Published online Jun 21, 2016. doi: 10.3748/wjg.v22.i23.5317
Peer-review started: March 25, 2016
First decision: April 14, 2016
Revised: April 23, 2016
Accepted: May 4, 2016
Article in press: May 4, 2016
Published online: June 21, 2016
Colorectal carcinoma (CRC) is one of the most common types of cancer worldwide and the prognosis for CRC patients with recurrence or metastasis is extremely poor. Although chemotherapy and radiation therapy can improve survival, there are still numerous efforts to be performed. Immunotherapy is frequently used, either alone or in combination with other therapies, for the treatment of CRC and is a safe and feasible way to improve CRC treatment. Furthermore, the significance of the immune system in the biology of CRC has been demonstrated by retrospective assessments of immune infiltrates in resected CRC tumors. MicroRNAs (miRNAs) are short, non-coding RNAs that can regulate multiple target genes at the post-transcriptional level and play critical roles in cell proliferation, differentiation and apoptosis. MiRNAs are required for normal immune system development and function. Nevertheless, aberrant expression of miRNAs is often observed in various tumor types and leads to immune disorders or immune evasion. The immunomodulatory function of miRNAs indicates that miRNAs may ultimately be part of the portfolio of anti-cancer targets. Herein, we will review the potential roles of miRNAs in the regulation of the immune response in CRC and then move on to discuss how to utilize different miRNA targets to treat CRC. We also provide an overview of the major limitations and challenges of using miRNAs as immunotherapeutic targets.
Core tip: Colorectal carcinoma (CRC) is one of the most common tumor types worldwide. Immunotherapy has been used to treat advanced CRC and has the potential to eradicate the disease by activating immune responses. MicroRNAs play critical roles in regulating anti-tumor immune responses. There is a need to summarize the current understanding of the diverse roles of microRNAs in the regulation of immune responses and their clinical applications for CRC immunotherapy.
- Citation: Li X, Nie J, Mei Q, Han WD. MicroRNAs: Novel immunotherapeutic targets in colorectal carcinoma. World J Gastroenterol 2016; 22(23): 5317-5331
- URL: https://www.wjgnet.com/1007-9327/full/v22/i23/5317.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i23.5317
Colorectal carcinoma (CRC) is the third most common malignancy in males and the second in females, posing a serious demographic and economic burden worldwide[1,2]. Although surgical resection for patients with localized disease has dramatically improved 5-year survival rates, more than half of all patients diagnosed with CRC eventually develop recurrence and metastasis[3,4]. Current strategies such as chemotherapy are approved for the treatment of CRC; however, these non-surgical therapeutics have only modest efficacy and are ineffective against distant metastasis[5]. Moreover, traditional strategies generate reverse events, such as cytotoxicity, that limit their use[6]. Therefore, the prognosis of CRC patients remains poor and presents an opportunity to study future therapeutic approaches to improve clinical outcomes. Recently, immunotherapy has been used to treat advanced CRC and has the potential to eradicate the disease by activating immune responses[6]. Some clinical trials utilizing immunotherapy have demonstrated objective responses in CRC patients[7,8]. Moreover, immunotherapy, such as monoclonal antibody cetuximab, has immune-modulating effects in combination with traditional approaches, such as chemotherapy, thus resulting in an increase of circulating dendritic cells (DCs), natural killers (NK), central memory T cells and T-helper 1 (Th1) cells[9]. However, CRC is able to evade detection and elimination by the immune system, and the objective responses have been manifested in only a small fraction of patients with CRC[6]. For example, the lack of CRC-associated antigens limits the development of immunotherapy[6]. The important role of the immune system in the biology of CRC has been emphasized by retrospective assessments of immune infiltrates, affecting all aspects of CRC from tumorigenesis to treatment. Within the tumor microenvironment (TME), immune infiltrates can act as suppressors of tumor initiation and progression, as well as promote the proliferation, infiltration and metastasis of CRC[10]. The presence of tumor infiltrating lymphocytes, including CD8+ T cells, in the TME is strongly associated with a better prognosis[11]. Regulatory T cells (Tregs) suppress the proliferation and activation of CD4+ and CD8+ T cells and increased Tregs activation is correlated with poorer prognosis[12]. However, higher Tregs infiltration scores are correlated with overall survival, treatment-relative survival and progression-free survival. Therefore, higher Tregs infiltration score is a favorable prognostic factor in advanced colon cancer patients undergoing chemo or chemoimmunotherapy[13]. The immune evasion of CRC has complex mechanisms. The induction of immune tolerance and resistance to activated immune effectors are two important mechanisms of immune evasion[14]. Programmed death 1 (PD-1) is an inhibitory receptor that is overexpressed in nonfunctional and exhausted T cells in cancers[15]. PD-1 and the ligands for PD-1 (PD-L1) have been implicated in escape from the immune system by directly suppressing antitumor CD8+ T cells[16]. Furthermore, the recognition of tumor-associated antigens by immune effectors is one of the most important steps in antitumor immune responses. Thus, the dysregulation of antigens limits specific immune responses. For example, NY-ESO-1 is silenced in many tumor types. As a result, the immune system cannot attack cancer cells through the recognition of the antigen[17].
MicroRNAs (miRNAs) are approximately 19 to 25 nt in length and post-transcriptionally regulate gene expression in a sequence-specific manner by binding to the 3’-untranslated region, leading to either degradation of mRNA or a translation blockade[18,19]. MiRNAs have emerged as crucial modulators in various biological processes such as proliferation, tumor initiation and development[20]. Global dysexpression of miRNAs has been identified in tumors and has become a potential biomarker for tumor diagnosis, therapy and prognosis[19,21]. MiRNAs were displayed to function as oncogenes or tumor suppressors according to their target mRNAs. For example, miR-484 is significantly decreased in microsatellite instability (MSI) CRC and functions as a tumor suppressor to inhibit MSI CRC cell viability[22]. MiR-21 is upregulated in several tumors such as lung cancer, which promotes tumor cell growth and metastasis[20]. Recent studies have demonstrated that miRNAs are coming to light as a critical regulator of immune responses and its aberrant expression or dysfunction in the immune system has been associated with cancers[23-25]. MiRNAs can either inhibit or enhance immune signals by regulating the expression of the positive or negative components of immune signaling pathways. MiR-181a expression appears to control T cell sensitivity to antigens during T cell development and maturation by repressing the expression of multiple phosphatases in the TCR signaling pathway[26]. Signal transducer and activator of transcription 3 (STAT3) has been shown to play an important role in tumor-mediated mechanisms of immunosuppression[27]. MiR-124 was found to target the STAT3 signaling pathway and exert potent therapeutic effects by enhancing immune effector responses in cancer[28]. Emerging evidence has demonstrated that immune-associated miRNAs are dysregulated in both tumor cells and immune cells, which suggests that miRNAs could be involved in communication between tumor cells and immune cells. MiR-124 is absent in all types of gliomas, and its upregulation in glioma cancer stem cells inhibits the STAT3 pathway and reverses stem cell-mediated immune suppression of T cell proliferation and Treg activation[28]. MiR-23a is upregulated in tumor-infiltrating cytotoxic T lymphocytes (CTLs) and impairs antitumor potential of patient CTLs by repressing the transcription factor B lymphocyte-induced maturation protein-1, which promotes CTL cytotoxicity and effector cell differentiation[29]. In addition to CTLs, miRNAs can regulate other immune cells, such as Th1 cells and NK cells. The members of miR-17-92 cluster miR-17 and miR-19b are the important regulators modulating Th1 responses through multiple coordinated biologic processes[30]. Moreover, miR-29 suppresses Th1 responses through the combined direct suppressive effect on T-bet, eomesodermin and interferon γ (IFN-γ)[31,32]. NK cells play critical roles in the innate immune system, contributing to the early detection and destruction of transformed cells[33]. MiRNAs have critical roles in controlling NK cell activation, survival and function[34]. Recent report has demonstrated that ovarian tumor-associated miR-20a decreases NK cell cytotoxicity by directly repressing MHC class I chain-related molecules A and B expression[35]. However, the immunomodulatory roles of miRNAs in CRC still need to be further explored. Therefore, in this review, we summarize the current understanding of the diverse roles of miRNAs involved in immune responses in CRC. We also discuss the clinical applications of miRNAs in CRC immunotherapy and the limitations and challenges in future use.
Primary miRNAs are processed into mature miRNAs through multiple biochemical steps. Our previous review[18] and other excellent reviews[19,20,36] have illustrated the mechanism of miRNA biogenesis. Thus, we will not discuss it further here. In the process of miRNA biogenesis, the RNase III endonuclease Dicer plays a critical role in the generation of miRNAs[37]. Dicer has been reported to regulate miRNA biogenesis in Tregs[38]. Depleting miRNAs by silencing Dicer generates functional miRNAs, reduces Treg cell numbers and results in immune pathology[38]. Previous studies have found that Dicer is required, in a cell-autonomous fashion, for the development of Tregs in the thymus[38]. However, the mechanism by with the expression of Dicer in tumor cells regulates immune surveillance through miRNA processing needs to be deeply explored. Indeed, Dicer depletion upregulates intercellular cell adhesion molecule 1 (ICAM-1) and enhances the susceptibility of CRC cells to antigen-specific lysis by CTLs[39]. ICAM-1 plays an important role in physical and functional interactions between cancer cells and CTLs[40]. Furthermore, Dicer-disrupted CRC cells have downregulated expression of miR-222/339, which directly target ICAM-1 mRNA. Upregulation of ICAM-1 by repressing Dicer or miR-222/339 in CRC cells contributes to increased susceptibility of tumor cells to antigen-specific CTLs[39]. Thus, Dicer is responsible for the generation of the mature miR-222/339, which repress the expression of ICAM by binding to the 3’UTR, thereby reducing the susceptibility of tumor cells to CTL-mediated cytolysis. These findings indicate the viability of the development of novel miRNA-targeted therapy to promote cytolysis of tumor cells in tumor immunotherapy.
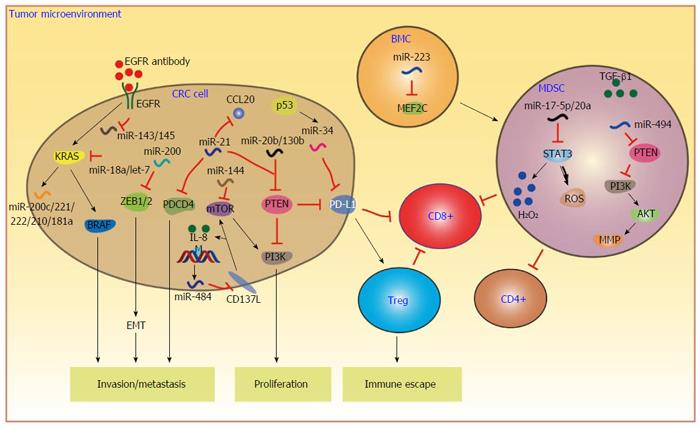
Multiple studies have demonstrated that miRNAs play key roles in CRC tumor biology including tumor initiation, progression, invasion and metastasis, and immune responses[41-43] (Figure 1). Several miRNAs have been found to directly regulate the primary pathways of CRC initiation and development. The development of CRC follows the sequential progression from adenoma to malignant adenocarcinoma[44]. Tumor protein 53 (TP53) is frequently lost during the progression of adenoma to adenocarcinoma[45]. TP53 loss mediates apoptosis and cell cycle arrest through the inhibition of a direct downstream target, miR-34a[46]. Increased miR-17-92 cluster expression from adenoma to adenocarcinoma is associated with the transcriptional activity of c-myc, which induces the expression of miR-17-92[47]. Uncontrolled growth and proliferation are key components in the colorectal tumorigenesis. The dysfunction of the PI3K pathway and its associated proteins plays important roles in the cell cycle regulation of CRC. MiR-144 has been found to inhibit the PI3K pathway by negatively regulating mTOR[48]. Emerging evidence has demonstrated that miRNAs are most frequently associated with the invasion and metastasis of CRC. Upregulation of miR-21 positively regulates invasion and metastasis by directly targeting several tumor suppressor genes, including programmed cell death 4 (PDCD4)[49] and phosphatase and tensin homolog (PTEN)[50] in CRC. MiRNAs could regulate CRC cell invasion and metastasis through the epithelial to mesenchymal transition (EMT). Activation of EMT allows CRC cells to migrate and metastasize to distant organs. Recent studies suggested that miRNAs are new EMT regulators and regulate CRC metastasis. Downregulation of the miR-200 family has been found to induce EMT by targeting zinc finger E-box binding homeobox 1/2 (ZEB1/2) in CRC cells, which results in the development of CRC metastasis[51-53]. Therefore, the dysregulation of miRNAs is involved in the full range of CRC development.
According to previous reports, the immune system plays complex and multi-faceted roles in CRC, affecting all aspects of CRC initiation and metastasis[10]. The body’s immune system can act both to suppress tumor initiation and progression, as well as promote malignancies. During tumorigenesis, normal cells transform into neoplastic cells through multifactorial processes that involve genetic and epigenetic factors, leading to the immune escape of tumor cells. Immune escape plays a key role for the success of tumor cell growth and survival through developing multiple suppressive mechanisms in the TME[54]. More recently, miRNAs have been observed to play important regulatory roles in the immune system. The abnormal expression of co-inhibitory molecules of PD-L1 in the TME has been identified as an important immunosuppressive mechanism in many tumor types, and contributes to tumor immune escape by inhibiting T cell activation and proliferation[55]. Furthermore, PD-L1 expression in tumor cells may be associated with the expansion of Tregs in the TME, which suppresses the activation and proliferation of effector T cells[16,56,57]. MiR-20b, -21 and -130b are upregulated in advanced CRC and inhibit PTEN expression, resulting in PD-L1 overexpression[58]. PTEN, a famous tumor suppressor gene, plays a critical role in CRC cell growth, proliferation and the occurrence of CRC[59]. This study revealed a novel mechanism of PD-L1 protein expression mediated by miRNAs indirectly, which leads to the immune escape of CRC. Moreover, miR-34 has been found to modulate tumor escape by directly targeting PD-L1 in lung cancer[60]. MiR-34a therapy elicits the activation of tumor-infiltrating immune cells (CD8+ T cells) and decreases exhausted tumor-infiltrating immune cells (PD1/CD8+), indicating an active immune response and reduced tumor tolerance[60]. These results suggest that the miRNA-PD-L1 axis may be a therapeutic target for CRC immunotherapy and miRNAs may be implicated in checkpoint blockade therapy.
Cancer immunotherapy has the potential role to eliminate tumor cells by eliciting immune responses through the recognition of tumor-specific antigens on tumor cells. Numerous studies have reported that monoclonal antibody (mAb)-based immunotherapy against tumor antigens is a promising strategy and has been clinically effective as CRC therapeutics[54]. Antibodies targeting tumor antigens block major pathways central to tumor cell proliferation and survival. Anti-human epidermal growth factor (anti-EGFR) antibodies, such as cetuximab and panitumumab, have been approved and routinely used for the treatment of CRC[54]. EGFR, commonly overexpressed in CRC, leads to the stimulation of the oncogene KRAS that subsequently activates BRAF, which contributes to the promotion and progression of a broad spectrum of CRC[61]. These two antibodies can trigger tumor antigen-specific cellular immunity to eliminate CRC cells. Recent studies have demonstrated that miRNAs could serve as predictive or prognostic biomarkers for anti-EGFR therapy in patients with wild-type KRAS (wt-KRAS) metastatic CRC (mCRC). Anti-tumor immune responses of antibody and EGFR signaling pathway have been shown to be regulated by miRNAs. MiR-31-3p/5p have been confirmed to be strongly associated with time to progression in wt-KRAS mCRC patients treated with cetuximab but not panitumumab, suggesting that miR-31-3p/5p should not be used as a predictor of response to panitumumab[62]. Two reasons could explain the predictive role of miR-31-3p/5p for cetuximab: one associated with concomitant chemotherapy, and the other with different immune responses linked to cetuximab and panitumumab[54]. Furthermore, the expression level of miR-31-3p is associated with progression-free survival, indicating that miR-31-3p seems to be a new mCRC biomarker for anti-EGFR therapy[63]. Significant miR-31* upregulation and miR-592 downregulation show a marked difference in progressive disease compared to disease control[64]. Moreover, the upregulation of some miRNAs, such as let-7 and miR-140-5p, as well as the downregulation of miR-1224-5p are significantly associated with poor overall survival, revealing that in mCRC patients with wt-KRAS/BRAF, miRNA profiling can efficiently predict the prognosis after anti-EGFR treatment[64]. Interestingly, miRNAs can also directly or indirectly target EGFR signaling pathway and its signaling components. Therefore, miRNAs-EGFR signaling has been validated as a therapeutic target in CRC. EGFR has been found to control and negatively regulate the expression of miR-143 and miR-145 in CRC cell lines, two famous tumor suppressive miRNAs, which suggests that inhibition of EGFR may upregulate the expression of these two miRNAs and suppress tumor growth and progression[65]. According to previous reports, wt-KRAS, a major factor that drives CRC progression, is a necessary condition for anti-EGFR treatment and it can be activated by the stimulation of EGFR[66]. Several miRNAs, miR-143, miR-145, let-7 and miR-18a*, act as tumor suppressors and repress the expression of KRAS[61]. In contrast to targeting KRAS, KRAS can regulate the expression of miRNAs. Recent studies have demonstrated that KRAS upregulates the expression of some oncogenic miRNAs, including miR-200c, miR-221/222, miR-210 and miR-181a[67]. These miRNAs act as oncogenic miRNAs and promote CRC progression. Therefore, the extensive involvement of miRNAs in regulating EGFR signaling pathway and its components suggests that miRNAs could serve as promising predictive biomarkers and therapeutic targets for anti-EGFR therapy in CRC.
Tumor tissues have two distinct regions: the tumor bed and TME. TME comprises fibroblasts, various immune cells and endothelial cells, along with extracellular matrix, proteases and cytokines[68]. TME represents an integral part of tumors and establishes dynamic interactions with tumor cells that influence tumor growth, metastasis and immune tolerance. Stromal cells, including cancer-associated fibroblasts (CAFs), myeloid-derived suppressor cells (MDSCs) and mesenchymal stem cells (MSCs), are important components of TME and can shape anti-tumor immunity and responsiveness to immunotherapy[68]. MDSCs are a heterogeneous group of myeloid cells in early differential stages and one of the major elements of the immunosuppressive network responsible for effector T cell defective response in cancer[69]. A previous report has found that the number of blood MDSCs correlates with the stage and metastasis in many tumor types, including CRC[70-72]. MDSCs could inhibit the host immune response in tumor cells by interfering with immune cell responsiveness to interferons α and γ in CT26 bearing mice[70]. Although MDSCs contribute to immune invasion via suppression of anti-tumor functions of CD4+ and CD8+ T cells, the molecular mechanisms involved in the expansion, function and infiltration of MDSCs are not fully understood. Numerous studies have demonstrated that miRNAs play key regulatory roles in MDSC development, expansion and function. Recently, miR-17-5p and miR-20a have been found to alleviate the suppressive potential of MDSCs in the CT26 colon carcinoma model[73]. The overexpression of miR-17-5p and miR-20a remarkably reduces the expression of reactive oxygen species (ROS) and the production of H2O2 by modulating the expression of STAT3. Furthermore, MDSCs transfected with miR-17-5p and miR-20a are less able to suppress antigen-specific CD4+ and CD8+ T cells. Thus, the modulation of miR-17-5p and miR-20a expression may play important roles for the process by which patients with a tumor can overcome the immune tolerance mediated by MDSCs and could potentially be applied in immunotherapy against CRC. Increased expression of miR-17-5p and miR-20a, which are members of the miR-17-92 cluster, can promote tumor development in cancer cells. However, miR-17-92 members block the immunosuppressive function of MDSCs by silencing STAT3 expression. These studies indicate that the functional diversity of miR-17-92 cluster may result from the different targets subjected to miR-17-92 post-transcriptional silencing in different cell types, different developmental or physiological contexts[73]. MiR-494, whose expression is mainly upregulated by TGF-β1, has been identified as an essential player in the regulation of the accumulation and activity of MDSCs by targeting PTEN and activating the PI3K/AKT pathway[74]. Upregulation of miR-494 represses PTEN expression, resulting in increased activity of AKT pathway, and facilitates tumor cell invasion and metastasis by upregulating of MMPs in murine breast and colon cancers. These findings revealed that TGF-β1-induced miR-494 expression in MDSCs plays a critical role in regulating the accumulation and functions of MDSCs and may be identified as a potential target in cancer immunotherapy. Emerging studies have demonstrated that CD11b+Gr1+ MDSCs play an important role in the suppression of T cell responses and the induction of T cell tolerance in tumors[75]. MiR-223 has been identified to be related to myeloid development in myeloid expansion and differentiation[76]. The expression of miR-223 was downregulated during the differentiation of CD11b+Gr+ MDSCs from bone marrow cells (BMCs) upon exposure to tumor-associated factors. Moreover, overexpression of miR-233 remarkably inhibits differentiation of BMCs into MDSCs by targeting myocyte enhancer factor 2c, which promotes the accumulation of MDSCs.
In addition to regulating stoma cells, miRNAs also play key roles in modulating the expression of cytokines or chemokines in TME. Chemokines, which are chemo-attractant cytokines and are primarily known as critical regulators of the immune system, have been implicated in malignant transformation and tumor progression[77]. In the TME, specific chemokines have been found to be responsible for the recruitment of leukocytes into tumor sites[78]. In addition, chemokines also play an important role in the functionally of tumor cells and other cells in TME, which together modulate the processes of tumor development and progression[79]. Emerging evidence has demonstrated that various miRNAs can regulate different chemokines in the TME of CRC. Chemokine CCL20 was shown to regulate CRC progression and metastasis through binding to its receptor CCR6[80]. Moreover, CCL20 and its receptor CCR6 are significantly increased in CRC[81]. Interestingly, miR-21, a well-known oncomiR, has been identified to target CCL20 in CRC cells[82,83]. Ectopic expression of miR-21 downregulates CCL20 gene expression in different cell lines. These results seem to indicate that miR-21 acts as a tumor suppressor in this situation. However, for functional interaction of miR-21 with CCL20, both the two molecules must be present in the same cell. Furthermore, miR-21 and CCL20 are both significantly upregulated in CRC tissues. It is well-known that TME is composed of tumor cells and various types of stromal cells, such as fibroblasts and endothelial cells, as well as various immune cells. Thus, this suggests that miR-21 and CCL20 may not be expressed in the same cell in the TME of CRC. Actually, CCL20 expression found in CRC tissues is largely restricted to mesenchymal cells, such as macrophage and lymphocytes, in the TME[83]. Likewise, miR-21 expression is primarily detected in stromal cells, such as CAFs, and immune cells, such as macrophages, but not within tumor cells and lymphocytes in the TME of CRC tissues[83]. These data clearly exclude the chance of miR-21 and CCL20 being expressed in the same cell of TME. It may also be speculated that CCL20 secreted by infiltrated-immune cells affects miR-21 expression in CAFs through the CCR6 signaling pathway. In our recent report, we found that methylation-induced loss of miR-484 in MSI CRC promotes both cell viability and IL-8 (also known as CXCL8) production by targeting CD137L[22]. CD137, a member of the TNF superfamily, can induce cell viability by triggering the activation of the PI3K and mTOR signaling pathways[84]. Moreover, CD137L can signal back to tumor cells and induce the production of IL-8, which can promote tumor invasion by the recruitment of tumor-associated macrophages and neutrophils. However, CD137L expressed in CRC cells also functions as a co-stimulatory molecule for the production of IFN-γ by human T cells. Thus, these results may indicate that miR-484 can play a suppressive role in anti-tumor immune responses in MSI CRC through repressing the expression of CD137L. It seems to suggest that MSI is detrimental to immunotherapy for CRC treatment. MSI is the molecular feature of a mismatch repair deficient (dMMR) system[85]. Interestingly, the immunotherapy PD-1 blockade is more effective against dMMR tumors[86]. The reason may be that the increased number of mutation-associated neoantigens in dMMR tumors stimulated antitumor immune responses.
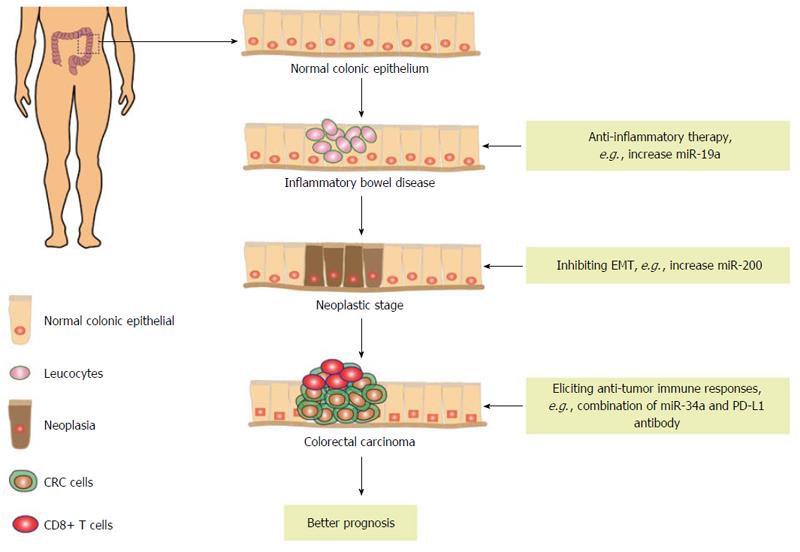
Chronic inflammatory disorders are often associated with an increased risk of CRC tumorigenesis[87,88]. CRC is preceded by clinically detectable inflammatory bowel disease (IBD), such as Crohn’s disease (CD) and ulcerative colitis (UC)[88]. Because of chronic damage to the colon and rectum in IBD, these patients are at increased risk for CRC. Increasing evidence has demonstrated that miRNA dysregulation is an important pattern in IBD. Elevated miR-150 was found in UC and reduced the expression of c-myb, a transcription factor known to regulate the anti-apoptotic protein, BCL-2[89]. Several studies have found that increased cytokines, such as TNF, IL and interferon, cause apoptosis and play important roles in intestinal epithelial cells[90]. Apoptosis can interrupt intestinal mucosal integrity and barrier function and eventually result in inflammation[91]. MiR-29a has been found to promote intestinal epithelial apoptosis in UC by directly targeting myeloid cell leukemia 1, which is an anti-apoptotic BCL-2 family member and essential for the survival of multiple cell lineages[92]. Although both CD and UC are subtypes of IBD, the expression patterns of miRNAs show region-specific differences and are expressed differently. Eight different miRNAs could be used to distinguish active CD from active UC[93]. Moreover, the expression of miR-23b/106/191 was increased in active UC tissues, while miR-19b/629 were decreased in Crohn’s colitis patients[94]. Given these common changes in miRNA expression profiles in IBD, these miRNAs may be involved in regulating central components of the immune system.
Numerous studies have demonstrated that miRNAs can regulate many inflammation associated pathways in IBD. The IL-6-STAT3 signaling pathway plays an important role in IBD and specifically STAT3 is up-regulated in UC[95,96]. This pathway regulates the development of UC and has been implicated in the progression of UC to CRC[97]. Recently, miRNAs have been identified as a potent regulator of STAT3 activity in human colonocytes. MiR-124 is downregulated in children with active UC, resulting in increased levels of STAT3 expression and the transcriptional activation of its downstream targets, which could promote inflammation and the pathogenesis of UC in children[98]. The vitamin D receptor (VDR) is highly expressed in the epithelial cells of the gut mucosal epithelial barrier and plays a critical role in maintaining the integrity of the mucosal epithelial barrier by suppressing inflammation-induced intestinal epithelial cell apoptosis[99]. Epithelial VDR levels are reduced by more than 50% in IBD and promote colitis[99]. MiRNAs have been identified to be involved in the downregulation of VDR. Pro-inflammatory cytokine TNF-α suppresses epithelial VDR expression through induction of miR-346[100]. TNF-α is one of the most important cytokines in regulating the development of colitis, and anti-TNF therapy is a standard therapy in the management of IBD[101,102]. Thus, the use of TNF-α, which as an inflammatory mediator inhibiting VDR, will have promising clinical applications. Further, TNF-α is markedly increased and may serve as an objective index of inflammatory activity in human UC. A recent report has found that miR-19a, which is downregulated in UC, directly targets the expression of TNF-α, and induces the expression of downstream inflammation targets IL-8 and granulocyte-macrophage colony-stimulating factor[103]. Immune dysregulation is thought to play a critical role in the pathogenesis of CD[104]. Cytokines are involved in immune regulation and the dysregulation of cytokines contributes to heightened inflammation[105]. Therefore, cytokine signaling pathways may be useful targets for CD treatment. MiR-19b has been reported to regulate cytokine signaling by directly targeting suppressors of cytokine signaling[106]. SOCS proteins are important physiological regulators of immune responses and negatively regulate cytokine expression through the JAK/STAT pathway[107]. Thus, this study will provide potential evidence for the development of miR-19b-based anti-inflammation therapies for CD treatment. Neurotensin (NT) is a neuropeptide expressed in the central nervous system and the intestine, while NT receptor 1 (NTR1), the high-affinity receptor of NT, is expressed in neurons and colonic epithelial cells[108]. In the colon, the NT/NTR1 signaling pathway can promote inflammation in acute colitis and their expression increases in patients with UC[109]. NT induces miR-133a expression in colonic epithelial cells[110]. Moreover, miR-133a modulates NT-induced MAP kinase and NF-κB activation, as well as transcription of pro-inflammatory cytokines, such as IL-6/8/1β, by inhibiting aftiphilin (AFTPH) expression[110]. This study may represent a promising strategy to inhibit miR-133a and restore AFTPH expression for the treatment of IBD.
Immune responses play important roles in connecting inflammation and CRC. IBD has been connected to predisposing genetic factors that contribute to abnormal immune responses. For example, adaptive immune system related genes that have been linked to IBD include components of the IL-23 signaling pathway, which are required for induction of Th17 cells[111,112]. Interestingly, a recent study has found that the intracellular sensor nucleotide-binding oligomerization domain protein 2 (NOD2) can induce miR-29 expression in human DCs to limit IL-23 release[113]. NOD2 is required for the induction of miR-29; miR-29 downregulates IL-23 and attenuates Th17 cell responses in CD. Activation of NOD2 has a key role in influencing the release of pro-/anti-inflammatory cytokines, particularly IL-1belta/6/8/23[114,115]. Furthermore, activation of NOD2-dependent pathways contributes to the induction of NF-κB, as well as the activation of JNK and p38 MAPK[116,117]. NOD2 not only induces miRNA expression but also has been identified as a functional downstream target of several miRNAs. MiR-122[118] and miR-192[119] can directly target NOD2 and suppress the expression of NOD2. Ectopic expression of these two miRNAs inhibits NOD2 expression, NF-κB activation and immune responses. Furthermore, miRNAs are also pivotal regulators of Th17 cells differentiation and function. MiR-210 has a particularly important role in regulating Th17 cell lineage, and is a signature miRNA of hypoxia[120]. The expression of miR-210 increases rapidly in hypoxia-challenged Th17 cells[120]. Hypoxic conditions inhibit degradation of the subunits of hypoxia-inducible factor 1 (HIF-1) and lead to their stabilization[121,122]. HIF-1 plays important roles in regulating Th17 differentiation[123]. Intriguingly, miR-210 directly targets HIF-1α, reduces HIF-1α transcript abundance and the proportion of cells that produce inflammatory cytokines, and controls IBD severity[120]. In addition to regulating Th cell differentiation, miRNAs can regulate leukocyte migration by targeting chemokines. MiR-141 regulates colonic leukocyte trafficking via directly targeting CXCL12β during intestinal inflammation of CD[124]. The miR-141-CXCL12β pathway contributes significantly to the development of CD, likely by affecting the recruitment of immune cells into the inflamed colonic tissue. Therefore, repressing colonic CXCL12β and immune cell recruitment with miRNAs represents a feasible and promising strategy that may be valuable for the treatment of CD. Moreover, the expression of a number of intestinal epithelial cell-derived chemokine is regulated by miRNAs. MiR-192 was dramatically upregulated in UC and regulated the expression of macrophage inflammatory peptide-2α (MIP-2α), which is a chemotactic chemokine secreted by colonic epithelial cells and macrophages[125]. Recently, miRNAs have been found to regulate gut mucosal immunity via epithelium-T-cell crosstalk. MiR-375 regulates gut mucosal immunity by modulating the differentiation of goblet cells, and targets the expression of thymic stromal lymphopoietin and resistin-like molecule-β, a Th-2-facilitating epithelial cytokine and an important goblet cell effector, respectively[126].
IBD and CRC are related to dysregulation of the intestinal immune system and of the microbiota[127]. The gut microbiota provides constant immunological signals to intestinal tissues, thus a variety of regulatory mechanisms have evolved to ensure proper development and function of the intestinal immune system. MiRNAs have been demonstrated to regulate expression of genes involved in microbial recognition and downstream immune activity. Thus, miRNAs play a key role within the intestinal immune system during its interactions with the gut microbiota[128]. Nine miRNAs are differentially expressed in the colon of germ-free mice colonized with the microbiota; the upregulation of miR-665 significantly downregulates the ATP-binding cassette sub-family C member 3[129], which belongs to the multidrug resistance-associated protein family[130]. MiR-146a represses a subset of gut barrier and inflammatory genes and restricts the expansion of intestinal T cell populations, including Tregs, Th17 and T follicular helper cells[131]. Furthermore, the regulatory effect of microbiota on miRNA expression and on the maintenance of intestinal homeostasis has been investigated. The expression of miR-10a and its targets IL-2/IL-23p40 play important roles in regulating innate immune responses to commensal bacteria in DC[132]. These studies open new perspectives for the investigation of miRNA regulation in intestinal diseases and microbiota. Furthermore, a variety of bacterial infection can trigger inflammasome formation and activation[133]. Inflammasomes are large cytosolic protein complexes that promote immediate inflammatory responses and regulate intestinal homeostasis through its effects on the intestinal microbiota[134,135]. The inflammasomes have potentially critical roles in the development and pathogenesis of IBD, because of their important role in intestinal immunity[135]. Emerging evidence has demonstrated that inflammasomes can influence intestinal microbiota. Some reports have exhibited that mice lacking various components of the inflammasome showed prototypical alterations in their microbiota, predisposing these mice to the development of IBD[136]. Inflammasomes have important roles in IBD, whereas the mechanisms of inflammasome formation and activation in the onset of intestinal diseases have not yet been fully established. MiRNAs may be regulators in inflammasomes formation and activation. Although only few reports explore the role of miRNAs in modulating inflammasomes in intestinal diseases, some works have been carried out in other disease types[137,138]. In the future, more investigations should illustrate novel understandings on miRNA and inflammasome interactions, which will potentially lead to the discovery of new treatment approaches for intestinal disease or CRC.
The limitations of surgery and chemo/radio therapies to treat CRC patients necessitate the development of novel strategies, including immunotherapy. Tremendous progress has been made in understanding the role of the immune system in driving the development of CRC. Immune cell markers could be used to predict CRC outcomes[11], while targeting various aspects of the immune system could be used to generate anti-tumor immune responses. Unfortunately, effective immunotherapy in CRC treatment remains elusive. The ultimate goal of immunotherapy is to recruit the immune effectors to destroy cancer cells. However, immune escape leads to the failure of immunotherapy. It has been proposed that cancer cells use immune escape to evade the recognition and killing by the host immune system. For example, the expression of B7, a co-stimulatory molecule, has been found to be involved in CRC progression and immune escape mechanism[139]. Furthermore, STAT3 regulates key pathways that mediate immune escape in the TME[140,141]. Therefore, novel approaches need to be identified to overcome immune escape. Although several trails utilizing cancer vaccines or immune-checkpoint therapies have demonstrated objective responses in immunized patients with mCRC, more work is needed.
MiRNAs are deregulated and play critical roles in CRC. Initially, miRNAs have been found to act as oncogenic or tumor suppressor genes and regulate tumorigenesis, angiogenesis, progression, invasion and metastasis. The discoveries of miRNAs have opened a new era of CRC research and the potential of novel therapies for CRC treatment (Figure 2). Recently, miRNAs have been identified to regulate immune responses in CRC. The fact that miRNAs regulate tumor behaviors through targeting multiple genes, including immune associated genes, makes them excellent candidates for gene therapy. Therefore, the ability to use miRNA-based therapy to restore a dysfunctional immune system to a healthy state needs to be deeply studied. Generally, restoring the expression of tumor suppressor miRNAs or inhibiting the expression of oncogenic miRNAs has been employed for the development of miRNA-based therapeutics. In this regard, an example such as inducing miR-34a expression has been found to elicit tumor-infiltrating CD8+ T cells activation by targeting PD-L1[60]. Actually, an miR-34 mimic (MRX34) has become the first miRNA to enter phase I clinical trial (NCT01829971). The study was started to evaluate the safety of MRX34 in patients with unresectable primary liver cancer or other selected solid tumors, such as renal cell carcinoma, non-small cell lung cancer and melanoma. Targeted therapy and immunotherapy, such as PD-1 antibodies, in melanoma have led to a marked improvement in patients’ survival and their quality of life[142]. Therefore, the miRNA-target mRNA axis may be another novel immunotherapeutic target for cancer treatment. For example, STAT3 has been identified to play a pivotal role in a wide variety of tumor-mediated immunosuppression and is one of the first candidates that researchers assessed for potential miRNA binding. MiR-124 can enhance immune effector responses by directly inhibiting the activation of the STAT3 signaling pathway[28]. Because aberrant activation of STAT3 pathway occurs in each stage of CRC tumorigenesis, including inflammation, adenoma and carcinogenesis, targeting diverse miRNA-STAT3 axes in specific pathological stages may improve therapeutic efficacy. In addition, combinational therapies are promising approaches for CRC immunotherapy. As shown above, PD-L1 is the target of miR-34. MiR-34 therapy and PD-L1 monoclonal antibody therapy combinations will elicit stronger anti-tumor immune responses. Furthermore, miRNAs are known to target networks of genes and this may provide a unique therapeutic advantage. Therefore, altering the expression of multiple miRNAs simultaneously by repressing distinct targets is a potential approach for CRC treatment.
MiRNAs exhibit diverse immunomodulatory effects through their action on multiple mRNA species because they control many important immunological processes. Although great efforts have been made to discover the precise role of miRNAs in anti-tumor immune response regulations, there are many challenges for the development of miRNA-based immunotherapy. These challenges include screening tissue-specific profiles, target identification, safe delivery to specific tissue, off-targets and biological safety. According to previous reports, miRNA expression is deregulated in tumor tissues compared to normal tissues and each patient has specific miRNA profiles. Thus, the ability to identify distinct miRNAs to treat CRC needs to be deeply explored. Moreover, although many computational algorithms have been developed to identify potential targets, many critical functional miRNA targets could not be identified due to the inability of bio-informatics predictions to find miRNA binding sites with seed mismatches[143]. Fortunately, novel approaches have been developed to overcome these limitations. The major challenge in miRNA-based immunotherapy is the safe delivery to specific tissues without side effects. Moreover, there are challenges to ensure miRNAs to reach the TME rather than other organs. To overcome these delivery and biological safety hurdles, some effective and marketable strategies, such as viral and non-viral strategies, have been developed[144,145]. However, novel approaches are needed to determine the efficacy of immune responses in clinical applications. In addition, another obstacle faced is whether exogenous miRNAs can precisely regulate their appeal targets in tumor cells. Therefore, there is a long way to go before we resolve many technical and biological challenges. Nevertheless, the successful development of miRNA-based immunotherapy has attracted the attention of many researchers.
The authors are grateful to the entire tumor group of Han’s Laboratory for help and support.
P- Reviewer: Botta C S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 320] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18694] [Cited by in F6Publishing: 20838] [Article Influence: 2315.3] [Reference Citation Analysis (2)] |
| 3. | Kerr D. Clinical development of gene therapy for colorectal cancer. Nat Rev Cancer. 2003;3:615-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1112] [Cited by in F6Publishing: 1147] [Article Influence: 81.9] [Reference Citation Analysis (0)] |
| 5. | Gallagher DJ, Kemeny N. Metastatic colorectal cancer: from improved survival to potential cure. Oncology. 2010;78:237-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 170] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 6. | Xiang B, Snook AE, Magee MS, Waldman SA. Colorectal cancer immunotherapy. Discov Med. 2013;15:301-308. [PubMed] [Cited in This Article: ] |
| 7. | Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, Suresh AS, Thomas A, Tjulandin S, Zhang K. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): a randomised, multicentre, open-label, non-inferiority phase 3 study. Lancet Oncol. 2014;15:569-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 315] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 8. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1314] [Cited by in F6Publishing: 1452] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 9. | Botta C, Bestoso E, Apollinari S, Cusi MG, Pastina P, Abbruzzese A, Sperlongano P, Misso G, Caraglia M, Tassone P. Immune-modulating effects of the newest cetuximab-based chemoimmunotherapy regimen in advanced colorectal cancer patients. J Immunother. 2012;35:440-447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Markman JL, Shiao SL. Impact of the immune system and immunotherapy in colorectal cancer. J Gastrointest Oncol. 2015;6:208-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 94] [Reference Citation Analysis (0)] |
| 11. | Oberg A, Samii S, Stenling R, Lindmark G. Different occurrence of CD8+, CD45R0+, and CD68+ immune cells in regional lymph node metastases from colorectal cancer as potential prognostic predictors. Int J Colorectal Dis. 2002;17:25-29. [PubMed] [Cited in This Article: ] |
| 12. | deLeeuw RJ, Kost SE, Kakal JA, Nelson BH. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18:3022-3029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 334] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 13. | Correale P, Rotundo MS, Del Vecchio MT, Remondo C, Migali C, Ginanneschi C, Tsang KY, Licchetta A, Mannucci S, Loiacono L. Regulatory (FoxP3+) T-cell tumor infiltration is a favorable prognostic factor in advanced colon cancer patients undergoing chemo or chemoimmunotherapy. J Immunother. 2010;33:435-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 14. | Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006;90:51-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 488] [Cited by in F6Publishing: 490] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 15. | Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006;27:195-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 506] [Cited by in F6Publishing: 530] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 16. | Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci USA. 2007;104:3360-3365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1041] [Cited by in F6Publishing: 1143] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 17. | Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Cited in This Article: ] |
| 18. | Mei Q, Li X, Guo M, Fu X, Han W. The miRNA network: micro-regulator of cell signaling in cancer. Expert Rev Anticancer Ther. 2014;14:1515-1527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2105] [Cited by in F6Publishing: 2246] [Article Influence: 132.1] [Reference Citation Analysis (0)] |
| 20. | Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1021] [Cited by in F6Publishing: 1089] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 21. | Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432-10439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 223] [Cited by in F6Publishing: 257] [Article Influence: 25.7] [Reference Citation Analysis (1)] |
| 22. | Mei Q, Xue G, Li X, Wu Z, Li X, Yan H, Guo M, Sun S, Han W. Methylation-induced loss of miR-484 in microsatellite-unstable colorectal cancer promotes both viability and IL-8 production via CD137L. J Pathol. 2015;236:165-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Yu HW, Sze DM, Cho WC. MicroRNAs Involved in Anti-Tumour Immunity. Int J Mol Sci. 2013;14:5587-5607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 346] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 25. | Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 804] [Cited by in F6Publishing: 823] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 26. | Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 898] [Cited by in F6Publishing: 913] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 27. | Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1322] [Cited by in F6Publishing: 1366] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 28. | Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, Yang Y, McEnery K, Jethwa K, Gjyshi O. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913-3926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 194] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 29. | Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, Wan Y, Sampson JH, Zhu B, Li QJ. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. 2014;124:5352-5367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 30. | Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487-5497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 236] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 31. | Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 282] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 32. | Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861-869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 484] [Cited by in F6Publishing: 502] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 33. | Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1260] [Cited by in F6Publishing: 1190] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 34. | Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DG, Lanier LL. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185:3835-3846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 35. | Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. 2014;11:495-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 36. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [Cited in This Article: ] |
| 37. | Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2851] [Cited by in F6Publishing: 2875] [Article Influence: 169.1] [Reference Citation Analysis (0)] |
| 38. | Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519-2527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 424] [Cited by in F6Publishing: 449] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 39. | Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze MT. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci USA. 2009;106:10746-10751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 138] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 40. | Bachmann MF, McKall-Faienza K, Schmits R, Bouchard D, Beach J, Speiser DE, Mak TW, Ohashi PS. Distinct roles for LFA-1 and CD28 during activation of naive T cells: adhesion versus costimulation. Immunity. 1997;7:549-557. [PubMed] [Cited in This Article: ] |
| 41. | Aslam MI, Patel M, Singh B, Jameson JS, Pringle JH. MicroRNA manipulation in colorectal cancer cells: from laboratory to clinical application. J Transl Med. 2012;10:128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Chi Y, Zhou D. MicroRNAs in colorectal carcinoma--from pathogenesis to therapy. J Exp Clin Cancer Res. 2016;35:43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 43. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5384] [Cited by in F6Publishing: 5479] [Article Influence: 304.4] [Reference Citation Analysis (0)] |
| 44. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4616] [Cited by in F6Publishing: 4368] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 45. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] [Cited in This Article: ] |
| 46. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1481] [Cited by in F6Publishing: 1531] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 47. | Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B, Meijer GA. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 207] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 48. | Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391-2397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 144] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 49. | Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2015;Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 100] [Article Influence: 12.5] [Reference Citation Analysis (1)] |
| 50. | Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 51. | Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-1326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 435] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 52. | Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062-2074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 288] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 53. | Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894-907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1717] [Cited by in F6Publishing: 1794] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 54. | Amin M, Lockhart AC. The potential role of immunotherapy to treat colorectal cancer. Expert Opin Investig Drugs. 2015;24:329-344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 55. | Singh PP, Sharma PK, Krishnan G, Lockhart AC. Immune checkpoints and immunotherapy for colorectal cancer. Gastroenterol Rep (Oxf). 2015;3:289-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 56. | Wang L, Pino-Lagos K, de Vries VC, Guleria I, Sayegh MH, Noelle RJ. Programmed death 1 ligand signaling regulates the generation of adaptive Foxp3+CD4+ regulatory T cells. Proc Natl Acad Sci USA. 2008;105:9331-9336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 57. | Hua D, Sun J, Mao Y, Chen LJ, Wu YY, Zhang XG. B7-H1 expression is associated with expansion of regulatory T cells in colorectal carcinoma. World J Gastroenterol. 2012;18:971-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 34] [Cited by in F6Publishing: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, Zhou H, Li R, Wang K, Wang W. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014;75:348-353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 142] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 59. | Sawai H, Yasuda A, Ochi N, Ma J, Matsuo Y, Wakasugi T, Takahashi H, Funahashi H, Sato M, Takeyama H. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol. 2008;8:56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 60. | Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst. 2016;108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 313] [Cited by in F6Publishing: 427] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 61. | Mlcochova J, Faltejskova P, Nemecek R, Svoboda M, Slaby O. MicroRNAs targeting EGFR signalling pathway in colorectal cancer. J Cancer Res Clin Oncol. 2013;139:1615-1624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 62. | Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, Zagatti B, Radova L, Svoboda M, Nemecek R, John S, Kiss I, Vyzula R. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget. 2015;6:38695-38704. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 63. | Manceau G, Imbeaud S, Thiébaut R, Liébaert F, Fontaine K, Rousseau F, Génin B, Le Corre D, Didelot A, Vincent M. Hsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 2014;20:3338-3347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 64. | Mosakhani N, Lahti L, Borze I, Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P, Knuutila S, Sarhadi VK. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer Genet. 2012;205:545-551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 65. | Zhu H, Dougherty U, Robinson V, Mustafi R, Pekow J, Kupfer S, Li YC, Hart J, Goss K, Fichera A. EGFR signals downregulate tumor suppressors miR-143 and miR-145 in Western diet-promoted murine colon cancer: role of G1 regulators. Mol Cancer Res. 2011;9:960-975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 66. | Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643-2648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 621] [Cited by in F6Publishing: 621] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 67. | Ota T, Doi K, Fujimoto T, Tanaka Y, Ogawa M, Matsuzaki H, Kuroki M, Miyamoto S, Shirasawa S, Tsunoda T. KRAS up-regulates the expression of miR-181a, miR-200c and miR-210 in a three-dimensional-specific manner in DLD-1 colorectal cancer cells. Anticancer Res. 2012;32:2271-2275. [PubMed] [Cited in This Article: ] |
| 68. | Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 575] [Cited by in F6Publishing: 710] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 69. | Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5121] [Cited by in F6Publishing: 4975] [Article Influence: 331.7] [Reference Citation Analysis (0)] |
| 70. | Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791-5802. [PubMed] [Cited in This Article: ] |
| 71. | Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2416] [Cited by in F6Publishing: 2469] [Article Influence: 123.5] [Reference Citation Analysis (0)] |
| 72. | Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 923] [Cited by in F6Publishing: 951] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 73. | Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su X, Liu J, Chen Y, Wang M, Zhang Y. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J Immunol. 2011;186:4716-4724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 74. | Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, Wang X, Wang J, Yu H, Cao X. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188:5500-5510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 209] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 75. | Kusmartsev S, Nagaraj S, Gabrilovich DI. Tumor-associated CD8+ T cell tolerance induced by bone marrow-derived immature myeloid cells. J Immunol. 2005;175:4583-4592. [PubMed] [Cited in This Article: ] |
| 76. | Liu Q, Zhang M, Jiang X, Zhang Z, Dai L, Min S, Wu X, He Q, Liu J, Zhang Y. miR-223 suppresses differentiation of tumor-induced CD11b+ Gr1+ myeloid-derived suppressor cells from bone marrow cells. Int J Cancer. 2011;129:2662-2673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 77. | Singh R, Lillard JW, Singh S. Chemokines: key players in cancer progression and metastasis. Front Biosci (Schol Ed). 2011;3:1569-1582. [PubMed] [Cited in This Article: ] |
| 78. | Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, Soucy G, Odze R, Glickman JN, Garrett WS. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015;12:244-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 222] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 79. | Kakinuma T, Hwang ST. Chemokines, chemokine receptors, and cancer metastasis. J Leukoc Biol. 2006;79:639-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 257] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 80. | Rubie C, Oliveira V, Kempf K, Wagner M, Tilton B, Rau B, Kruse B, Konig J, Schilling M. Involvement of chemokine receptor CCR6 in colorectal cancer metastasis. Tumour Biol. 2006;27:166-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 81. | Ghadjar P, Rubie C, Aebersold DM, Keilholz U. The chemokine CCL20 and its receptor CCR6 in human malignancy with focus on colorectal cancer. Int J Cancer. 2009;125:741-745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 82. | Vicinus B, Rubie C, Faust SK, Frick VO, Ghadjar P, Wagner M, Graeber S, Schilling MK. miR-21 functionally interacts with the 3’UTR of chemokine CCL20 and down-regulates CCL20 expression in miR-21 transfected colorectal cancer cells. Cancer Lett. 2012;316:105-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 83. | Vicinus B, Rubie C, Stegmaier N, Frick VO, Kölsch K, Kauffels A, Ghadjar P, Wagner M, Glanemann M. miR-21 and its target gene CCL20 are both highly overexpressed in the microenvironment of colorectal tumors: significance of their regulation. Oncol Rep. 2013;30:1285-1292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 84. | Kim DK, Lee SC, Lee HW. CD137 ligand-mediated reverse signals increase cell viability and cytokine expression in murine myeloid cells: involvement of mTOR/p70S6 kinase and Akt. Eur J Immunol. 2009;39:2617-2628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 85. | Li X, Li X, Liao D, Wang X, Wu Z, Nie J, Bai M, Fu X, Mei Q, Han W. Elevated microRNA-23a Expression Enhances the Chemoresistance of Colorectal Cancer Cells with Microsatellite Instability to 5-Fluorouracil by Directly Targeting ABCF1. Curr Protein Pept Sci. 2015;16:301-309. [PubMed] [Cited in This Article: ] |
| 86. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6096] [Cited by in F6Publishing: 6542] [Article Influence: 726.9] [Reference Citation Analysis (0)] |
| 87. | McConnell BB, Yang VW. The Role of Inflammation in the Pathogenesis of Colorectal Cancer. Curr Colorectal Cancer Rep. 2009;5:69-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 88. | Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 380] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 89. | Bian Z, Li L, Cui J, Zhang H, Liu Y, Zhang CY, Zen K. Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol. 2011;225:544-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 90. | Edelblum KL, Yan F, Yamaoka T, Polk DB. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm Bowel Dis. 2006;12:413-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 124] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 91. | Zeissig S, Bojarski C, Buergel N, Mankertz J, Zeitz M, Fromm M, Schulzke JD. Downregulation of epithelial apoptosis and barrier repair in active Crohn’s disease by tumour necrosis factor alpha antibody treatment. Gut. 2004;53:1295-1302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 243] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 92. | Lv B, Liu Z, Wang S, Liu F, Yang X, Hou J, Hou Z, Chen B. MiR-29a promotes intestinal epithelial apoptosis in ulcerative colitis by down-regulating Mcl-1. Int J Clin Exp Pathol. 2014;7:8542-8552. [PubMed] [Cited in This Article: ] |
| 93. | Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 94. | Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16:1729-1738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 95. | Sugimoto K. Role of STAT3 in inflammatory bowel disease. World J Gastroenterol. 2008;14:5110-5114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 83] [Cited by in F6Publishing: 93] [Article Influence: 5.8] [Reference Citation Analysis (2)] |
| 96. | Musso A, Dentelli P, Carlino A, Chiusa L, Repici A, Sturm A, Fiocchi C, Rizzetto M, Pegoraro L, Sategna-Guidetti C. Signal transducers and activators of transcription 3 signaling pathway: an essential mediator of inflammatory bowel disease and other forms of intestinal inflammation. Inflamm Bowel Dis. 2005;11:91-98. [PubMed] [Cited in This Article: ] |
| 97. | Atreya R, Neurath MF. Involvement of IL-6 in the pathogenesis of inflammatory bowel disease and colon cancer. Clin Rev Allergy Immunol. 2005;28:187-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 230] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 98. | Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M, Pothoulakis C, Winter HS, Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842-852.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 179] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 99. | Liu W, Chen Y, Golan MA, Annunziata ML, Du J, Dougherty U, Kong J, Musch M, Huang Y, Pekow J. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest. 2013;123:3983-3996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 244] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 100. | Chen Y, Du J, Zhang Z, Liu T, Shi Y, Ge X, Li YC. MicroRNA-346 mediates tumor necrosis factor α-induced downregulation of gut epithelial vitamin D receptor in inflammatory bowel diseases. Inflamm Bowel Dis. 2014;20:1910-1918. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 101. | Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462-2476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2744] [Cited by in F6Publishing: 2694] [Article Influence: 141.8] [Reference Citation Analysis (2)] |
| 102. | Colombel JF, Sandborn WJ, Rutgeerts P, Enns R, Hanauer SB, Panaccione R, Schreiber S, Byczkowski D, Li J, Kent JD. Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: the CHARM trial. Gastroenterology. 2007;132:52-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1598] [Cited by in F6Publishing: 1523] [Article Influence: 89.6] [Reference Citation Analysis (0)] |
| 103. | Chen B, She S, Li D, Liu Z, Yang X, Zeng Z, Liu F. Role of miR-19a targeting TNF-α in mediating ulcerative colitis. Scand J Gastroenterol. 2013;48:815-824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 104. | Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1352] [Cited by in F6Publishing: 1444] [Article Influence: 103.1] [Reference Citation Analysis (0)] |
| 105. | Zídek Z, Anzenbacher P, Kmonícková E. Current status and challenges of cytokine pharmacology. Br J Pharmacol. 2009;157:342-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 106. | Cheng X, Zhang X, Su J, Zhang Y, Zhou W, Zhou J, Wang C, Liang H, Chen X, Shi R. miR-19b downregulates intestinal SOCS3 to reduce intestinal inflammation in Crohn’s disease. Sci Rep. 2015;5:10397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 107. | Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, Kubo M, Yamashita A, Okabe M, Takeda K. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193:471-481. [PubMed] [Cited in This Article: ] |
| 108. | Castagliuolo I, Wang CC, Valenick L, Pasha A, Nikulasson S, Carraway RE, Pothoulakis C. Neurotensin is a proinflammatory neuropeptide in colonic inflammation. J Clin Invest. 1999;103:843-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 100] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 109. | Brun P, Mastrotto C, Beggiao E, Stefani A, Barzon L, Sturniolo GC, Palù G, Castagliuolo I. Neuropeptide neurotensin stimulates intestinal wound healing following chronic intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2005;288:G621-G629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 110. | Law IK, Bakirtzi K, Polytarchou C, Oikonomopoulos A, Hommes D, Iliopoulos D, Pothoulakis C. Neurotensin--regulated miR-133α is involved in proinflammatory signalling in human colonic epithelial cells and in experimental colitis. Gut. 2015;64:1095-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 111. | Cătană CS, Berindan Neagoe I, Cozma V, Magdaş C, Tăbăran F, Dumitraşcu DL. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2015;21:5823-5830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 126] [Cited by in F6Publishing: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 112. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1894] [Cited by in F6Publishing: 1924] [Article Influence: 137.4] [Reference Citation Analysis (0)] |
| 113. | Brain O, Owens BM, Pichulik T, Allan P, Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity. 2013;39:521-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 114. | van Beelen AJ, Zelinkova Z, Taanman-Kueter EW, Muller FJ, Hommes DW, Zaat SA, Kapsenberg ML, de Jong EC. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 2007;27:660-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 395] [Cited by in F6Publishing: 397] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 115. | Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1334] [Cited by in F6Publishing: 1302] [Article Influence: 68.5] [Reference Citation Analysis (2)] |
| 116. | Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 584] [Cited by in F6Publishing: 573] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 117. | Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8061] [Cited by in F6Publishing: 8304] [Article Influence: 461.3] [Reference Citation Analysis (0)] |
| 118. | Chen Y, Wang C, Liu Y, Tang L, Zheng M, Xu C, Song J, Meng X. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem Biophys Res Commun. 2013;438:133-139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 119. | Chuang AY, Chuang JC, Zhai Z, Wu F, Kwon JH. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm Bowel Dis. 2014;20:126-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 120. | Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. 2014;15:393-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 193] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 121. | Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3728] [Cited by in F6Publishing: 3670] [Article Influence: 146.8] [Reference Citation Analysis (0)] |
| 122. | Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468-472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4121] [Cited by in F6Publishing: 4097] [Article Influence: 178.1] [Reference Citation Analysis (0)] |
| 123. | Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3754] [Cited by in F6Publishing: 3614] [Article Influence: 240.9] [Reference Citation Analysis (0)] |
| 124. | Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, Dong L, Zhang C, Zeng K, Chen J. miR-141 Regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn’s disease. Gut. 2014;63:1247-1257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 125. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 365] [Cited by in F6Publishing: 383] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 126. | Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, Kredo-Russo S, Avnit-Sagi T, Cojocaru G, Zreik F. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12:239-246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 127. | Hold GL. Gastrointestinal Microbiota and Colon Cancer. Dig Dis. 2016;34:244-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 128. | Runtsch MC, Round JL, O’Connell RM. MicroRNAs and the regulation of intestinal homeostasis. Front Genet. 2014;5:347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 129. | Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. Microbiota modulate host gene expression via microRNAs. PLoS One. 2011;6:e19293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 130. | Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881-884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1565] [Cited by in F6Publishing: 1424] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 131. | Runtsch MC, Hu R, Alexander M, Wallace J, Kagele D, Petersen C, Valentine JF, Welker NC, Bronner MP, Chen X. MicroRNA-146a constrains multiple parameters of intestinal immunity and increases susceptibility to DSS colitis. Oncotarget. 2015;6:28556-28572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Xue X, Feng T, Yao S, Wolf KJ, Liu CG, Liu X, Elson CO, Cong Y. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J Immunol. 2011;187:5879-5886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 133. | de Zoete MR, Flavell RA. Interactions between Nod-Like Receptors and Intestinal Bacteria. Front Immunol. 2013;4:462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 134. | Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821-832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3880] [Cited by in F6Publishing: 4233] [Article Influence: 302.4] [Reference Citation Analysis (0)] |
| 135. | Gagliani N, Palm NW, de Zoete MR, Flavell RA. Inflammasomes and intestinal homeostasis: regulating and connecting infection, inflammation and the microbiota. Int Immunol. 2014;26:495-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 136. | Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745-757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1392] [Cited by in F6Publishing: 1476] [Article Influence: 113.5] [Reference Citation Analysis (0)] |
| 137. | Yang Z, Zhong L, Xian R, Yuan B. MicroRNA-223 regulates inflammation and brain injury via feedback to NLRP3 inflammasome after intracerebral hemorrhage. Mol Immunol. 2015;65:267-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 138. | Zhou Y, Lu M, Du RH, Qiao C, Jiang CY, Zhang KZ, Ding JH, Hu G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener. 2016;11:28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 329] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 139. | Tirapu I, Huarte E, Guiducci C, Arina A, Zaratiegui M, Murillo O, Gonzalez A, Berasain C, Berraondo P, Fortes P. Low surface expression of B7-1 (CD80) is an immunoescape mechanism of colon carcinoma. Cancer Res. 2006;66:2442-2450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 140. | Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314-1321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 762] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 141. | Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113-1125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 472] [Cited by in F6Publishing: 472] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 142. | Jacobsoone-Ulrich A, Jamme P, Alkeraye S, Dzwiniel V, Faure E, Templier C, Mortier L. Ipilimumab in anti-PD1 refractory metastatic melanoma: a report of eight cases. Melanoma Res. 2016;26:153-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 143. | Lee HM, Nguyen DT, Lu LF. Progress and challenge of microRNA research in immunity. Front Genet. 2014;5:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 144. | Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 638] [Cited by in F6Publishing: 618] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 145. | Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin Drug Deliv. 2009;6:673-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |