AIH-1 patients.
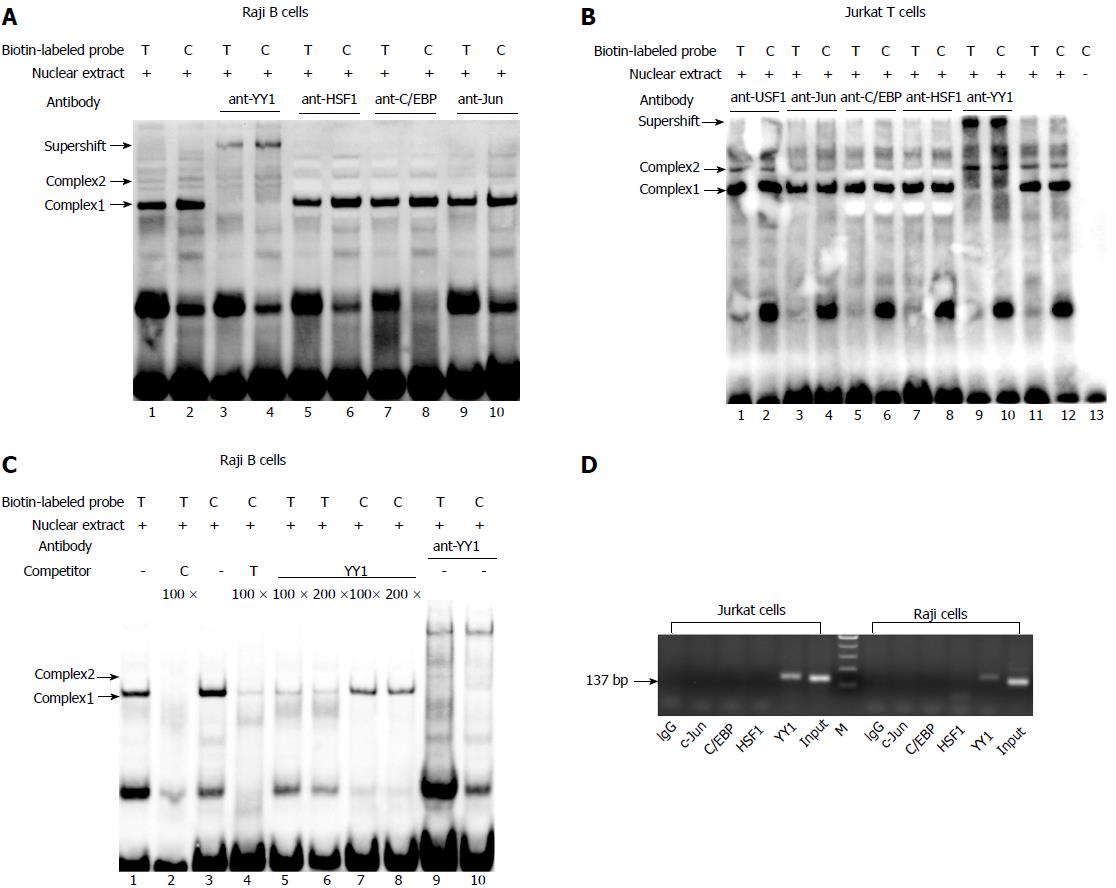
The results of EMSA showed the formation of two specific DNA-protein complexes (1 and 2) for both -1993T and -1993C allelic probes (Figure 1A and B). The addition of antibody against YY1 led to abrogation of only complex 2 in both the -1993T and -1993C probes (Figure 1A, lanes 3 and 4; Figure 1B, lanes 9 and 10), suggesting that YY1 participates in complex 2 formation. AP1, HSF1 and C/EBPβ did not seem to be involved in the formation of complexes 1 and 2 (Figure 1A, lanes 5-10; Figure 1B, lanes 3-8). A competition assay was performed to confirm whether the -1993C probe interacted more efficiently with the YY1 protein compared with the -1993T. It was found that the complexes formed with the -1993C probe were consistently more intense than these with -1993T (Figure 1C), which was consistent with the results of our study using Jurkat T cells[22]. In comparison with complex 1, we also observed that complex 2 required higher concentrations of competitors to diminish its band intensity. These data indicate that the complexes contain sequence-specific DNA-binding proteins and that the T-1993C SNP resides in a functionally important nucleotide, and is perhaps more relevant for complex 2.
ChIP assay was performed to further determine if YY1 binds to the T-1993C element of the TBX21 promoter in vivo. PCR amplification with the primers specific to the TBX21 promoter region, wherein the YY1-binding sites exist at the T-1993C locus, demonstrated a 137-bp product in both Jurkat and Raji chromatin after immunoprecipitation with the anti-YY1 antibody, but not with the ant-C/EBPβ and ant-C-Jun antibodies, or rabbit IgG (Figure 1D). These results demonstrate the direct interaction of YY1 with the TBX21 promoter in a non-cell-specific manner.
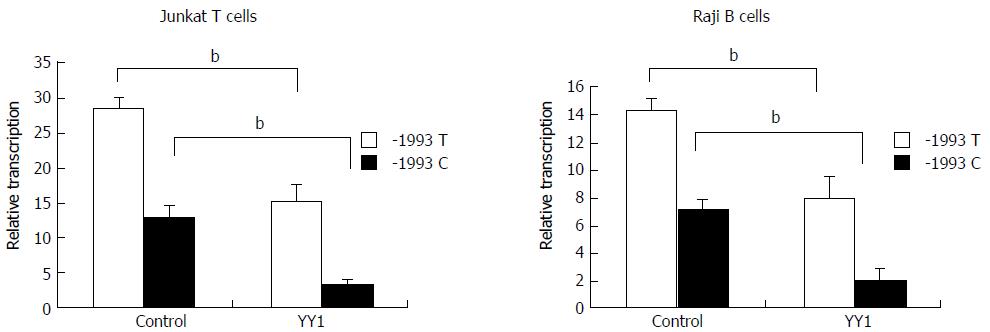
Luciferase reporter assay was performed to study transcriptional regulation of the TBX21 promoter constructs carrying either the -1993T or -1993C allele in the Jurkat T cell and Raji B cell lines. The luciferase activity derived from the -1993T promoter was significantly higher than that from the -1993C promoter in both cell lines (P < 0.01) (Figure 2). These data indicate that TBX21-1993C is significantly less active than the -1993T allele in a non-cell-specific manner. We further examined whether the transfection of a YY1 expression vector induced changes in the promoter activity of the -1993T and -1993C constructs in both Jurkat T cells and Raji B cells. We found that the luciferase activity derived from the -1993T promoter was 4-fold lower in the presence of overproduced YY1, and that from the -1993C promoter decreased only by approximately 2-fold (P < 0.01) (Figure 2). These results demonstrated that the YY1 motif upstream of the T-1993C polymorphism plays a strong negative regulatory role in the activity of the TBX21 promoter, and the -1993C promoter responds to YY1 better than the -1993T promoter.
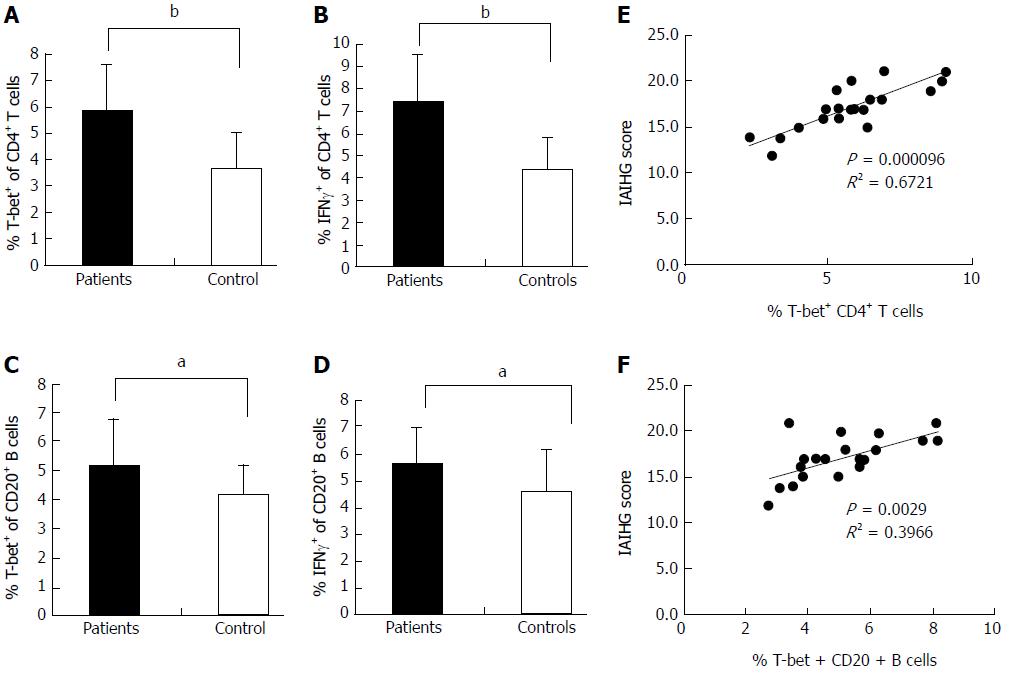
