Published online Jun 28, 2016. doi: 10.3748/wjg.v22.i24.5578
Peer-review started: February 29, 2016
First decision: March 31, 2016
Revised: April 13, 2016
Accepted: April 20, 2016
Article in press: April 20, 2016
Published online: June 28, 2016
AIM: To perform sequencing analysis in patients with very early-onset inflammatory bowel disease (VEO-IBD) to determine the genetic basis for VEO-IBD in Chinese pediatric patients.
METHODS: A total of 13 Chinese pediatric patients with VEO-IBD were diagnosed from May 2012 and August 2014. The relevant clinical characteristics of these patients were analyzed. Then DNA in the peripheral blood from patients was extracted. Next generation sequencing (NGS) based on an Illumina-Miseq platform was used to analyze the exons in the coding regions of 10 candidate genes: IL-10, IL-10RA, IL-10RB, NOD2, FUT2, IL23R, GPR35, GPR65, TNFSF15, and ADAM30. The Sanger sequencing was used to verify the variations detected in NGS.
RESULTS: Out of the 13 pediatric patients, ten were diagnosed with Crohn’s disease, and three diagnosed with ulcerative colitis. Mutations in IL-10RA and IL-10RB were detected in five patients. There were four patients who had single nucleotide polymorphisms associated with IBD. Two patients had IL-10RA and FUT2 polymorphisms, and two patients had IL-10RB and FUT2 polymorphisms. Gene variations were not found in the rest four patients. Children with mutations had lower percentile body weight (1.0% vs 27.5%, P = 0.002) and hemoglobin (87.4 g/L vs 108.5 g/L, P = 0.040) when compared with children without mutations. Although the age of onset was earlier, height was shorter, and the response to treatment was poorer in the mutation group, there was no significant difference in these factors between groups.
CONCLUSION: IL-10RA and IL-10RB mutations are common in Chinese children with VEO-IBD. Patients with mutations have an earlier disease onset, lower body weight and hemoglobin, and poorer prognosis.
Core tip: In this small-sample size study, we performed next generation sequencing for 10 candidate genes in Chinese pediatric patients with very early onset inflammatory bowel disease. We found that IL-10RA and IL-10RB mutations were common. There were five patients harbouring mutations in these two genes and accounted for 38.5% of all samples. Besides, there were four patients who had single nucleotide polymorphisms associated with inflammatory bowel disease. Pediatric patients with mutations had an earlier disease onset, lower body weight, markedly lower hemoglobin, and poorer prognosis.
- Citation: Xiao Y, Wang XQ, Yu Y, Guo Y, Xu X, Gong L, Zhou T, Li XQ, Xu CD. Comprehensive mutation screening for 10 genes in Chinese patients suffering very early onset inflammatory bowel disease. World J Gastroenterol 2016; 22(24): 5578-5588
- URL: https://www.wjgnet.com/1007-9327/full/v22/i24/5578.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i24.5578
Inflammatory bowel disease (IBD) is a chronic and recurrent gastrointestinal inflammatory disease in children. Based on clinical characteristics, laboratory tests, and endoscopic and pathological presentations, IBD can be subdivided into Crohn’s disease (CD), ulcerative colitis (UC), and IBD-unclassified (IBD-U)[1]. Our previous study showed that the annual incidence of IBD in the 0- to 14-year age group of Shanghai residents steadily increased from 2000 to 2010[2]. Although pediatric IBD mainly occurs in adolescence[2], approximately 15% of IBD pediatric patients have very early-onset IBD (VEO-IBD) that begins before 6 years of age, and 1% of children develop this disease before reaching 1 year of age[3,4]. The majority of VEO-IBD cases have clinical characteristics that are distinct from those of classic IBD with adult and adolescent onset. VEO-IBD has more severe clinical symptoms, resistance to a variety of immunosuppressive therapies, and a poor prognosis after conventional treatments. Some scholars even consider VEO-IBD to be a completely different disease from classic IBD[5].
Previous studies suggested that persistent intestinal immune dysfunction in a genetically susceptible individual exposed to adverse environmental factors is an important mechanism for IBD development. Genome-wide association studies (GWAS) have discovered a total of 163 loci associated with the risk for IBD development[6]. However, disease onset at an early stage of life suggests a leading role for rare gene variations in VEO-IBD patients, especially in children with a disease onset before the age of 1 year. These low frequency mutations are difficult to detect using GWAS. Next generation sequencing technology allows for the high-throughput sequencing of exons in a series of genes concurrently; therefore, rare gene variations can be discovered[7]. Since Glocker et al[8] first discovered in 2009 that mutations in genes encoding the α subunit (IL-10R1, encoding gene IL-10RA) and the β subunit (IL-10R2, encoding gene IL-10RB) of the interleukin-10 (IL-10) receptor could induce VEO-IBD development, a few studies have continuously discovered mutations in genes encoding IL-10R1, IL-10R2, and IL-10[5,9-12]. However, current reports are limited, and the majority of studies are small-size case studies. Reports on the Han Chinese population are scarcer[13,14].
This study used the Illumina-Miseq platform to sequence candidate genes in Han Chinese children diagnosed with VEO-IBD. The candidate genes included genes involved in the IL-10 signaling pathway, such as IL-10, IL-10RA, and IL-10RB, and genes highly associated with the development of CD in previous studies, including NOD2, FUT2, IL-23R, GPR35, GPR65, TNFSF15, and ADAM30. This study furthers our understanding of the genetic factors associated with VEO-IBD development in Han Chinese children.
Verbal and written consent was obtained from the parents of all of children included this study. Ethic committee approval for the study was granted by Institutional Review Boards of Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
A total of 13 pediatric patients with repeated diarrhea, mucus and bloody stool, or abdominal pain who were diagnosed by laboratory tests and digestive endoscopy with VEO-IBD in the Pediatric Department of Ruijin Hospital of Shanghai Jiao Tong University School of Medicine between May 2012 and August 2014 were included in this study. All of the patients were Han Chinese. VEO-IBD was defined as IBD onset before the age of 6 years, and a disease onset before 2 years of age was called infantile-onset IBD[15,16]. The clinical characteristics of these pediatric patients, including gender, age of disease onset, body height, body weight, family history, clinical symptoms, complications, major laboratory examinations, endoscopic presentations, and therapeutic effects, were retrospectively analyzed.
Relevant laboratory examinations, including complete blood count (CBC), C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), tumor necrosis factor α (TNF-α) level, immunoglobulins G, A, M, and E, vitamin D, human Immunodeficiency virus (HIV) and human cytomegalovirus (CMV) antibody detection in serum, T lymphocyte flow cytometry sorting, stool parasite tests, stool culture, and stool Clostridium difficile toxin detection, were performed when the patients were admitted to the hospital. Common infectious diseases and primary immunodeficiency diseases were excluded.
All patients received a colonoscopy under general anesthesia. A biopsy of the colonic mucosa under endoscopy was performed for pathological examination.
Genomic DNA extraction: After obtaining verbal and written informed consent from the patients’ parents, genomic DNA in the peripheral blood from 13 pediatric patients was extracted using a FlexiGene DNA Kit (Qiagen Inc., Germany). Another 100 copies of DNA extracted from patients suffering from idiopathic short stature (ISS) in previous research were used to test frequency of mutant sites which were newly detected in our study.
Multiplex PCR primer design: Based on the stability of the Illumina-Miseq experiment and the operability of subsequent steps, the length requirement of target fragments for sequencing was < 400 bp. If the length of an exon was longer than 400 bp, an additional pair of primers was designed with overlapping bases of adjacent fragments. To avoid a high number of non-target fragment products, primers were grouped and suspended in a primer mix before the multiplex PCR was performed. The concentration of each primer in the primer mix was 10 mmol/L. The basic requirement for grouping was the lack of matching sequences between two of the amplified products. Oligo 7 software was used to design primers for exons of the encoding region of the 10 candidate genes: IL-10, IL-10RA, IL-10RB, NOD2, FUT2, IL23R, GPR35, GPR65, TNFSF15, and ADAM30. A total of 86 pairs of primers were designed. The sequences are shown in supplementary Table 1.
Multiplex PCR amplification of candidate genes: A Qiagen Multiple PCR Kit was used in this study. The PCR amplification reaction system had a total volume of 21 μL, including 4 μL of ddH2O, 2 μL of Q-solution (5 ×), 4 μL of 10 mmol/L primer mix, 10 μL of buffer mix, and 1 μL of the DNA template (20 ng/μL). The reaction procedure consisted of pre-denaturation at 94 °C for 15 min, denaturation at 94 °C for 40 s, annealing at 63 °C for 1 min, and extension at 72 °C for 40 s. After each cycle, the annealing temperature was reduced by 0.5 °C for 10 cycles until the annealing temperature reached 58 °C. Next, the amplification was continued for 30 cycles with a constant annealing temperature of 58 °C. The final extension at 72 °C lasted for 10 min. The PCR products were stained with 100 × GelRed and subjected to 1% agarose electrophoresis (120 V for 60 min).
The purified multiplex PCR products were sent to Shanghai South Gene Technology Co., Ltd. for sequencing analysis with the Illumina-Miseq platform. After sequencing, the nucleotide sequence information was compared with the standard gene sequences available in GenBank. The obtained gene mutation sites were compared with information in the dbSNP, HGMD, and OMIM databases to determine if the mutations had been previously reported.
To confirm the accuracy of the results, the corresponding gene sequences for the mutations discovered using the Illumina-Miseq platform were sequenced again using the Sanger sequencing method.
The newly discovered gene variation sites were analyzed to predict their influence on protein functions using two online databases: SIFT (http://http://sift.jcvi.org/) and PolyPhen 2 (http://genetics.bwh.harvard.edu/pph2/).
According to the sequencing results, the 13 pediatric patients were divided into two groups. The patients who harboured pathogenic mutations were in group 1. Those without pathogenic mutations (including presence of polymorphisms only or wild type) were in group 2. The differences in diagnosis, age of disease onset, growth indicators (percentiles of body weight and height were calculated according to WHO standards), complications (perianal diseases and recurrent infection), and therapeutic effects among all groups were compared. Because the sample size was small, quantitative and ranked ordinal data were subjected to nonparametric statistics. The Mann-Whitney Test was performed, and the difference was statistically analyzed using exact probability. SPSS13.0 for Windows software was used for the statistical analysis. P < 0.05 indicated a significant difference.
IL-10RA, IL-10RB, and IL-10 mutations:IL-10RA mutations were detected in four patients, an IL-10RB mutation was detected in one patient, and an IL-10 mutation was not detected in any of the 13 patients.
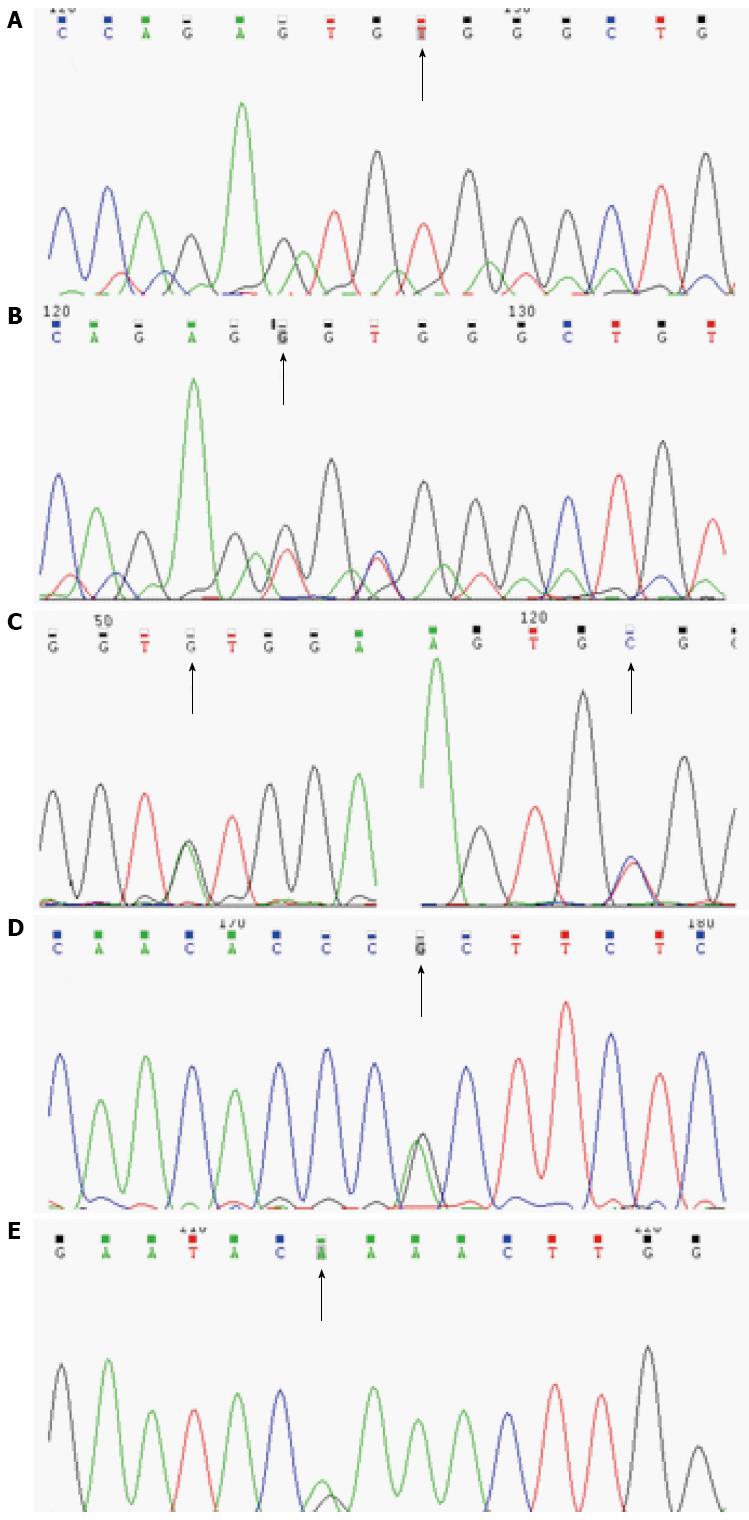
The detected IL-10RA mutations were all in exon 3: c.A191G (p.Y64C), c.T299G (p.V100G), c.C301T (p.R101W), and c.G350A (rs199989396) (p.R117H). The p.R101W mutation was the most common and was detected in three patients (patients 1-3). The other mutations were detected in only one patient. Patient 1 had a homozygous mutation, patients 2 and 3 had compound mutations, and patient 4 had a heterozygous mutation (Table 1 and Figure 1).
| Patient | Gene | Variation | Homo/Heterozygote | Function defect |
| 1 | IL-10RA | p.R101W | Homozygote | Yes |
| 2 | IL-10RA | p.R101W | Compound heterozygote | Yes |
| p.V100G (novel mutation) | Pathogenic supporting by Polyphen 2 and SIFT | |||
| 3 | IL-10RA | p.R101W | Compound heterozygote | Yes |
| p.Y64C (novel mutation) | Pathogenic supporting by Polyphen 2 and SIFT | |||
| 4 | IL-10RA | p.R117H (rs199989396) | Heterozygote | Yes |
| NOD2 | p.R703C (rs5743277) | Heterozygote | Susceptibility to CD recorded in HGMD | |
| FUT2 | p.I140F (rs1047781) | Heterozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] | |
| 5 | IL-10RB | p.K47E (rs2834167) | Homozygote | SNP in a VEO-UC child reported by Galatola et al[29] |
| p.E141K (rs387907326) | Heterozygote | Pathogenic supporting by Polyphen 2 and SIFT | ||
| IL-10RA | p.P115P (rs22280554) | Homozygote | Susceptibility to VEO-IBD reported by Moran et al[30] | |
| p.I224V (rs22280555) | Homozygote | |||
| FUT2 | p.I140F (rs1047781) | Heterozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] | |
| 6 | IL-10RA | p.P115P (rs22280554) | Homozygote | Susceptibility to VEO-IBD reported by Moran et al[30] |
| p.I224V (rs22280555) | Homozygote | |||
| FUT2 | p.I140F (rs1047781) | Homozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] | |
| 7 | IL-10RA | p.P115P (rs22280554) | Homozygote | Susceptibility to VEO-IBD reported by Moran et al[30] |
| p.I224V (rs22280555) | Homozygote | |||
| FUT2 | p.I140F (rs1047781) | Heterozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] | |
| 8 | IL-10RB | p.K47E (rs2834167) | Homozygote | SNP in a VEO-UC child reported by Galatola et al[29] |
| FUT2 | p.I140F (rs1047781) | Heterozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] | |
| 9 | IL-10RB | p.K47E (rs2834167) | Heterozygote | SNP in a VEO-UC child reported by Galatola et al[29] |
| FUT2 | p.I140F (rs1047781) | Heterozygote | Susceptibility to CD in Chinese population reported by Hu et al[31] |
Among detected IL-10RA mutations, p.Y64C and p.V100G were new mutations that were predicted to be deleterious by SIFT and Polyphen 2. These novel mutant sites were not found in 100 ISS children. The other two mutations had been confirmed to be deleterious in several studies[5,12,14,17].
An IL-10RB heterozygous mutation was detected in one patient (patient 5) (Table 1 and Figure 1). This c.G421A (p.E141K) (rs387907326) mutation was located in exon 4 and was also predicted as a deleterious mutation by SIFT and Polyphen 2. A nonsense mutation in the same site was detected in previous studies[11,18].
After the sequence analysis of the coding regions of 10 candidate genes, we found that six patients (patient 4, 5, 6, 7, 8 and 9) had many IBD-associated single nucleotide polymorphisms (SNPs) in IL-10RA, IL-10RB, NOD2, and FUT2. The SNP loci in IL-10RA were rs22280554: c.G525A, p. P175P and rs22280555: c.A670G, p.I224V; the SNP locus in IL-10RB was rs2834167: c.A139G, p.K47E; the SNP locus in NOD2 was rs5743277: c.C2107T, p.R703C; and the SNP locus in FUT2 was rs1047781: c.A418T, p.I140F (Table 1).
In addition to the detected p.R117H heterozygous mutation in IL-10RA, patient 4 also had heterozygous SNPs in NOD2 and FUT2.
Patient 5 had a heterozygous p.E141K mutation (rs387907326) in IL-10RB and SNPs in IL-10RA, IL-10RB, and FUT2. The SNP loci in IL-10RA were rs22280554 and rs22280555. The homozygous SNP loci for IL-10RB were rs2834167. The SNP in FUT2 was heterozygous.
Patients 6 and 7 had SNPs in IL-10RA and FUT2. Patients 8 and 9 had SNPs in IL-10RB and FUT2.
Four patients did not show any IBD-associated variations in the coding regions of the 10 candidate genes.
There was no IBD-associated variation discovered in the coding regions of six genes: IL-10, IL-23R, GPR35, GPR65, TNFSF15, and ADAM30.
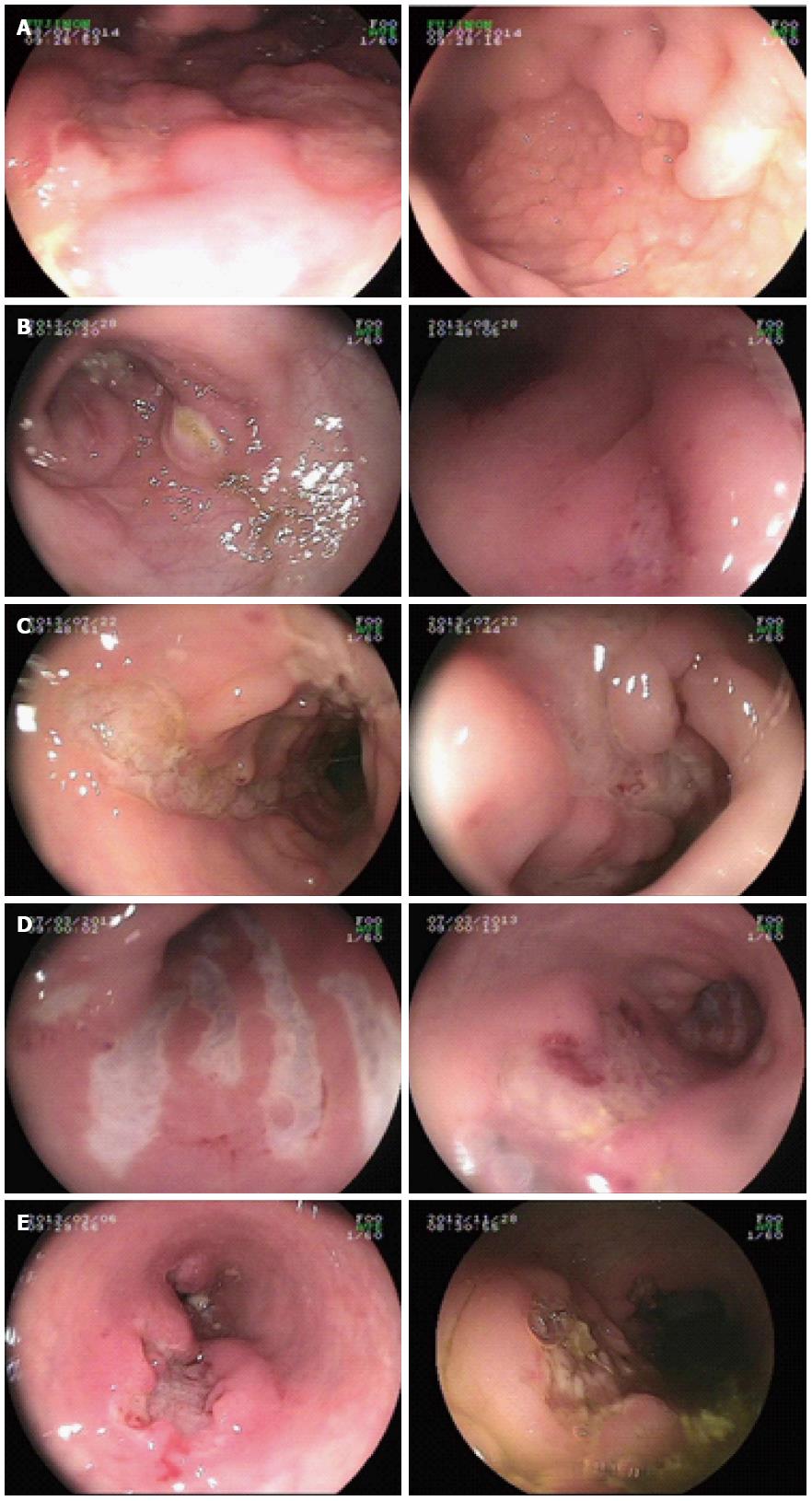
Out of the 13 VEO-IBD pediatric patients in this study, ten were diagnosed with CD (M:F = 9:1) and three had UC (M:F = 1:2). The mean age of disease onset was 5.8 ± 9.7 mo (range: birth to 3 years of age). None of the parents of the patients had a consanguineous marriage. Patient 8 had a brother that died as a neonate because of repeated diarrhea after birth. There was no clear diagnosis made at that time. The clinical symptoms of the pediatric patients included repeated abdominal pain (13/13), diarrhea (11/13), mucus and bloody stool (11/13), failure to thrive (8/13), recurrent infection (7/13), and perianal fistulas and abscesses (5/13). The colonoscopic presentation of patients with causative mutations showed pancolitis, cobblestone-like changes in mucosa, and deep and large ulcers (Figure 2). All patients received immunosuppressive treatment with glucocorticoids, 6-mercaptopurine and/or infliximab, and thalidomide; however, varying therapeutic effects were observed. Two patient died from severe sepsis or intestinal failure, 2 patients showed no change, 4 patients showed a partial alleviation of symptoms, and 5 patients showed complete clinical remission (Table 2).
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | Patient 11 | Patient 12 | Patient 13 | |
| Gender | F | M | M | M | M | M | M | M | M | M | M | F | F |
| Age of onset (mo) | 8 | 1 | 0.3 | 0.3 | 4 | 0.2 | 9 | 2 | 0.5 | 3 | 0.7 | 10 | 36 |
| Height percentile | 19% | 1% | 3% | 1% | 1% | 1% | 52% | 1% | 15% | 1% | 19% | 16% | 20% |
| Weight percentile | 1% | 1% | 1% | 1% | 1% | 20% | 55% | 13% | 8% | 15% | 16% | 60% | 33% |
| Diarrhea (times/d) | > 10 | 7-8 | > 10 | 10 | 5-10 | 5-6 | 7-8 | 2-4 | 7-8 | No diarrhea | 7-8 | No diarrhea | 2-3 |
| Bloody stool | + | + | + | + | + | - | + | + | + | - | + | + | + |
| Infection | Sepsis | Pneumonia | No | Pneumonia, Clostridium difficile infection | Sepsis, oral candidiasis, fungemia, Clostridium difficile infection | Recurrent respiratory infection | No | No | No | Repeated fever of unknow origin | Oral candidiasis, gingivitis | No | No |
| Perianal lesion | Fistulae | No | No | Excrescence | Fistulae, abscess, excrescence | Fistulae, ulcer | No | No | No | No | Fistulae, abscess, excrescence | No | No |
| Clinical diagnosis | CD | CD | CD | CD | CD | CD | CD | CD | UC | CD | CD | UC | UC |
| Medication | GC, 6-MP | IFX, THD | GC, THD | GC, IFX1, THD | GC, IFX, THD | GC, IFX1 | GC, IFX, MES | GC, IFX1, THD, 6-MP | GC, MES | GC, 6-MP, THD | GC, IFX, THD, 6-MP | MES | GC, MES |
| Clinical status | NR | PR | Died at 2 yr because of severe sepsis | PR | Died at 3 yr because of intestinal failure | NR | CR | PR | CR | CR | PR | CR | CR |
Based on the presence of causative mutations in IL-10RA and IL-10RB, 13 patients were divided into two groups for analysis (group 1: causative mutations in IL-10RA or IL-10RB; group 2: polymorphisms and no causative mutations). The five patients in group 1 were all diagnosed with CD (100%). Four of these patients had recurrent infections (80%), and three patients had perianal diseases (60%). In group 2 (eight patients), five patients were diagnosed with CD (62.5%), and the other three patients were diagnosed with UC (37.5%). There were only three (37.5%) and two (25%) patients that had recurrent infections and perianal diseases, respectively. Patients in group 1 had lower body weight percentile (1.0% vs 27.5%, P = 0.002) and hemoglobin concentrations (87.4 g/L vs 108.5 g/L, P = 0.040) when compared with group 2. Although patients in group 1 had a younger age of disease onset (2.7 mo), lower body height percentile (5.0%), and higher CRP (60.7 mg/L), there were no significant differences when compared with group 2 (Table 3).
| Group 1 | Group 2 | |
| Size of sample | 5 | 8 |
| Age of onset (mo) | 2.7 | 7.7 |
| Height percentile | 5.0% | 15.6% |
| Weight percentilea | 1.0% | 27.5% |
| WBC (× 10-9) | 15.2 | 16.3 |
| Hemoglobin (g/L)a | 87.4 | 108.5 |
| Platelets (× 10-9) | 538.4 | 424.0 |
| C reactive protein (mg/L) | 60.7 | 35.9 |
| ESR (mm/H) | 32.2 | 16.6 |
| TNFα (pg/mL) | 44.5 | 51.6 |
| Diagnosis of CD | 100.0% | 62.5% |
| Recurrent infection | 80.0% | 25.0% |
| Perianal disease | 60.0% | 25.0% |
The currently recognized pathogenetic mechanism of IBD is the involvement of many environmental triggers and genetic susceptibility that causes intestinal immune dysfunction. However, the influence of genes are likely more important than environmental factors for VEO-IBD patients with a disease onset prior to 6 years of age, especially for patients with an infantile onset prior to 1 year of age[19]. GWAS studies suggested that SNPs of IL-10 and STAT3 were associated with IBD[20-23]. Previous studies confirmed that IL-10 or IL-10 receptor gene knockout mice had severe chronic inflammation of the intestinal tract[24]. IL-10 forms a complex with two molecules of IL-10R1 and two molecules of IL-10R2 to activate Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2). This activation results in the phosphorylation of signal transducer and activator of transcription 3 (STAT3), which regulates the transcription of specific genes. Studies suggested that IL-10-mediated signals effectively reduced the number of Th17 cells and relieved intestinal inflammation in CD[25]. These data indicated that the anti-inflammatory IL-10 signaling pathway plays a critical role in the regulation of intestinal immune homeostasis.
Since Glocker et al[8] first reported in 2009 that gene mutations in IL-10RA and IL-10RB caused infantile onset IBD[8], studies have continuously reported mutations in IL-10, IL-10RA, and IL-10RB in patients with infantile onset IBD[9-12,18,26]. In these limited data, the majority of patients were Arabian or Caucasian and the offspring of a consanguineous marriage. There are few reports on the Han Chinese population, which included only three pediatric patients to date[13,14].
In this study, we used high-throughput next generation sequencing technology to sequence 10 IBD-associated genes, IL-10, IL-10RA, IL-10RB, NOD2, FUT2, IL-23R, GPR35, GPR65, TNFSF15, and ADAM30, in 13 Han Chinese children diagnosed with VEO-IBD. A total of four mutations were discovered in IL-10RA, including two novel mutations. There was one mutation in IL-10RB. These pathogenic mutations were found in five patients, which accounted for 38.5% of all VEO-IBD cases. Among these patients, one had an IL-10RA homozygous mutation, two had IL-10RA compound heterozygous mutations, one had an IL-10RA heterozygous mutation, and one had an IL-10RB heterozygous mutation. All IL-10RA mutations were in exon 3, and c.C301T (p.R101W) showed the highest frequency. The c.C301T (p.R101W) and c.G350A (p.R117H) mutations in IL-10RA were previously reported in similar pediatric patients. These mutations may disrupt signal transduction after activation of the IL-10 receptor; therefore, STAT3 is not phosphorylated and intractable inflammatory reactions in the intestinal tract of pediatric patients develop[5,12]. The two novel mutations in IL-10RA discovered in this study were c.A191G (p.Y64C) and c.T299G (p.V100G). Because of condition limitations, we did not perform functional studies on these mutations. However, the SIFT prediction results for these two mutations were deleterious (scores of 0 and 0.002, respectively), and the Polyphen 2 prediction results were probably damaging (both scores were 1.000). These predictions suggest that these two mutations are pathogenic. According to the recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology[27], these two mutations were defined as pathological supporting. Therefore, we speculate that these two novel mutations individually formed compound heterozygotes with the c.C301T (p.R101W) mutation to cause the disease symptoms observed in patients 2 and 3.
Previous analyses showed that the colitis caused by gene mutations in IL-10 and its receptor exhibited an autosomal recessive inheritance pattern. In the current study, patients 4 and 5 were carriers of heterozygous mutations in IL-10RA and IL-10RB, respectively. The c.G350A (p.R117H) mutation in IL-10RA carried by patient 4 was a pathogenic mutation[5,12,17]. The c.G421A (p.E141K) mutation in IL-10RB carried by patient 5 may affect protein function as predicted by SIFT (score = 0.026) and Polyphen 2 (score = 0.946). However, the clinical presentation of these two patients was similar to the symptoms of patients with other IL-10 receptor mutations: disease onset within 1 year of age, the presence of perianal diseases and recurrent infection, and resistance to conventional medication treatment. Based on currently available knowledge, there are at least 50 single-gene genetic conditions that induce IBD-like diseases, and the majority of conditions are related to immunodeficiency[4,6]. Therefore, the two patients that did not conform to a Mendelian genetic pattern might also carry abnormal sites on other genes that cause the disease symptoms. In addition to carrying a pathogenic mutation in IL-10RA, patient 4 also had a non-synonymous SNP (nsSNP): rs5743277 in NOD2. SIFT prediction results suggest that the nsSNP is deleterious (score = 0), and the Polyphen 2 prediction results suggest the nsSNP is probably damaging (score = 0.999). This polymorphism was already present in the HGMD database and has been considered to cause susceptibility to CD[28]. Patient 5 had a similar condition. In addition to carrying an IL-10RB mutation, patient 5 also had multiple polymorphisms: rs22280554 (homozygous) and rs22280555 (homozygous) in IL-10RA, rs2834167 (homozygous) in IL-10RB, and rs1047781 (heterozygous) in FUT2. There are previous reports on the pathogenicity of these SNPs. For example, Galatola et al[29] reported that the heterozygous rs2834167 in IL-10RB and the heterozygous mutation in the promoter region of IL-10RA caused the development of UC in an 18-month-old patient. Although rs22280554 did not cause a change in the amino acid sequence of IL-10R1, a study by Moran et al[30] showed that rs22280554 and rs2228055 in IL-10RA may increase the risk for VEO-IBD, especially VEO-UC. Furthermore, in the Han Chinese population, the rs1047781 polymorphism in FUT2 may increase the risk for CD development[31]. The above SNPs were also detected in four patients in this study. Therefore, their disease development may be due to “trans-heterzygous”: the collective effects of a variety of detected mutations. Another possible cause is that the pathogenic genes were not detected in this study.
When genotypes and phenotypes were combined for analyses, the results showed that the disease phenotype in patients with mutations were more severe. The age of disease onset was earlier, the patients were more likely to have combined recurrent infections and perianal diseases, their body weight and height were low, anemia was more severe, inflammatory indicators were high, and the prognosis was much poorer. These results are in accordance with previous studies[5,8-14,18,32]. However, the sample size of this study was small, and significant differences were only found in body weight and hemoglobin parameters. Because of the influence of cultural ideas, family members find difficulty in accepting an ileostomy as a disease treatment. Past literature reported that pediatric patients with IL-10RA and IL-10RB mutations could be cured through hematopoietic stem cell transplantation[4,6,8,17]; therefore, some patients are waiting for a donor match.
In this study, we found that mutations in IL-10RA and IL-10RB were more common in Han Chinese VEO-IBD patients and accounted for 38.5% of all VEO-IBD cases. The high percentage is probably due to the small number of patients in the cohort as most of our patients who were referred by other clinical IBD centers were very ill. There was a selection bias. Because VEO-IBD is relatively rare, multi-center studies on the relationship between genotypes and phenotypes in VEO-IBD patients in China are necessary. The implementation of hematopoietic stem cell transplantation therapy is the focus in research agenda.
Very early-onset inflammatory bowel disease (VEO-IBD) may have stronger genetic contribution. Recently, a few studies on genetic defects in the IL-10 signaling pathway have provided new insights into IBD, especially in VEO-IBD. Furthermore, a lot of genes associated with IBD were identified, such as NOD2, FUT2, IL-23R, GPR35, GPR65, TNFSF15, and ADAM30. Because of different genetic background, this study was set to disclose whether mutations in these genes contributed to VEO-IBD in Chinese children.
In addition to the polygenic variants associated with IBD, there are rare monogenic disorders, including many immunodeficiencies that can present with IBD-like intestinal inflammation, especially in early life.
To our knowledge, this is the first cohort study to apply NGS in 13 Chinese pediatric patients with VEO-IBD to discover gene variations in these children. The result revealed that IL-10RA and IL-10RB mutations were common in Chinese VEO-IBD, especially in infantile IBD. These monogenic IBD patients had more severe clinical features.
According to the results of this study and previous studies of VEO-IBD, the authors suggest that screening for gene mutations in IL-10 signaling pathway is necessary.
The clinical study is focused on gene mutation analysis in VEO-IBD by NGS. The authors conclude that mutations in the IL-10 pathway are common in VEO-IBD.
P- Reviewer: Gassler N S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Ma S
| 1. | Levine A, Koletzko S, Turner D, Escher JC, Cucchiara S, de Ridder L, Kolho KL, Veres G, Russell RK, Paerregaard A. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr. 2014;58:795-806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 822] [Cited by in F6Publishing: 811] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 2. | Wang XQ, Zhang Y, Xu CD, Jiang LR, Huang Y, Du HM, Wang XJ. Inflammatory bowel disease in Chinese children: a multicenter analysis over a decade from Shanghai. Inflamm Bowel Dis. 2013;19:423-428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 3. | Heyman MB, Kirschner BS, Gold BD, Ferry G, Baldassano R, Cohen SA, Winter HS, Fain P, King C, Smith T. Children with early-onset inflammatory bowel disease (IBD): analysis of a pediatric IBD consortium registry. J Pediatr. 2005;146:35-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 346] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 4. | Uhlig HH, Schwerd T, Koletzko S, Shah N, Kammermeier J, Elkadri A, Ouahed J, Wilson DC, Travis SP, Turner D. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990-1007.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 452] [Cited by in F6Publishing: 427] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 5. | Shim JO, Seo JK. Very early-onset inflammatory bowel disease (IBD) in infancy is a different disease entity from adult-onset IBD; one form of interleukin-10 receptor mutations. J Hum Genet. 2014;59:337-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Uhlig HH. Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut. 2013;62:1795-1805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 7. | Christodoulou K, Wiskin AE, Gibson J, Tapper W, Willis C, Afzal NA, Upstill-Goddard R, Holloway JW, Simpson MA, Beattie RM. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut. 2013;62:977-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 8. | Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033-2045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1121] [Cited by in F6Publishing: 1015] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 9. | Beser OF, Conde CD, Serwas NK, Cokugras FC, Kutlu T, Boztug K, Erkan T. Clinical features of interleukin 10 receptor gene mutations in children with very early-onset inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2015;60:332-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 10. | Lee CH, Hsu P, Nanan B, Nanan R, Wong M, Gaskin KJ, Leong RW, Murchie R, Muise AM, Stormon MO. Novel de novo mutations of the interleukin-10 receptor gene lead to infantile onset inflammatory bowel disease. J Crohns Colitis. 2014;8:1551-1556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Pigneur B, Escher J, Elawad M, Lima R, Buderus S, Kierkus J, Guariso G, Canioni D, Lambot K, Talbotec C. Phenotypic characterization of very early-onset IBD due to mutations in the IL10, IL10 receptor alpha or beta gene: a survey of the Genius Working Group. Inflamm Bowel Dis. 2013;19:2820-2828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Shim JO, Hwang S, Yang HR, Moon JS, Chang JY, Ko JS, Park SS, Kang GH, Kim WS, Seo JK. Interleukin-10 receptor mutations in children with neonatal-onset Crohn’s disease and intractable ulcerating enterocolitis. Eur J Gastroenterol Hepatol. 2013;25:1235-1240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Lu D, Xu Y, Chen Y, Zeng P, Chen H, Zeng H. [Interleukin-10 receptor mutations in children with neonatal onset inflammatory bowel disease: genetic diagnosis and pathogenesis]. Zhonghua Erke Zazhi. 2015;53:348-354. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Mao H, Yang W, Lee PP, Ho MH, Yang J, Zeng S, Chong CY, Lee TL, Tu W, Lau YL. Exome sequencing identifies novel compound heterozygous mutations of IL-10 receptor 1 in neonatal-onset Crohn’s disease. Genes Immun. 2012;13:437-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Hyams JS. Standardized recording of parameters related to the natural history of inflammatory bowel disease: from Montreal to Paris. Dig Dis. 2014;32:337-344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Levine A, Griffiths A, Markowitz J, Wilson DC, Turner D, Russell RK, Fell J, Ruemmele FM, Walters T, Sherlock M. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314-1321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 932] [Cited by in F6Publishing: 1008] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 17. | Engelhardt KR, Shah N, Faizura-Yeop I, Kocacik Uygun DF, Frede N, Muise AM, Shteyer E, Filiz S, Chee R, Elawad M. Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2013;131:825-830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 18. | Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544-1555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 19. | Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335-1340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 365] [Cited by in F6Publishing: 379] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 20. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1894] [Cited by in F6Publishing: 1924] [Article Influence: 137.4] [Reference Citation Analysis (0)] |
| 21. | Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 296] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 22. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955-962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2041] [Cited by in F6Publishing: 1984] [Article Influence: 124.0] [Reference Citation Analysis (0)] |
| 23. | Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319-1323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 434] [Cited by in F6Publishing: 452] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 24. | Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263-274. [PubMed] [Cited in This Article: ] |
| 25. | Paul G, Khare V, Gasche C. Inflamed gut mucosa: downstream of interleukin-10. Eur J Clin Invest. 2012;42:95-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Glocker EO, Frede N, Perro M, Sebire N, Elawad M, Shah N, Grimbacher B. Infant colitis--it’s in the genes. Lancet. 2010;376:1272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 459] [Reference Citation Analysis (1)] |
| 27. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13374] [Cited by in F6Publishing: 17932] [Article Influence: 1992.4] [Reference Citation Analysis (0)] |
| 28. | Lesage S, Zouali H, Cézard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O’Morain C, Gassull M, Binder V. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845-857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 712] [Cited by in F6Publishing: 701] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 29. | Galatola M, Miele E, Strisciuglio C, Paparo L, Rega D, Delrio P, Duraturo F, Martinelli M, Rossi GB, Staiano A. Synergistic effect of interleukin-10-receptor variants in a case of early-onset ulcerative colitis. World J Gastroenterol. 2013;19:8659-8670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 25] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, Wolters VM, Bandsma RH, Mouzaki M, Zachos M. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 31. | Hu DY, Shao XX, Xu CL, Xia SL, Yu LQ, Jiang LJ, Jin J, Lin XQ, Jiang Y. Associations of FUT2 and FUT3 gene polymorphisms with Crohn’s disease in Chinese patients. J Gastroenterol Hepatol. 2014;29:1778-1785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Shah N, Kammermeier J, Elawad M, Glocker EO. Interleukin-10 and interleukin-10-receptor defects in inflammatory bowel disease. Curr Allergy Asthma Rep. 2012;12:373-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |