Published online Jun 21, 2017. doi: 10.3748/wjg.v23.i23.4158
Peer-review started: February 7, 2017
First decision: March 16, 2017
Revised: April 12, 2017
Accepted: June 1, 2017
Article in press: June 1, 2017
Published online: June 21, 2017
Helicobacter pylori (H. pylori) as a causative agent of gastric complications, is well adapted for the colonization of gastric mucosa. Although the infectious process depends on several factors, the adhesion to the gastric mucosa is the first and important step. Among several outer membrane proteins, BabA is one of the significant protein involving in many inflammatory processes in addition to its role in the attachment for the persistent colonization. We performed a PubMed search using the key words: “babA”, “pylori”, “gastric complications”, “homologous recombination”, “slipped strand mispairing”; a total of 249 articles were displayed. Of these we mainly focused on articles with the full text in English and published between 2005 and 2016. H. pylori BabA is involved in binding with receptors; however, its synthesis is regulated by phase variation. In this review we confirm that H. pylori babA can be modulated at the molecular and functional levels to adapt to the stress within the gastro-intestinal tract.
Core tip:Helicobacter pylori are well adapted to colonize the gastric mucosa. Although the infectious process depends on several factors, adhesion to the gastric mucosa is the first and critical step. Among outer membrane proteins, BabA is an important protein involved in many inflammatory processes in addition to playing a role in the aforementioned attachment process. In this review, we have summarized the current, published knowledge regarding its functional and molecular aspects by which the pathogen can attach to the host cell receptors.
- Citation: Ansari S, Yamaoka Y. Helicobacter pylori BabA in adaptation for gastric colonization. World J Gastroenterol 2017; 23(23): 4158-4169
- URL: https://www.wjgnet.com/1007-9327/full/v23/i23/4158.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i23.4158
The Helicobacter pylori (H. pylori) is a helical, Gram-negative bacterium that chronically infects the stomach of half of the world’s population[1]. Since this bacterium is strongly associated with the development of duodenal ulcer and gastric cancer, International Agency for Research on Cancer categorized this bacterium under strong carcinogen (group I carcinogen) in 1994[2,3]. The global burden of gastric cancer is high which accounts for 6.2% of cancer burden worldwide[4]. However, the appearance of new cases of gastric cancer has been found variable in developing (8.4%) and developed countries (4.5%)[5]. Mortality rate due to the gastric cancer is the third most common cause among all cancer related deaths[6,7].
Although the exact means of transmission of this bacterium is not understood clearly the evidences suggest that this bacterium is transmitted from person to person contact between family members via fecal-oral or oral-oral route and mostly the infections are acquired in childhood[8-10]. The data from epidemiological studies conducted support the transmission of infection by exposure to contaminated water or food as well[11,12]. The socio-economic status of family highly reflects the infection rate among peoples as the infection rate tends to be higher in peoples belonging to family with low socio-economic status[13]. This contribution highly suggests the higher prevalence rate of H. pylori infection in developing countries than the developed countries. The infection rate ranging from 70%-90% and 25%-50% in developing and developed countries respectively[14].
H. pylori related clinical outcomes also depend on the ancestral relation between human and bacterium as suggested recently. The individuals infected with non-ancestral strains that are not evolutionarily adapted to their host develop severe histo-pathological damage and clinical outcomes than infected with co-evolutionarily developed strains[15]. However, being a truly opportunistic pathogen, the H. pylori utilizes the various virulence factors such as CagA and VacA as the effector protein for the development of gastric diseases[16] and the outer membrane proteins (OMPs) such as blood group antigen binding adhesion (BabA), sialic acid binding adhesin (SabA), and outer inflammatory protein (OipA) found on the surface (adhesins) of bacterial cell envelop for the attachment to the mucus layer of the gastric epithelium playing an important and initial step for the colonization and development of persistent infection for decades or for entire life time[17-19].
We conducted a PubMed search using the key terms “babA”, “pylori”, “gastric complications”, “homologous recombination”, “slipped strand mispairing”; 249 articles were found during the search. We focused on, but did not limit ourselves to, articles with the full text in English and published between 2005 and December 2016. For greater clarity and insight we considered few articles published between 1989 and 2005. We retrieved a total of 98 articles that studied BabA and its paralogs, presence, function, production, and role in the development of gastric complications, attachment to host cells, and adaptation. Abstract only publications, case reports, editorials, and review articles were excluded. While performing the search, irrelevant articles with authors’ family names as ‘Baba’ were also excluded. We also performed a literature search in Google using the terms: “pylori”, “gastric cancer”, and “slipped strand mispairing”. Through this, we selected three reports that were not indexed in PubMed.
H. pylori colonization elicits humoral and cellular immune responses although, ineffective in the bacterial clearance. However, despite of the peristaltic movement of the intestinal tract and movement of the chyme, the bacterium establishes the strong interaction with the epithelial surfaces. The bacterial attachment on the epithelial surface is the interaction between the receptor molecules on the host cell surface and adhesin molecules found on the bacterial cell envelope and this interaction is the first and important step of the H. pylori related pathogenicity. The Gram-negative bacterial cell envelope consists of two concentric lipid bilayers, the inner membrane (cytoplasmic membrane) and the outer membrane. More than 50% of the outer membrane mass consists of OMPs. H. pylori genome encodes a large number of OMPs and most of them are surface exposed and consists of transport channels (porins), and adhesins. The closely related (at least partially redundant) proteins are grouped into paralogous families[20].
Among the large group of adhesins identified BabA, SabA, adherence associated lipoprotein A and B (AlpA/B), OipA, and HopZ[17,18,21-24], BabA (or HopS or OMP28) is around 75-80 kDa major OMP which was the first identified member of adhesin family[25]. Two other closely related paralogs to BabA has been found, the BabB (HopT or OMP19) and BabC (HopU or OMP9)[26]. The function of BabB and BabC is not known yet and needs to be determined. The bab gene sequence analysis revealed that there is extensive 5’ and 3’ region homology particularly between babA and babB but the variability in middle region of sequence suggests that the middle divergent region of babA likely mediates the binding function[26,27].
The babA, babB and babC genes can be located in at least 3 different chromosomal loci. The three marker genes hypD, s18 and heme oxygenase gene hp0318 represent the chromosomal location of bab genes. The bab gene found downstream of hypD, s18 and hp0318 is said to be located at locus A, locus B and locus C respectively[28]. For example; in strain J99 (where babC is absent), babA (jhp0833) and babB (jhp1164) are located downstream of hypD and s18 (locus A and locus B) respectively whereas in strain 26695, the locations for babA and babB are reversed; babA (hp1243), babB (hp0896) and babC (hp0317) are located downstream of s18, hypD and hp0318 (locus B, locus A and locus C) respectively. Similarly, in strain HPAG1, the babA, babB and babC are located downstream of hypD, hp0318 and s18 (locus A, locus C and locus B) respectively whereas in strain G27 (where babB is absent), babA and babC are located downstream of hp0318 and hypD (locus C and locus A) respectively and in strain 51 (where babC is absent), the babA and babB are located downstream of hypD and s18 (locus A and locus B) respectively[29-31] as shown in Table 1. We made an attempt to find the bab genes in respective to their genomic locus in different strains deposited in gene bank. Among H. pylori deposited in gene bank by 30 November 2016, we randomly selected the 33 strains for the characterization of chromosomal locations of bab genes (babA, babB and babC) (Table 1). Although, the chromosomal locations of babA, babB and babC in few strains such as J99, 26695, HPAG1, G27 and 51 already have been described yet in other strains we made an effort to document the chromosomal location of these genes. In strain P12 there is a babB homologous gene at locus C but due to the insertion of one adenine (A) at position 679 resulted the shift in the nucleotide frame and the stop codon at position 230 and premature termination of the amino acid sequence. The reports of several studies from various parts of the world depicts that in majority of the strains babA and babB prefers to be located at locus A and locus B respectively[28-30,32]. Despite of the three identified loci there can be an unidentified locus. In a study, Hennig et al[29] found babA sequences from 2 strains not located at locus A, B or C and they hypothesized that there may be a yet not identified chromosomal locus for babA.
| Strain | Locus A | Locus B | Locus C |
| 26695 | babB | babA | babC |
| J99 | babA | babB | No corresponding gene |
| HPAG1 | babA | babC | babB |
| G27 | babC | No corresponding gene | babA |
| 51 | babA | babB | No corresponding gene |
| Shi470 | babA | babB | No corresponding gene |
| P12 | babA | babB | babB homolog1 |
| 52 | babA | babB | No corresponding gene |
| B38 | babA | No corresponding gene | No corresponding gene |
| v225d | babA | babB | No corresponding gene |
| SJM180 | babB | babC | babA |
| PeCan4 | babA | babB | No corresponding gene |
| Sat464 | babA | babB | No corresponding gene |
| 35A | babA | babB | No corresponding gene |
| India7 | babA | babB | No corresponding gene |
| Gambia94/24 | babA | babB | No corresponding gene |
| SouthAfrica7 | babA | babA | No corresponding gene |
| Oki112 | babA | babA | No corresponding gene |
| Oki154 | babA | babB | No corresponding gene |
| Oki828 | babA | babA | No corresponding gene |
| J166 | babA | babB | No corresponding gene |
| SNT49 | babA | babB | No corresponding gene |
| Puno120 | babA | babB | No corresponding gene |
| Puno135 | babA | babB | No corresponding gene |
| B8 | babA | babA | No corresponding gene |
| 83 | babA | babB | No corresponding gene |
| ELS37 | babB | babA | No corresponding gene |
| HUP-B14 | babA | babC | No corresponding gene |
| F16 | babA | babB | No corresponding gene |
| F30 | babA | babB | No corresponding gene |
| Hp238 | babA | babB | No corresponding gene |
| BM013A | babB | babB | No corresponding gene |
| 29CaP | babA | babA | No corresponding gene |
As discussed in previous section, the babA can be located in at least three different identified loci. The BabA production has been claimed to be influenced by its genomic location. In strain J99, babA located at locus A is expressed and binds with Lewis blood group (Leb) antigen whereas in strain 26695; babA located at locus B is not expressed and does not bind to Leb antigen[33,34]. The detailed mechanisms involved in the expression of babA have not been depicted in detail. However, we have summarized the possible mechanisms published in several literatures.
The strain CCUG17875 was reported to possess two alleles of babA denoted as babA1 found at locus B and babA2 found at locus A[34]. The allelic form babA1 does not contain the translation initiation codon ATG because of the deletion of 10 bp including start codon in its signal sequence and is not expressed whereas the babA2 containing the translation initiation codon ATG is expressed and involved in the Leb binding activity[34]. The expression of babA can be influenced on transcriptional as well as translational level. The absence of translation initiation codon ATG in babA1 influence the translation, but this phenomenon is quite uncommon. Nonetheless, unfortunately, several epidemiological studies still use the methods to characterize babA2 as the functional status of babA by using primers specific for the signal region to differentiate babA1 and babA2. The presence of variable number of cytosine-thiamidine (CT) dinucleotide in the 5’-region of the babA sequence due to the intra-genomic recombination with babB leads to the phase variation and affects the expression of babA (discussed in latter section). The characteristics of the promoter sequence also play a critical role in the expression of babA. During protein expression all mRNAs are not synthesized in the same amount because of the promoter characteristics which controls the expression on transcriptional level. In a study, the presence of additional 4 adenines between the -10 and -35 motif in babA located at locus B could diminished the strength of the promoter of babA[34]. The nucleotide structure of -10 and -35 motifs sequence and the characteristics of nucleotides present between the -10 and -35 motifs tend to make the promoter strong or weak. For example, the promoter region of one of the OMP gene sabA contains a thymidine (T) nucleotide repeat tract adjacent to the -35 motif and the length of this T-tract varies between strain to strain and this variation have been suggested to affect the expression of sabA[35-37]. Recently it has been also suggested that the T-tract modulates the binding of RNA polymerase σ-factor and α-factor resulting in the alteration of SabA expression[38]. The nucleotide repeat motifs located between the -35 and -10 promoter motifs influence the docking of RNA polymerase σ-factor and the nucleotide repeat motif located upstream of -35 motif alters the binding of regulatory factors of RNA polymerase[38]. However, the detailed studies are needed to evaluate the characteristics of promoter regions responsible for the variation in expression of babA in loci A/B/C.
H. pylori babA is highly polymorphic being susceptible for the changes in their regulatory and coding sequence that leads to the loss of BabA expression in multiple animal models including mice, gerbils and macaques; therefore, it is difficult to study the role of BabA contribution in the development of gastric damages[39]. However, it has been shown that in H. pylori isolated from the chronically infected persons, the BabA expression was maintained in three quarter of the isolates surveyed over time and it suggests that there is selective advantage for BabA expression in human host[40].
Most epidemiological studies have reported the babA functional status using the babA2 specific primers designed to detect the 10-bp deletion in the signal region of the babA1 allele and its association with increased risk for gastric inflammation[41]. The reported prevalence of H. pylori containing babA shows great variation across different parts of the world[28,32,42-55] as shown in Table 2. However, we previously reported the BabA expression level by immunoblotting and Leb binding in diverse collection of H. pylori strains and divided in to high, low, and non BabA expressing strains[52]. The strains expressing low level of BabA were associated with higher incidence of gastrointestinal damages as compared to BabA with high expression and BabA negative strains. It was concluded that although, the babA2 detection provides an indication of functional babA status, it may not reliably reflect the complete information of genetic variants for babA expression.
| Ref. | Countries | Criteria for babA positive | Prevalence | Year |
| Asia | ||||
| Abdullah et al[43] | Iraq | Positive with babA2 specific primers | 33.7% in NUD and 58.8% in PUD | 2012 |
| Karabiber et al[44] | Turkey | Positive with babA2 specific primers | 49% in gastritis | 2014 |
| Abadi et al[45] | Iran | Positive with babA2 specific primers | Overall-40.6% | 2013 |
| 95%-GC, 18.1-DU, 26.1-NUD | ||||
| Yadegar et al[46] | Iran | Positive with babA2 specific primers | Overall-96.7% | 2014 |
| 94.4%-NUD, 100%-PUD, 100%-GE, 100%-GC | ||||
| Saberi et al[42] | Iran | BabA expression | Overall-62% | 2016 |
| Osman et al[47] | Malaysia | Positive with babA2 specific primers | 41%-NUD | 2015 |
| Boonyanugomol et al[48] | Thailand | Positive with babA2 specific primers | Overall-66.2%, | 2012 |
| 70.7%-CCA, 54.5%-Cholelithiasis | ||||
| Chomvarin et al[49] | Thailand | Positive with babA2 specific primers | Overall-92% | 2008 |
| 92%-NUD, 85%-GU, 100%-DU, 94%-GC | ||||
| Ghosh et al[50] | India | Positive with babA2 specific primers | Overall-67.5% | 2016 |
| 65.6%-NUD, 70%-DU | ||||
| Con et al[51] | Japan | Positive with babA2 specific primers | Overall-96.8% | 2010 |
| Fujimoto et al[52] | Japan | Leb binding activity | Over all-88.0% | 2007 |
| Kim et al[32] | South Korea | Positive with babA specific primers | Overall-47.5% | 2015 |
| Other countries | ||||
| Kim et al[32] | United States | Positive with babA specific primers | Overall-90% | 2015 |
| Biernat et al[53] | Poland | Positive with babA2 specific primers | Overall-23.1% | 2014 |
| 18%-NUD, 30%-PUD, 31.8%-GERD | ||||
| Homan et al[54] | Slovenia | Positive with babA2 specific primers | Overall-47.9% | 2014 |
| Boyanova et al[55] | Bulgaria | Positive with babA2 specific primers | Overall-48.8% | 2010 |
| 59.3%-PUD, 43.5%-NUD | ||||
| Matteo et al[28] | Argentina | Positive with babA specific primers | Overall-67% | 2011 |
| Con et al[51] | Costa-Rica | Positive with babA2 specific primers | Overall-73.7% | 2010 |
| Fujimoto et al[52] | Colombia | Leb binding activity | Overall-83.0% | 2007 |
Adherence of H. pylori to the host cell receptor provides several benefits to the bacteria such as protection from the washing out during mucus shedding, provides nutrient access from damaged host epithelial cells, promotes delivery of the bacterial toxins and other effector molecules to the host cells for development of pathogenicity and facilitates the development of persistent infections[56-58]. The attachment of bacteria to the host cells also elicits immune response; however, no longer protective in H. pylori infection. Several functional molecules serving as a receptor for the binding of H. pylori through BabA has been reported (Table 3).
| Receptors identified | Localization | Ref. |
| Mucin MUC5B | Saliva | Walz et al[59] and Prakobphol et al[60] |
| Agglutinin glycoprotein-340 (gp-340) | Saliva | Prakobphol et al[61] |
| Prolin rich glycoprotein containing Fucα1-2Galβ motif | Saliva | Walz et al[59] |
| Secretory immunoglobulin A containing fucose-oligosaccharide motifs | Saliva | Borén et al[17] and Royle et al[62] |
| Salivary agglutinin DMBT1 | Saliva | Issa et al[63] |
| Lewis b blood group antigen (Leb) and terminal fucose, H1-antigen, A-antigen and B-antigen | Gastric epithelia | Borén et al[17] and McGuckin et al[64] |
| Mucin MUC5AC with N-acetylgalactosamine-β-1,4-N-acetylglucosamine | Gastric mucus | Lindén et al[68] and Kenny et al[69] |
| Mucin MUC1 | Gastric mucus | Lindén et al[71] |
| Mucin MUC2 | Gastric mucus | Cohen et al[72] |
Since the postulated transmission for the H. pylori infection takes place through oral route, the possible initial attachment could be occurred with the salivary proteins. The BabA mediates the attachment by binding with difucosylated glycans found on Leb antigens of the salivary mucin MUC5B serving as a receptor for BabA[59,60]. The surface immobilized salivary agglutinin; glycoproteins-340 (gp-340) also provides the platform for the attachment of BabA[61]. Salivary prolin rich glycoprotein (PRG) is the major component of parotid and submandibular saliva containing several repeats of short prolin rich sequence. Salivary PRG containing Fucα1-2Galβ motif also provides the access for the attachment of BabA[59]. The secretory immunoglobulin A (s-IgA) is the major immunoglobulin found in the mucus secretions which protects the mucus membrane from invading organisms. However, the fucose-containing oligosaccharide motifs on s-IgA could play a role in the binding phenomena of BabA[17]. However, in a study this phenomenon was not confirmed suggesting that the variation in the glycosylation and sialylation between the salivary and gastric s-IgA or the proportions of s-IgA1 and s-IgA2 subclasses which are known to be differently glycosylated may play this role[62]. Similarly, a salivary agglutinin called the deleted in malignant brain tumors 1 (DMBT1) is expressed in saliva and composed of highly fucosylated oligosaccharide. The salivary DMBT1 was found to act as the receptor to interact with BabA[63].
After transit to the stomach, the bacterium localizes to the specific locations and mediates adaptation through several ways. The gastrointestinal epithelium is covered by a semi-permeable mucus layer primarily consists of secreted mucins that protects the gastric epithelial surface by trapping the invading materials[64]. However, the attachment of bacteria to the epithelial surface mediates the survival adaptation and persistent infections. The di-fucosylated glycan found on Leb and mono-fucosylated glycans found on H1-antigen, A-antigen and B-antigen of blood group O, A and B respectively binds with BabA[17,33,65]. However, the binding affinity of BabA with H1 antigen is low because it lacks the Leb Fuc4 residue that forms a hydrogen bond with amino acid asparagine (N) at 206 position of BabA[66] as described below in next section. These antigens are expressed on the mucus surface and gastric epithelial surface of which the Leb antigen is the dominant antigen found on the surface of gastric epithelial cells and it is also the most studied receptor for BabA attachment[67].
Similarly, other molecules have also been reported to play a crucial role in the binding of BabA. The gastric mucin MUC5AC with N-acetylgalactosamine-β-1,4-N-acetylglucosamine residues on O-linked oligosaccharides from gastric MUC5AC has also been reported to act as receptor for BabA[68,69]. Most recent immunohistochemistry laden tissue profiling assay using mice model showed that the expression of Leb structure is a mucin α 1,2-fucosyltransferase (FUT2) enzyme dependent and consequently identified MUC5AC as the carrier molecule of the Leb structure[70].
The MUC1 is also notable to discuss. In case when bacteria do not bind to MUC1, an extracellular mucin domain of 200-500 nm long appears that physically distances the bacteria from the host cell surface and in case when it does bind MUC1, the extracellular mucin domain is released from the epithelial surface[71]. In another study by Cohen et al[72] conducted in children, the gastric mucin MUC2 was reported to be expressed in few foleolar cells and it was shown to participate in binding with BabA but only in 11.11% of children. Therefore, fucosylated glycans found on several glycoprotein molecules serve as the receptor.
The detailed molecular determination of BabA binding with its receptor Leb was studied recently by Hage et al[66]. They depicted that the Leb oligosaccharide molecules involving in the binding with BabA consists of two fucose residues (Fuc1 and Fuc4), two galactose residues (Gal2 and Gal5), an N-acetylglucosamine residue (GlcNAc3), and a glucose residue (Glc6). Using strain J99 as an experimental model they found that 8 highly conserved amino-acids of BabA involved in the interaction with Leb by hydrogen bond formation. The bond formation occurs between amino acid cysteine (C) at position 189 and Fuc1 (C189:Fuc1), glycine (G) at position 191 and Fuc1 (G191:Fuc1), asparagine (N) at position 194 and Fuc1 (N194:Fuc1), asparagine (N) at position 206 and Fuc4 (N206:Fuc4), aspartic acid (D) at position 233 and GlcNAc3 (D233:GlcNAc3), aspartic acid (D) at position 233 and Gal5 (D233:Gal5), serine (S) at position 234 and Gal5 (S234:Gal5), serine (S) at position 244 and GlcNAc3 (S244:GlcNAc3, Gal5), serine (S) at position 244 and Gal5 (S244:Gal5) and threonine (T) at position 246 and Fuc1 (T246:Fuc1) (Table 4). They also elaborated that the binding of N at position 206 with Fuc4 determines the binding affinity and substitution of N with alanine (A) at position 206 resulted in the reduction of binding affinity by about 2.3-fold and the binding of BabA with H-1 which lacks Fuc4 residue of Leb showed about 2.4 fold reduction in binding affinity[66]. Despite of the role of Leb for providing the binding receptor and enhancing the colonization, the several reports indicate the antagonistic effect of Leb. In a study led by Lindén et al[73] found that the individuals with Leb-negative showed significantly higher H. pylori density than Leb-positive, which is in accordance with the result obtained in a study using Rhesus monkey[74]. Therefore, it can be said that although, the attachment of bacterium via BabA-Leb is important but not elusive.
| Oligosaccharide in Leb | Amino acid in BabA | Position of amino acid in BabA |
| Fuc1 | Cysteine (C) | 189 |
| Fuc1 | Glycine (G) | 191 |
| Fuc1 | Asparagine (N) | 194 |
| Fuc4 | Asparagine (N) | 206 |
| GlcNAc3 or Gal5 | Aspartic acid (D) | 233 |
| Gal5 | Serine (S) | 234 |
| GlcNAc3 or Gal5 | Serine (S) | 244 |
| Fuc1 | Threonine (T) | 246 |
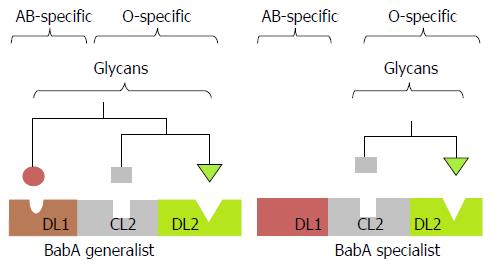
Based on their ABO blood group antigen binding preferences, BabA bearing H. pylori strains can be classified as “specialists” and “generalists”. The specialist strains prefer to bind only on blood group O specific glycans whereas generalist strains bind blood group O as well as blood group A and B specific glycans[65]. Till recently the detailed insight concept about the binding of specialists and generalists with the mono-fucosylated glycan on A, B and O blood group was unknown. However, recently the X-ray structures of the BabA specialist and generalist adhesins have been revealed according to the O and ABO blood group preferences[75]. The preference shifting was found to be due to the single amino acid substitution in its carbohydrate binding domain. The carbohydrate binding domain contains one conserved loop (CL2) and two diversity loop (DL1 and DL2). The loop DL1 is responsible for making the BabA to be either specialist or generalist. Binding of specialists with blood group O antigen involves only CL2 and DL2 loops while the binding of generalists involve CL2, DL2 as well as DL1 loops. In specialists BabA, the domain DL1 contains the amino acid leucin (L) at 198 position which makes the DL1 to be inaccessible for binding with larger glycans present on blood group A and B. The replacement of L to serine (S) in generalists makes the DL1 to be accessible for the glycans found on blood group A and B as well as O (Figure 1). In this way the generalists can bind with glycans found on blood group A, B as well as O antigen whereas the specialists can bind the glycan found on blood group O antigen only[75]. Therefore, this capability of binding explains why the peoples of blood group O are more prone to develop duodenal ulcers[65]. The fucosylated oligosaccharide residue found on the H1 antigen is abundantly expressed by the healthy gastric mucosa of most Western peoples which makes them more susceptible to colonize by generalists as well as specialists and for the development of peptic ulcer[17,76].
The binding of BabA to the Leb receptor is severely affected by the level of BabA production which is mediated on transcriptional and translational level. We detected that the strains expressing low levels (less than 10% to that of J99) of BabA protein did not mediate the enough Leb binding activity whereas, the strains expressing high level (more than 20% to that of J99) of BabA did enough Leb binding[52]. However, Saberi et al[42] categorized the H. pylori strains in four types depending on the BabA expression and Leb-binding, strains expressing high level BabA showed high Leb-binding, strains expressing low level BabA showed low Leb-binding whereas strains with no BabA expression showed no Leb-binding (8%) as well as low Leb-binding (30%) activity.
Recombination between similar sequences is called the homologous recombination. Homologous recombination takes part in the double strand DNA repair caused by environmental stress and it aids adaptation advantage for the development of persistent colonization to H. pylori to the changing gastric environment within a single host or to the new host[77-79]. H. pylori exhibits the highest genetic recombination rate than any other known bacterial species[80] and it suggests that the bacteria has a selective advantage of genetic recombination for long term survival and colonization in human stomach[81].
The allelic diversity in H. pylori is remarkable and it exhibits high transformation mediated homologous recombination[77]. Recent analyses of sequential sampling of H. pylori from the same individual elaborated the OMPs futB, babA and hopZ to be the genes with high recombination events whereas the hopQ with low recombination[82,83]. Although the function of the most closely related babA paralogs; the babB and babC, is unknown yet but extensive sequence similarity shows the intra-genomic recombination between these genes and the evolution of bab chimera[35]. The RecA-dependent intragenomic recombination between homologous genes causing fusion of portions of two or more coding regions results in the formation of chimeric genes. In H. pylori the recombinational gene conversion occurs in homologous genes babA, babB and babC. The formation of chimeric babB/A can lead to a non Leb binding strains regain Leb binding activity or it can abolish babA-dependent adhesion[35,84]. Varying proportions of chimeras have been reported from different studies across the world. In a study by Matteo et al[28], the chimeric babA/B gene was observed in 20% whereas chimeric babB/A gene was observed in 15%. Similarly, in another study among American and Korean strains, the babA/B chimera, babB/A chimera and babB/C chimera was identified in 28.7%, 2.5% and 1.25% respectively among American strains whereas babA/B chimera and babB/A chimera was identified from 6.25% and 1.25% respectively from Korean strains. The babA/B was most prevalent type of bab chimera among all types of chimera reported[32].
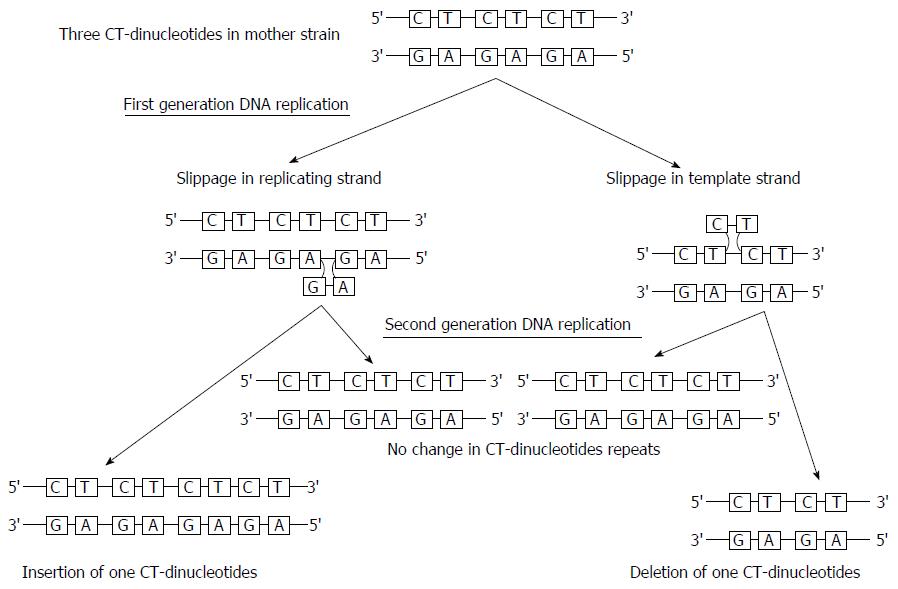
Slipped strand mispairing (SSM) is a phenomenon of either deletion or insertion of nucleotides during DNA replication because of the short, contiguous homogenous or heterogeneous repetitive DNA sequence of 6 base pairs or less. This repetitive sequence, susceptible for mispairing is designated as short sequence repeats (SSR), microsatellites or variable number of tandem repeats (VNTR)[85,86]. SSM is one of the mechanisms resulting in the phase variation that produces nucleotide mispairing between the mother and daughter strand during the DNA replication[87,88]. The OMP genes such as babB, hopZ, oipA that possess the dinucleotide CT-repeats in the 5’-coding region, likely undergo SSM to regulate their expression by phase variation[18,89,90]. Nucleotide slippage can be occurred in replicating or template strand. If slippage occurs in replicating strand, there is insertion of one CT-dinuleotide and if slippage occurs in template strand, there is deletion of one CT-dinucleotide (Figure 2)[91]. This phenomenon leading to the variation in the number of CT-dinucleotides over time may provide certain advantages for adaptation to different niches and stomach micro-environments via immune evasion or adhesion. In H. pylori, the fragments of babB containing CT repeats may recombine with babA or babC resulting in the formation of chimeric genes. According to the Bäckström et al[34] the strains with in-frame protein expression (ON) status contain 8-CT repeats whereas out of frame protein expression (OFF) phenotypes contained 7 or 9 CT repeats. The gain or loss of one CT-dinucleotide creates frame shifts and loss of expression of babA.
The number of CT-repeats in the 5’coding region of nucleotide sequence decides whether the protein expression is ON or OFF. The protein expression becomes in-frame if the number of CT-repeats is 5, 8, 11 and 14 whereas if the number of CT-repeats is 6, 7, 9, 10, 12, 13 it makes protein expression out of frame[30,34,84]. Out of frame status of protein expression evolved due to the insertion or deletion of one nucleotide resulting in the truncated (immature) proteins whereas if the status is in-frame it results in the full-length protein expression[92].
The genes containing variable number of nucleotide repeats may go to the phase variation. Phase variation is an adaptation mechanism that provides advantages for colonization[93]. In a study, the BabA expression was lost during the experimental infection in rhesus macaques by phase variation or allele replacement with BabB and in subsequent follow up study in mouse, the BabA expression was found to be lost due to phase variation in 5’-CT repeat regions of H. pylori strain J166[39]. The Leb binding clones from OFF to ON phase conversion express BabA adhesin that are functionally equivalent to the wild type but the quantity of BabA adhesin less abundant than the wild type[34].
The attachment of H. pylori via BabA mediates several outcomes. H. pylori BabA binds with Leb and induces the double strand breaks in the host cells, which is thought to be independent of VacA, γ-glutamyl transpeptidase and the cag PAI[94]. However, it has been also confirmed that BabA-positive status of infecting strains is associated with CagA-positive, OipA-positive (oipA “on”) and vacA s1 genotype for the development of intestinal metaplasia[95]. The infection with BabA-positive strains has been associated with the increased risk for development of peptic ulcer diseases in Western countries[41,96]. However, the association of BabA with peptic ulcer diseases has not been confirmed in patients from East Asian or some other Western countries[97-99]. It has been claimed that the BabA-mediated attachment to gastric epithelial cells might enhance CagA translocation and the enhancement of inflammation[100]. Furthermore, the triple-positive (BabA, CagA and vacA s1-positive) strains of H. pylori shows greater colonization densities, elevated levels of gastric inflammation and a higher incidence of intestinal metaplasia in patients in contrast with the only CagA and vacAs1 positive strains[101,41]. Despite of the association of high level BabA with severe clinical outcomes, surprisingly, it has been also found that low level BabA expressing strains could more likely be associated with increased mucosal inflammation and severe clinical outcomes compared to that of high level BabA-positive and Leb binding strains[42,52]. Although, the underlying mechanisms remains unclear, it has been hypothesized that during adulthood, the induced hyperacidity may enhance the development of gastric metaplastic patches in the duodenum and the H. pylori expressing low levels of BabA and Leb attachment are able to detach from the gastric niche and reattached to the gastric metaplastic patches in the duodenum and develop the ulcers at the gastric-duodenal tissue border[42]. The BabA adhesin expression also seems tightly associated with the onset of type 4 secretory system (T4SS)-related host response in vivo[100]. In an in vivo study, using the Mongolian gerbils, the BabA-mediated adherence of H. pylori to the epithelial cells has been shown to augment the cag PAI dependent H. pylori pathogenicity. The BabA-Leb interaction can trigger the production of pro-inflammatory cytokines and some factors that can enhance the cancer development[101].
H. pylori infection is a major cause of gastric complications including gastric cancer which is the third leading cause for mortality among all cancer related deaths. H. pylori being a Gram-negative organism, the OMP present in the cell envelope provides the initial step binding with the Leb antigen for the persistent colonization. BabA is capable of binding with receptors found on the oral mucosa and gastric mucus layer. The closer insight of bab genes revealed the presence of dinucleotide (CT) repeats in 5’-region which causes SSM leading to the phase variation. The formation of chimera with the homologous genes babB and babC causes the regulation of protein expression on transcriptional level. In addition to the high level BabA expression associated with the severe clinical outcomes many reports have also suggested the low level expression with severe outcomes where the underlying mechanism is not clear yet. Even though many roles of BabA in disease process have been evaluated, the role of BabB and BabC has not been assessed yet. Further study is under demand for the assessment of the role of BabB and BabC as well as to elaborate the underlying mechanism of low expression BabA and severe clinical outcomes.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kuo FC, Safaei HG, Yamaoka Y S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF
| 1. | Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1446] [Cited by in F6Publishing: 1410] [Article Influence: 78.3] [Reference Citation Analysis (1)] |
| 2. | Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1-241. [PubMed] [Cited in This Article: ] |
| 3. | IARC Helicobacter pylori Working Group. Helicobacter pylori Eradication as a Strategy for Preventing Gastric Cancer. Lyon, France 2014: International Agency for Research on Cancer (IARC Working Group Reports, No. 8). Available from: http://www.iarc.fr/en/publications/pdfsonline/wrk/wrk8/index.php. [Cited in This Article: ] |
| 4. | Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 643] [Cited by in F6Publishing: 594] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 5. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20108] [Cited by in F6Publishing: 19780] [Article Influence: 2197.8] [Reference Citation Analysis (17)] |
| 6. | GLOBOCON 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. [Cited in This Article: ] |
| 7. | Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 817] [Cited by in F6Publishing: 886] [Article Influence: 63.3] [Reference Citation Analysis (1)] |
| 8. | Mamishi S, Eshaghi H, Mahmoudi S, Bahador A, Hosseinpour Sadeghi R, Najafi M, Farahmand F, Khodadad A, Pourakbari B. Intrafamilial transmission of Helicobacter pylori: genotyping of faecal samples. Br J Biomed Sci. 2016;73:38-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Goh KL, Chan WK, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16 Suppl 1:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 232] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 10. | Bui D, Brown HE, Harris RB, Oren E. Serologic Evidence for Fecal-Oral Transmission of Helicobacter pylori. Am J Trop Med Hyg. 2016;94:82-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Ranjbar R, Khamesipour F, Jonaidi-Jafari N, Rahimi E. Helicobacter pylori isolated from Iranian drinking water: vacA, cagA, iceA, oipA and babA2 genotype status and antimicrobial resistance properties. FEBS Open Bio. 2016;6:433-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Talaei R, Souod N, Momtaz H, Dabiri H. Milk of livestock as a possible transmission route of Helicobacter pylori infection. Gastroenterol Hepatol Bed Bench. 2015;8:S30-S36. [PubMed] [Cited in This Article: ] |
| 13. | Graham DY, Malaty HM, Evans DG, Evans DJ, Klein PD, Adam E. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Effect of age, race, and socioeconomic status. Gastroenterology. 1991;100:1495-1501. [PubMed] [Cited in This Article: ] |
| 14. | Rowland M, Daly L, Vaughan M, Higgins A, Bourke B, Drumm B. Age-specific incidence of Helicobacter pylori. Gastroenterology. 2006;130:65-72; quiz 211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 71] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, Sicinschi LA, Shaffer CL, Romero-Gallo J, de Sablet T. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA. 2014;111:1455-1460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 163] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 16. | Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629-641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 420] [Cited by in F6Publishing: 428] [Article Influence: 30.6] [Reference Citation Analysis (1)] |
| 17. | Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892-1895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 840] [Cited by in F6Publishing: 777] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 18. | Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573-578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 639] [Cited by in F6Publishing: 626] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 19. | Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533-7538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 305] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 20. | Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155-4168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 258] [Article Influence: 10.8] [Reference Citation Analysis (1)] |
| 21. | Yamaoka Y. Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis. World J Gastroenterol. 2008;14:4265-4272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 67] [Cited by in F6Publishing: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Moore ME, Borén T, Solnick JV. Life at the margins: modulation of attachment proteins in Helicobacter pylori. Gut Microbes. 2011;2:42-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537-1548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 176] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325-3333. [PubMed] [Cited in This Article: ] |
| 25. | Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373-377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 872] [Cited by in F6Publishing: 814] [Article Influence: 31.3] [Reference Citation Analysis (1)] |
| 26. | Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Pride DT, Meinersmann RJ, Blaser MJ. Allelic Variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160-1171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 28. | Matteo MJ, Armitano RI, Romeo M, Wonaga A, Olmos M, Catalano M. Helicobacter pylori bab genes during chronic colonization. Int J Mol Epidemiol Genet. 2011;2:286-291. [PubMed] [Cited in This Article: ] |
| 29. | Hennig EE, Allen JM, Cover TL. Multiple chromosomal loci for the babA gene in Helicobacter pylori. Infect Immun. 2006;74:3046-3051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Colbeck JC, Hansen LM, Fong JM, Solnick JV. Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect Immun. 2006;74:4375-4378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | National Center for Biotechnology Information (NCBI): Helicobacter pylori. Available from: https://www.ncbi.nlm.nih.gov/genome/genomes/169. [Cited in This Article: ] |
| 32. | Kim A, Servetas SL, Kang J, Kim J, Jang S, Cha HJ, Lee WJ, Kim J, Romero-Gallo J, Peek RM. Helicobacter pylori bab Paralog Distribution and Association with cagA, vacA, and homA/B Genotypes in American and South Korean Clinical Isolates. PLoS One. 2015;10:e0137078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect Immun. 2004;72:3429-3435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Bäckström A, Lundberg C, Kersulyte D, Berg DE, Borén T, Arnqvist A. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding. Proc Natl Acad Sci USA. 2004;101:16923-16928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Goodwin AC, Weinberger DM, Ford CB, Nelson JC, Snider JD, Hall JD, Paules CI, Peek RM, Forsyth MH. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology. 2008;154:2231-2240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 36. | Kao CY, Sheu SM, Sheu BS, Wu JJ. Length of thymidine homopolymeric repeats modulates promoter activity of sabA in Helicobacter pylori. Helicobacter. 2012;17:203-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 37. | Åberg A, Gideonsson P, Vallström A, Olofsson A, Öhman C, Rakhimova L, Borén T, Engstrand L, Brännström K, Arnqvist A. A repetitive DNA element regulates expression of the Helicobacter pylori sialic acid binding adhesin by a rheostat-like mechanism. PLoS Pathog. 2014;10:e1004234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2014;38:119-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 39. | Styer CM, Hansen LM, Cooke CL, Gundersen AM, Choi SS, Berg DE, Benghezal M, Marshall BJ, Peek RM, Borén T. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect Immun. 2010;78:1593-1600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Nell S, Kennemann L, Schwarz S, Josenhans C, Suerbaum S. Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans. MBio. 2014;5:pii: e02281-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 41. | Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778-12783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 482] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 42. | Saberi S, Schmidt A, Eybpoosh S, Esmaili M, Talebkhan Y, Mohajerani N, Oghalaie A, Eshagh Hosseini M, Mohagheghi MA, Bugaytova J. Helicobacter pylori Strains from Duodenal Ulcer Patients Exhibit Mixed babA/B Genotypes with Low Levels of BabA Adhesin and Lewis b Binding. Dig Dis Sci. 2016;61:2868-2877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 43. | Abdullah SM, Hussein NR, Salih AM, Merza MA, Goreal AA, Odeesh OY, Majed HS, Assafi MA, Hawrami K. Infection with Helicobacter pylori strains carrying babA2 and cagA is associated with an increased risk of peptic ulcer disease development in Iraq. Arab J Gastroenterol. 2012;13:166-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 44. | Karabiber H, Selimoglu MA, Otlu B, Yildirim O, Ozer A. Virulence factors and antibiotic resistance in children with Helicobacter pylori gastritis. J Pediatr Gastroenterol Nutr. 2014;58:608-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Talebi Bezmin Abadi A, Taghvaei T, Mohabbati Mobarez A, Vaira G, Vaira D. High correlation of babA 2-positive strains of Helicobacter pylori with the presence of gastric cancer. Intern Emerg Med. 2013;8:497-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Yadegar A, Mobarez AM, Alebouyeh M, Mirzaei T, Kwok T, Zali MR. Clinical relevance of cagL gene and virulence genotypes with disease outcomes in a Helicobacter pylori infected population from Iran. World J Microbiol Biotechnol. 2014;30:2481-2490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Osman HA, Hasan H, Suppian R, Hassan S, Andee DZ, Abdul Majid N, Zilfalil BA. Prevalence of Helicobacter pylori cagA, babA2, and dupA genotypes and correlation with clinical outcome in Malaysian patients with dyspepsia. Turk J Med Sci. 2015;45:940-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Boonyanugomol W, Chomvarin C, Sripa B, Chau-In S, Pugkhem A, Namwat W, Wongboot W, Khampoosa B. Molecular analysis of Helicobacter pylori virulent-associated genes in hepatobiliary patients. HPB (Oxford). 2012;14:754-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, Tor-Udom S, Vilaichone RK. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 50. | Ghosh P, Sarkar A, Ganguly M, Raghwan J, De R, Mukhopadhyay AK. Helicobacter pylori strains harboring babA2 from Indian sub population are associated with increased virulence in ex vivo study. Gut Pathog. 2016;8:1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 51. | Con SA, Takeuchi H, Nishioka M, Morimoto N, Sugiura T, Yasuda N, Con-Wong R. Clinical relevance of Helicobacter pylori babA2 and babA2/B in Costa Rica and Japan. World J Gastroenterol. 2010;16:474-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 9] [Cited by in F6Publishing: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 52. | Fujimoto S, Olaniyi Ojo O, Arnqvist A, Wu JY, Odenbreit S, Haas R, Graham DY, Yamaoka Y. Helicobacter pylori BabA expression, gastric mucosal injury, and clinical outcome. Clin Gastroenterol Hepatol. 2007;5:49-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Biernat MM, Gościniak G, Iwańczak B. Prevalence of Helicobacter pylori cagA, vacA, iceA, babA2 genotypes in Polish children and adolescents with gastroduodenal disease. Postepy Hig Med Dosw (Online). 2014;68:1015-1021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 54. | Homan M, Šterbenc A, Kocjan BJ, Luzar B, Zidar N, Orel R, Poljak M. Prevalence of the Helicobacter pylori babA2 gene and correlation with the degree of gastritis in infected Slovenian children. Antonie Van Leeuwenhoek. 2014;106:637-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 55. | Boyanova L, Yordanov D, Gergova G, Markovska R, Mitov I. Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristics. Antonie Van Leeuwenhoek. 2010;98:343-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 56. | Odenbreit S. Adherence properties of Helicobacter pylori: impact on pathogenesis and adaptation to the host. Int J Med Microbiol. 2005;295:317-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 57. | Rhen M, Eriksson S, Clements M, Bergström S, Normark SJ. The basis of persistent bacterial infections. Trends Microbiol. 2003;11:80-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 58. | Aspholm M, Kalia A, Ruhl S, Schedin S, Arnqvist A, Lindén S, Sjöström R, Gerhard M, Semino-Mora C, Dubois A. Helicobacter pylori adhesion to carbohydrates. Methods Enzymol. 2006;417:293-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 59. | Walz A, Odenbreit S, Stühler K, Wattenberg A, Meyer HE, Mahdavi J, Borén T, Ruhl S. Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics. 2009;9:1582-1592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Prakobphol A, Borén T, Ma W, Zhixiang P, Fisher SJ. Highly glycosylated human salivary molecules present oligosaccharides that mediate adhesion of leukocytes and Helicobacter pylori. Biochemistry. 2005;44:2216-2224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 61. | Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, Frängsmyr L, Holmskov U, Leffler H, Nilsson C. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275:39860-39866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 177] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 62. | Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, Redwan el-RM, Wilson IA, Daha MR, Dwek RA, Rudd PM. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem. 2003;278:20140-20153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 245] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 63. | Issa S, Moran AP, Ustinov SN, Lin JH, Ligtenberg AJ, Karlsson NG. O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine. Glycobiology. 2010;20:1046-1057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265-278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 907] [Cited by in F6Publishing: 928] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 65. | Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305:519-522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 290] [Cited by in F6Publishing: 291] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 66. | Hage N, Howard T, Phillips C, Brassington C, Overman R, Debreczeni J, Gellert P, Stolnik S, Winkler GS, Falcone FH. Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA. Sci Adv. 2015;1:e1500315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 67. | Sakamoto S, Watanabe T, Tokumaru T, Takagi H, Nakazato H, Lloyd KO. Expression of Lewisa, Lewisb, Lewisx, Lewisy, siayl-Lewisa, and sialyl-Lewisx blood group antigens in human gastric carcinoma and in normal gastric tissue. Cancer Res. 1989;49:745-752. [PubMed] [Cited in This Article: ] |
| 68. | Lindén S, Mahdavi J, Hedenbro J, Borén T, Carlstedt I. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem J. 2004;384:263-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 69. | Kenny DT, Skoog EC, Lindén SK, Struwe WB, Rudd PM, Karlsson NG. Presence of terminal N-acetylgalactosamineβ1-4N-acetylglucosamine residues on O-linked oligosaccharides from gastric MUC5AC: involvement in Helicobacter pylori colonization? Glycobiology. 2012;22:1077-1085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 70. | Magalhães A, Rossez Y, Robbe-Masselot C, Maes E, Gomes J, Shevtsova A, Bugaytsova J, Borén T, Reis CA. Muc5ac gastric mucin glycosylation is shaped by FUT2 activity and functionally impacts Helicobacter pylori binding. Sci Rep. 2016;6:25575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 71. | Lindén SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, Sutton P, McGuckin MA. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5:e1000617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 72. | Cohen M, Drut R, Cueto Rúa E. SIALYL-Tn antigen distribution in Helicobacter pylori chronic gastritis in children: an immunohistochemical study. Pediatr Pathol Mol Med. 2003;22:117-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 73. | Lindén S, Semino-Mora C, Liu H, Rick J, Dubois A. Role of mucin Lewis status in resistance to Helicobacter pylori infection in pediatric patients. Helicobacter. 2010;15:251-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 74. | Lindén S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, Borén T, Dubois A. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 75. | Moonens K, Gideonsson P, Subedi S, Bugaytsova J, Romaõ E, Mendez M, Nordén J, Fallah M, Rakhimova L, Shevtsova A. Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori. Cell Host Microbe. 2016;19:55-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 76. | Van de Bovenkamp JH, Mahdavi J, Korteland-Van Male AM, Büller HA, Einerhand AW, Borén T, Dekker J. The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach. Helicobacter. 2003;8:521-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 77. | Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 264] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 78. | Borst P, Greaves DR. Programmed gene rearrangements altering gene expression. Science. 1987;235:658-667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 217] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 79. | Dworkin J, Blaser MJ. Molecular mechanisms of Campylobacter fetus surface layer protein expression. Mol Microbiol. 1997;26:433-440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 65] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 80. | Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619-12624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 473] [Cited by in F6Publishing: 440] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 81. | Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun. 2003;71:2022-2031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 82. | Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108:5033-5038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 83. | Yahara K, Kawai M, Furuta Y, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M. Genome-wide survey of mutual homologous recombination in a highly sexual bacterial species. Genome Biol Evol. 2012;4:628-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 84. | Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101:2106-2111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 193] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 85. | Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci USA. 1998;95:1647-1652. [PubMed] [Cited in This Article: ] |
| 86. | van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275-293. [PubMed] [Cited in This Article: ] |
| 87. | Henderson IR, Owen P, Nataro JP. Molecular switches--the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919-932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 316] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 88. | Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203-221. [PubMed] [Cited in This Article: ] |
| 89. | Kennemann L, Brenneke B, Andres S, Engstrand L, Meyer TF, Aebischer T, Josenhans C, Suerbaum S. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect Immun. 2012;80:4364-4373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 90. | Zhang J, Qian J, Zhang X, Zou Q. Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pylori. Mol Biol Rep. 2014;41:7807-7814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 91. | Myers P. Tandem repeats and morphological variation. Nature Education. 2007;1 Available from: http://www.nature.com/scitable/topicpage/tandem-repeats-and-morphological-variation-40690. [Cited in This Article: ] |
| 92. | de Vries N, Duinsbergen D, Kuipers EJ, Pot RG, Wiesenekker P, Penn CW, van Vliet AH, Vandenbroucke-Grauls CM, Kusters JG. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J Bacteriol. 2002;184:6615-6623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 93. | van der Woude MW, Bäumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581-611, table of contents. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 494] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 94. | Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci USA. 2011;108:14944-14949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 95. | Azevedo M, Eriksson S, Mendes N, Serpa J, Figueiredo C, Resende LP, Ruvoën-Clouet N, Haas R, Borén T, Le Pendu J. Infection by Helicobacter pylori expressing the BabA adhesin is influenced by the secretor phenotype. J Pathol. 2008;215:308-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 96. | Olfat FO, Zheng Q, Oleastro M, Voland P, Borén T, Karttunen R, Engstrand L, Rad R, Prinz C, Gerhard M. Correlation of the Helicobacter pylori adherence factor BabA with duodenal ulcer disease in four European countries. FEMS Immunol Med Microbiol. 2005;44:151-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 97. | Sheu BS, Sheu SM, Yang HB, Huang AH, Wu JJ. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52:927-932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 98. | Mizushima T, Sugiyama T, Komatsu Y, Ishizuka J, Kato M, Asaka M. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463-2465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 99. | Oleastro M, Cordeiro R, Yamaoka Y, Queiroz D, Mégraud F, Monteiro L, Ménard A. Disease association with two Helicobacter pylori duplicate outer membrane protein genes, homB and homA. Gut Pathog. 2009;1:12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 100. | Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Borén T, Haas R, Sasakawa C, Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem. 2011;286:25256-25264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 101. | Rad R, Gerhard M, Lang R, Schöniger M, Rösch T, Schepp W, Becker I, Wagner H, Prinz C. The Helicobacter pylori blood group antigen-binding adhesin facilitates bacterial colonization and augments a nonspecific immune response. J Immunol. 2002;168:3033-3041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 127] [Article Influence: 5.8] [Reference Citation Analysis (0)] |