Flow cytometric analysis revealed that the T-bet expression of circulating CD4+ T cells and CD20+ B cells in active AIH-1 patients was significantly greater than that in controls (Z = -3.92, P < 0.01, Z = 2.43, P < 0.05,) (Figure 3A and C). The levels of IFN-γ production in circulating CD4+ T cells and CD20+ B cells from active AIH-1 patients were significantly higher than that from controls (Z = -4.89, P < 0.01; Z = 2206, P < 0.05) (Figure 3B and D). These data indicate an involvement of Th1 inflammation in Chinese patients with active AIH-1. The disease activity, represented by the IAIHG score, was positively correlated with T-bet expression in CD4+ T cells and CD20+ B cells from active AIH-1 patients (Figure 3E and F). Together, these results suggest an association between AIH-1 development and T-bet expression of circulating T cells or B cells.
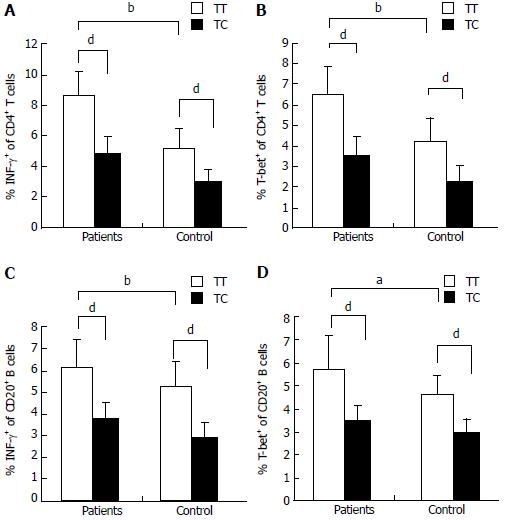
Flow cytometry was performed to analyze T-bet expression and IFN-γ production in circulating T cells and B cells from AIH-1 patients carrying -1993TC and -1993TT genotypes, and from healthy controls carrying -1993TC and -1993TT genotypes (Figure 4). The T-bet expression in CD4+ T cells and CD20+ B cells was significantly higher in the -1993TT patients than in the -1993TC patients and the -1993TT controls (Z = -2.75, P < 0.01; Z = -4.33, P < 0.01; Z = -4.21, P < 0.01; Z = -2.75, P < 0.05 respectively) (Figure 5A-D). No significant difference in T-bet expression was observed between in the -1993TC patients and -1993TC controls in both CD4+ T cells and CD20+ B cells. The levels of IFN-γ production was significantly higher in CD4+ T cells and CD20+ B cells from the -1993TT patients than from the -1993TC patients and the -1993TT controls (Z = -4.16, P < 0.01; Z = -5.07, P < 0.01; Z = -3.274, P < 0.01; Z = -2.529, P < 0.05 respectively) (Figure 5A-D). No significant difference in IFN-γ production was observed between in the -1993TC patients and -1993TC controls. The results demonstrated that increased IFN-γ and T-bet expression in circulating T and B cells from AIH-1 patients might result primarily from the -1993TT homozygotous patents with AIH-1.
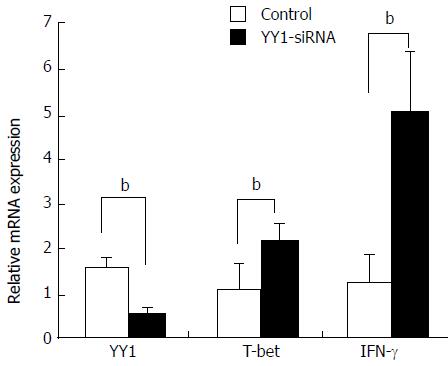
Knockdown of YY1 with siRNA was performed to analyze whether YY1 affects T-bet and IFN-γ expression of circulating CD4+ T and B cells in AIH-1 patients. Real-time PCR assay showed that YY1 siRNA significantly lowered the YY1 mRNA level in PBMCs from AIH patients (t = 13.83, P < 0.01) (Figure 6). The levels of T-bet and IFN-γ mRNA were up-regulated in the YY1-siRNA PBMCs compared with that in the control-siRNA PBMCs (t = -7.61; t = 12.11, P < 0.01 respectively) (Figure 6). These results suggested that YY1 affects type 1 immunity of AIH-1 patients by T-bet-mediated regulation.
Alterations in T-cell function and cytokines have been demonstrated to play a central role in AIH[2,27]. In human, liver tissue injury in AIH is also mediated by CD4+ and CD8+ T cells[28]. T-bet plays a critical role in both the development and maintenance of Th1 cells, and has been proved to be a positive transcriptional regulator of IFN-γ production in CD4+ T cells, CD8+ T cells, and B effector cells[15]. Prominent T-bet-positive lymphocytes have been found in both the lobular and portal infiltrate of human AIH[29]. T-bet together with IFN-γ were highly expressed in the inflamed liver tissue of a mouse model of fatal AIH[30]. To date there are no data concerning the expression of T-bet in circulating lymphomononuclear cells from AIH patients. Previous studies have indicated that the Th1/Th2 balance in peripheral blood of AIH patients is shifted towards Th1 cytokines and circulating cytochrome P4502D6-specific CD4+ T cells producing IFN-γ are present in AIH patients[31].
In this study, we first showed that active AIH-1 patients exhibited higher T-bet and IFN-γ expression in peripheral blood CD4+ T cells than healthy controls. The increased expression of T-bet was associated with an increased production of IFN-γ by peripheral blood CD4+ T cells, thus supporting the involvement of Th1 polarization in the pathogenesis of AIH-1. We found that the T-bet expression and IFN-γ production of circulating CD20+ B cells in AIH-1 patients increased significantly compared with that in the controls, which was consistent with other studies that demonstrated an elevation of T-bet and IFN-γ expression in circulating B cells from patients with other autoimmune diseases, such as coeliac disease and SLE[32,33]. Moreover, T-bet expression levels in peripheral blood T and B cells were directly correlated with the IAIHG score, suggesting an association between circulating T-bet-expressing T cells or B cells and disease activity. T-bet is expressed at low levels in naive B cells, but B cells in the presence of polarized Th1 cells and antigens were able to develop into high-IFN-γ-producing B cells that also express high levels of T-bet and ware capable of promoting the differentiation of naive T cells into Th1 effectors[14]. The cognate interactions that occur between T-bet-expressing and IFN-γ-producing T cells and B cells may result in the amplification of type 1 immune responses. Whether this mechanism contributes to AIH development requires further investigations.
Recent studies have shown that TBX21 T-1993C polymorphism is associated with susceptibility to autoimmune diseases including AIH[23,34]. To eliminate the possible effect of the T-1993C polymorphism on the regulation of the extensive activation of immune responses in AIH-1, we determined the expression of T-bet and production of IFN-γ in peripheral blood CD4+ T cells and B cells from the -1993TC heterozygote and -1993TT homozygote AIH-1 patients, compared with -1993TC heterozygote and -1993TT homozygote controls. Increased T-bet and IFN-γ expression was detected in the activated CD4+ T cells and B cells of AIH patients carrying the TBX21-1993 TT genotype compared with the patients carrying the -1993 TC genotype and controls with the -1993 TT genotype. The increased expression of T-bet correlated with an increased production of IFN-γ, demonstrating a higher biological effects of T-bet in Th1 and B cells from the patients with the TBX21-1993TT genotype. To verify whether the immunity of AIH patients is related to the regulation of TBX21 gene expression by YY1, we knocked down YY1 in PBMCs from AIH patients using siRNA. YY1 knockdown increased mRNA levels of T-bet and IFN-γ. This result highlights the increase of Th1 dominant response by repressing the production of YY1. These data suggested that the T-1993C variant in the TBX21 promoter, which represses T-bet expression and the size of the Th1 and B1 cell compartments by high-affinity binding of YY1 to the TBX21 promoter, may serve as a protective marker for AIH-1. To elucidate the relationship between the T-1993C polymorphism and the development of AIH, further detailed analysis of a large number of individuals including AIH patients carrying the CC genotype will be required.
YY1 is involved in repression and activation of several genes that play roles in various biological processes depending on the promoter sequence surrounding the YY1 binding site. Studies have shown YY1 positively regulated expression of several oncogenes, such as c-Myc and c-Fos[35]. In addition, YY1 can positively regulate some tumor suppressor genes, such as p21 and p16[36,37]. However, one study showed that YY1 was a negative cell growth regulator by inhibiting c-Myc function[38]. Interestingly, the promoter regions of genes that encode cytokines related to AIH, such as IL-6, IL-13 and IFN-γ, contain consensus YY1 binding sequences and some of them can be regulated by this transcription factor[30,39]. The transcriptional regulation of human TBX21 gene is complex, and several transcription factors such as SMAD4, SP-1, and NF-κB are involved[40,41]. One potential YY1 binding site has been previously identified in the -2211 to +1 region of the TBX21 promoter, wherein T/CCCATTTT was the critical base sequence for YY1 binding and the T-to-C mutation at the 5’ terminal of this sequence resulted in an increase of YY1 binding. These regions have also been associated with silencing of promoter activity in transient transfection assays of transformed cell lines. Our studies indicate that the silencing activity of this region of theTBX21 promoter is also operational in native T cells and B cells in vivo and is probably regulated by YY1. Thus, we propose that YY1 could be a crucial transcription factor for T-bet expression which play a critical role in the inflammation in AIH.
In summary, we showed that the TBX21-1993C allele created a strong YY1-binding site and decreased T-bet expression in CD4+ T and B cells. Reduced T-bet and IFN-γ expression of circulating CD4+ T cells and B cells existed in the individuals carrying the -1993C allele compared with those without the -1993C allele, and played a protective role in AIH-1 development. Furthermore, we propose a schematic model showing high-affinity binding of YY1 to the -1993C allele site leading to down-regulation of the TBX21 promoter activity, a mechanism for a promoter polymorphism affecting T-bet expression in AIH. We recognized that our study had limitations, such as a relative small sample size and the lack of functional study of T-bet-expressing T and B cells in the pathogenic process of AIH, as well as no information about T-bet-expressing T and B cells infiltrates in the liver. Therefore, further longitudinal studies are necessary to analyze the numbers and function of T-bet-expressing T and B cells in the pathogenic process of AIH -1 with a bigger population.
Autoimmune hepatitis (AIH) is an autoimmune disease that involves aberrant B and T lymphocyte responses. T-box transcription factor (T-bet) is a key regulator for the lineage commitment in CD4+ T hepler 1 and B effector 1 cells by activating the hallmark production of interferon-γ (IFN-γ). Although the role of T-bet has been studied in a variety of autoimmune diseases in mice, its relative importance in human AIH is not characterized. Detailed knowledge about T-bet-medicated immune responses will therefore enhance our understanding of the pathogenesis of AIH and might support the development of new immunomodulatory treatment approaches.
TBX21, which encodes T-bet, harbors many common polymorphisms at a strong linkage disequilibrium, and distinct TBX21 haplotypes are associated with autoimmune diseases in ethnically distinct populations. Our case-control study (84 cases and 318 control subjects) demonstrated a commone single nucleotide polymorphism (SNP) at the -1993 site of the TBX21 gene promoter that was associated with susceptibility to type 1 AIH (AIH-1) in a chinese population. Individuals carrying the -1993C allele had a decreased risk to AIH-1 compared with those without the -1993C allele (OR = 0.22; 95%CI: 0.09-0.56; P = 0.0016). Functional studies are necessary to dissect the mechanisms underlying the contribution of natural genetic variations to TBX21 dysregulation and the susceptibility to AIH-1 development.
In this study, the authors determined the characterization of transcription factor binding to the T-1993C SNP site of the TBX21 promoter in CD4+ T and B cell lines, and measured the expression of T-bet and IFN-γ in peripheral blood CD4+ T cells and B cells from active AIH-1 patients carrying -1993TC and -1993TT genotypes, compared with healthy controls carrying -1993TC and -1993TT genotypes, in an attempt to provide functional evidence for the association of the TBX21 T-1993C promoter polymorphism and AIH-1 development.
In vivo, in vitro, and reporter analyses were performed to determine the function of transcription factor Yin-Yang 1(YY1) binding to the T-1993C element of the TBX21 promoter in human CD4+ T and B cell lines. Flow cytometry and quantitative real-time PCR were used to analyze T-bet and IFN-γ expressions in CD4+ T cells, B cells and monocytes from the peripheral blood of AIH-1 patients including 5-1993TC and 15-1993TT genotype carriers, and healthy controls including 10-1993TC and 25-1993TT genotype carriers. Furthermore, knockdown of YY1 with siRNA was performed to investigate T-bet-medicated regulation of immune response in peripheral blood of AIH-1 patients. The difference between the groups was analyzed by parametric Student’s t-test or nonparametric Mann-Whitney U test. Correlations of T-bet expression in CD4+ T cells and B cells from active AIH-1 patients with AIH disease activity, represented by the International Autoimmune Hepatitis Group score, were examined by Pearson correlation.
TBX21-1993C allele created a strong Yin-Yang 1 (YY1)-binding site and decreased TBX21 promoter activity in human CD4+ T and B cells. Higher levels of T-bet and IFN-γ were detected in the circulating CD4+ T cells and B cells of AIH-1 patients carrying the TBX21-1993 TT genotype compared with the patients carrying the -1993 TC genotype and controls with the -1993 TC genotype. T-bet expression levels of circulating T cells and B cells were also positively correlated with AIH-1 disease activity. Knockdown of YY1 with siRNA caused increased expression of T-bet and IFN-γ in peripheral blood mononuclear cells in AIH-1 patients. This study has limitations, such as a relative small sample size and the lack of functional study of T-bet-expressing T and B cells in the pathogenic process of AIH, as well as no information about T-bet-expressing T and B cells infiltrates in the liver.
Reduced T-bet and IFN-γ expression of circulating CD4+ T cells and B cells existed in the individuals carrying the -1993C allele compared with those without the -1993C allele, and played a protective role in AIH-1 development. Moreover, the authors propose a schematic model showing high-affinity binding of YY1 to the -1993C allele site leading to down-regulation of the TBX21 promoter activity, a mechanism for a promoter polymorphism affecting T-bet expression in AIH.
Further studies are necessary to identify polymorphisms in other loci of the genome and to analyze their association with the T-1993C polymorphism for further elucidation of the genomic background of AIH. Furthermore, further longitudinal studies are necessary to analyze the numbers and function of T-bet-expressing T and B cells in the pathogenic process of AIH -1 with a bigger population.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Shimizu Y, Sipos F S- Editor: Ma YJ L- Editor: A E- Editor: Ma YJ
| 1. | Czaja AJ, Freese DK; American Association for the Study of Liver Disease. Diagnosis and treatment of autoimmune hepatitis. Hepatology. 2002;36:479-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 380] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 2. | Gregorio GV, Portmann B, Reid F, Donaldson PT, Doherty DG, McCartney M, Mowat AP, Vergani D, Mieli-Vergani G. Autoimmune hepatitis in childhood: a 20-year experience. Hepatology. 1997;25:541-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 475] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 3. | Verma S, Maheshwari A, Thuluvath P. Liver failure as initial presentation of autoimmune hepatitis: clinical characteristics, predictors of response to steroid therapy, and outcomes. Hepatology. 2009;49:1396-1397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 4. | Krawitt EL. Autoimmune hepatitis. N Engl J Med. 2006;354:54-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 636] [Cited by in F6Publishing: 578] [Article Influence: 32.1] [Reference Citation Analysis (1)] |
| 5. | Ichiki Y, Aoki CA, Bowlus CL, Shimoda S, Ishibashi H, Gershwin ME. T cell immunity in autoimmune hepatitis. Autoimmun Rev. 2005;4:315-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 6. | Löhr HF, Schlaak JF, Gerken G, Fleischer B, Dienes HP, Meyer zum Büschenfelde KH. Phenotypical analysis and cytokine release of liver-infiltrating and peripheral blood T lymphocytes from patients with chronic hepatitis of different etiology. Liver. 1994;14:161-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 104] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Lapierre P, Johanet C, Alvarez F. Characterization of the B cell response of patients with anti-liver cytosol autoantibodies in type 2 autoimmune hepatitis. Eur J Immunol. 2003;33:1869-1878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Edwards JC, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, Stevens RM, Shaw T. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350:2572-2581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1803] [Cited by in F6Publishing: 1762] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 9. | Zhong X, Tumang JR, Gao W, Bai C, Rothstein TL. PD-L2 expression extends beyond dendritic cells/macrophages to B1 cells enriched for V(H)11/V(H)12 and phosphatidylcholine binding. Eur J Immunol. 2007;37:2405-2410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Finnegan A, Ashaye S, Hamel KM. B effector cells in rheumatoid arthritis and experimental arthritis. Autoimmunity. 2012;45:353-363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 575] [Cited by in F6Publishing: 585] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 12. | Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2577] [Cited by in F6Publishing: 2597] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 13. | Lametschwandtner G, Biedermann T, Schwärzler C, Günther C, Kund J, Fassl S, Hinteregger S, Carballido-Perrig N, Szabo SJ, Glimcher LH. Sustained T-bet expression confers polarized human TH2 cells with TH1-like cytokine production and migratory capacities. J Allergy Clin Immunol. 2004;113:987-994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Harris DP, Goodrich S, Gerth AJ, Peng SL, Lund FE. Regulation of IFN-gamma production by B effector 1 cells: essential roles for T-bet and the IFN-gamma receptor. J Immunol. 2005;174:6781-6790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 142] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Durali D, de Goër de Herve MG, Giron-Michel J, Azzarone B, Delfraissy JF, Taoufik Y. In human B cells, IL-12 triggers a cascade of molecular events similar to Th1 commitment. Blood. 2003;102:4084-4089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Siebler J, Wirtz S, Klein S, Protschka M, Blessing M, Galle PR, Neurath MF. A key pathogenic role for the STAT1/T-bet signaling pathway in T-cell-mediated liver inflammation. Hepatology. 2003;38:1573-1580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Bettelli E, Sullivan B, Szabo SJ, Sobel RA, Glimcher LH, Kuchroo VK. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 370] [Cited by in F6Publishing: 385] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 18. | Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci USA. 2002;99:5545-5550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 359] [Cited by in F6Publishing: 361] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 19. | Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152-1156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 756] [Cited by in F6Publishing: 700] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 20. | Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25 Suppl 2:74-78; discussion 95-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 380] [Cited by in F6Publishing: 404] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 21. | Suttner K, Rosenstiel P, Depner M, Schedel M, Pinto LA, Ruether A, Adamski J, Klopp N, Illig T, Vogelberg C. TBX21 gene variants increase childhood asthma risk in combination with HLX1 variants. J Allergy Clin Immunol. 2009;123:1062-1068, 1068.e1-1068.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Li JR, Li JG, Deng GH, Zhao WL, Dan YJ, Wang YM, Chen S. A common promoter variant of TBX21 is associated with allele specific binding to Yin-Yang 1 and reduced gene expression. Scand J Immunol. 2011;73:449-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Chen S, Zhao W, Tan W, Luo X, Dan Y, You Z, Kuang X, Wang Y, Deng G. Association of TBX21 promoter polymorphisms with type 1 autoimmune hepatitis in a Chinese population. Hum Immunol. 2011;72:69-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Fyall KM, Fong AM, Rao SB, Ibrahim JG, Waxweiler WT, Thomas NE. The TBX21 transcription factor T-1993C polymorphism is associated with decreased IFN-γ and IL-4 production by primary human lymphocytes. Hum Immunol. 2012;73:673-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 25. | Alvarez F, Berg PA, Bianchi FB, Bianchi L, Burroughs AK, Cancado EL, Chapman RW, Cooksley WG, Czaja AJ, Desmet VJ. International Autoimmune Hepatitis Group Report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatol. 1999;31:929-938. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2003] [Cited by in F6Publishing: 1886] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 26. | de Goër de Herve MG, Durali D, Dembele B, Giuliani M, Tran TA, Azzarone B, Eid P, Tardieu M, Delfraissy JF, Taoufik Y. Interferon-alpha triggers B cell effector 1 (Be1) commitment. PLoS One. 2011;6:e19366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Ma Y, Bogdanos DP, Hussain MJ, Underhill J, Bansal S, Longhi MS, Cheeseman P, Mieli-Vergani G, Vergani D. Polyclonal T-cell responses to cytochrome P450IID6 are associated with disease activity in autoimmune hepatitis type 2. Gastroenterology. 2006;130:868-882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 154] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 28. | Murthy A, Shao YW, Defamie V, Wedeles C, Smookler D, Khokha R. Stromal TIMP3 regulates liver lymphocyte populations and provides protection against Th1 T cell-driven autoimmune hepatitis. J Immunol. 2012;188:2876-2883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Hadžić N, Quaglia A, Cotoi C, Hussain MJ, Brown N, Vergani D, Mieli-Vergani G. Immunohistochemical phenotyping of the inflammatory infiltrate in de novo autoimmune hepatitis after liver transplantation in children. Pediatr Transplant. 2012;16:501-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Ikeda A, Aoki N, Kido M, Iwamoto S, Nishiura H, Maruoka R, Chiba T, Watanabe N. Progression of autoimmune hepatitis is mediated by IL-18-producing dendritic cells and hepatic CXCL9 expression in mice. Hepatology. 2014;60:224-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Longhi MS, Ma Y, Mieli-Vergani G, Vergani D. Aetiopathogenesis of autoimmune hepatitis. J Autoimmun. 2010;34:7-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 137] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 32. | Lit LC, Wong CK, Li EK, Tam LS, Lam CW, Lo YM. Elevated gene expression of Th1/Th2 associated transcription factors is correlated with disease activity in patients with systemic lupus erythematosus. J Rheumatol. 2007;34:89-96. [PubMed] [Cited in This Article: ] |
| 33. | Frisullo G, Nociti V, Iorio R, Patanella AK, Plantone D, Bianco A, Marti A, Cammarota G, Tonali PA, Batocchi AP. T-bet and pSTAT-1 expression in PBMC from coeliac disease patients: new markers of disease activity. Clin Exp Immunol. 2009;158:106-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Zhang D, Zhang X, Ge M, Xuan M, Li H, Yang Y, Fu R, Zhou F, Zheng Y, Yang R. The polymorphisms of T cell-specific TBX21 gene may contribute to the susceptibility of chronic immune thrombocytopenia in Chinese population. Hum Immunol. 2014;75:129-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13:7487-7495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 98] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 36. | Wang X, Feng Y, Xu L, Chen Y, Zhang Y, Su D, Ren G, Lu J, Huang B. YY1 restrained cell senescence through repressing the transcription of p16. Biochim Biophys Acta. 2008;1783:1876-1883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Ishii H, Hulett MD, Li JM, Santiago FS, Parish CR, Khachigian LM. Yin Yang-1 inhibits tumor cell growth and inhibits p21WAF1/Cip1 complex formation with cdk4 and cyclin D1. Int J Oncol. 2012;40:1575-1580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 38. | Austen M, Cerni C, Lüscher-Firzlaff JM, Lüscher B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene. 1998;17:511-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 62] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Soutto M, Zhang F, Enerson B, Tong Y, Boothby M, Aune TM. A minimal IFN-gamma promoter confers Th1 selective expression. J Immunol. 2002;169:4205-4212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 40. | Yu J, Wei M, Boyd Z, Lehmann EB, Trotta R, Mao H, Liu S, Becknell B, Jaung MS, Jarjoura D. Transcriptional control of human T-BET expression: the role of Sp1. Eur J Immunol. 2007;37:2549-2561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | McCracken SA, Hadfield K, Rahimi Z, Gallery ED, Morris JM. NF-kappaB-regulated suppression of T-bet in T cells represses Th1 immune responses in pregnancy. Eur J Immunol. 2007;37:1386-1396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |