Published online Aug 21, 2006. doi: 10.3748/wjg.v12.i31.4986
Revised: November 11, 2004
Accepted: November 23, 2004
Published online: August 21, 2006
AIM: To find the relationship between hepatitis B virus (HBV) and hepatocytes during the initial state of infection by cDNA microarray.
METHODS: Primary normal human hepatocytes (PNHHs) were isolated and infected with HBV. From the PNHHs, RNA was isolated and inverted into complement DNA (cDNA) with Cy3- or Cy5- labeled dUTP for microarray analysis. The labeled cDNA was hybridized with microarray chip, including 4224 cDNAs. From the image of the microarray, expression profiles were produced and some of them were confirmed by RT-PCR, immunoblot analysis, and NF-κB luciferase reporter assay.
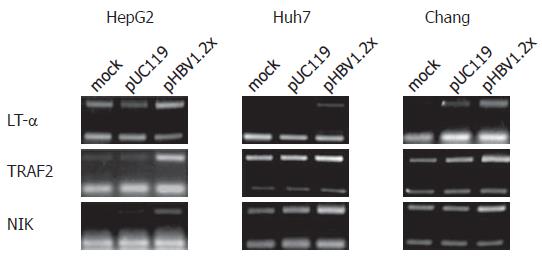
RESULTS: From the cDNA microarray, we obtained 98 differentially regulated genes. Of the 98 genes, 53 were up regulated and 45 down regulated. Interestingly, in the up regulated genes, we found the TNF signaling pathway-related genes: LT-α, TRAF2, and NIK. By using RT-PCR, we confirmed the up-regulation of these genes in HepG2, Huh7, and Chang liver cells, which were transfected with pHBV1.2×, a plasmid encoding all HBV messages. Moreover, these three genes participated in HBV-mediated NF-κB activation.
CONCLUSION: During the initial state of HBV infection, hepatocytes facilitate the activation of NF-κB through up regulation of LT-α, TRAF2, and NIK.
- Citation: Ryu HM, Park SG, Yea SS, Jang WH, Yang YI, Jung G. Gene expression analysis of primary normal human hepatocytes infected with human hepatitis B virus. World J Gastroenterol 2006; 12(31): 4986-4995
- URL: https://www.wjgnet.com/1007-9327/full/v12/i31/4986.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i31.4986
Human hepatitis B virus (HBV) is a causative agent for liver diseases such as cirrhosis and hepatocellular carcinoma (HCC)[1]. Chronic infection of HBV affects approximately 800 million people and is the principal cause of chronic liver diseases[2]. Moreover, HBV carriers have a much higher frequency of developing liver cancer than uninfected people[3].
HBV has a small, partially double-stranded DNA genome. After viral infection of hepatocytes, the partially double-stranded DNA genome converts into covalently closed circular DNA (cccDNA) in nuclei[4-7]. Several kinds of viral transcripts are then produced by the host RNA polymerase. The transcripts encode for viral polymerase, viral oncogene HBx protein, and viral structural proteins such as surface proteins and core proteins[3].
Many efforts have been made to investigate the process of liver disease by HBV. Traditional techniques such as Northern blot and reverse transcription polymerase chain reaction (RT-PCR) for identification of genes differentially expressed by HBV infection have shown limited success, because only one gene or at best a handful of genes can be studied in one experiment. However, complementary DNA (cDNA) microarray allows the study of several thousands of genes at one time. To evaluate the relationship between HBV infection and liver diseases, recent studies have analyzed the gene expression profiles at tissue level. In these studies, the effects of HBV infection are analyzed by cDNA microarray analysis of HCC tissue samples[8-10]. Through the analyses, many differentially expressed genes can be identified[11]. The analyses, however, have mainly focused on the gene expression profiles of already transformed cells or long-term infected HBV hepatocytes. Therefore, these analyses mostly stem from the analysis of the end result of pathogenesis of HBV in hepatocytes rather than the analysis of ongoing pathogenesis of HBV infection in hepatocytes. In this report, however, we focused on the gene expression profile analysis of the early stage HBV infection, thereby excluding factors such as responses to host immune surveillance. To mimic the early stage HBV infection of hepatocytes, we isolated primary normal human hepatocytes (PNHHs) and the cells were infected with HBV in culture. These conditions were chosen as they could represent the most similar conditions to those in vivo, except for the absence of other types of cells such as immunocytes. Therefore, gene expression profiles in this report could show the result of interaction only between HBV and PNHHs. In this study we have identified 45 down-regulated genes and 53 up-regulated genes.
pHBV1.2×, a plasmid which provides all HBV transcripts, was used to infect PNHHs as previously described[12]. This construct was similar to that as described by Guidotti et al[13]. Mammalian expression vector for NIK and NIK DN (aa 624-947) was provided by DV Goeddel (Turarik Inc.)[14], for TRAF2 and TRAF2 DN (aa 241-501) by Dr. SY Lee[15].
We transfected HepG2 cells with pHBV1.2× constructs for generation of HBV using Fugene 6 transfection reagent (Roche) as instructed by the manufacturer. After transfection, the cells were cultured for 5 d and harvested. HBV particles in the harvested media were cleared and concentrated through ultracentrifugation using PST55Ti rotor (Hitachi) for 1 h at 220 000 g with 3 mL cushion buffer containing 20 g/L sucrose, 50 mmol/L Tris-HCl pH 7.5, and 30 mmol/L NaCl. After ultracentrifugation, the pellet was resuspended with 1 × PBS. The resuspended viral solution was filtered with a 0.2 μm pore filter (Millipore). The titer of HBV solution was adjusted to 109 virus genome equivalent (v.g.e.) per mL. PNHHs were infected with the above virus solution at about 100 v.g.e. per cell. Using this method, the efficiency of HBV infection to PNHHs was generally 50%[16].
Healthy parts of a liver from a patient who underwent hepatic resection for an intrahepatic stone at the Inje University Paik Hospital, Pusan, Korea was obtained and used as the source of hepatocytes. The removed tissue was immediately placed in Hank’s balanced salt solution (HBSS) and processed for cell culture. Isolation of hepatocytes was performed using a two-step collagenase perfusion technique[17,18]. The isolated hepatocytes were resuspended in a nutrient medium containing 90 mL/L Williams’ E and 10 mL/L Medium 199, supplemented with 10 μg/mL insulin, 5 μg/mL transferrin, 10-7 mol/L sodium selenite, and 50 mL/L FBS.
A test was performed with the isolated DNA to determine whether the HBV- infected PNHHs formed cccDNA. Amplification with primers specific for both outside regions was performed with the isolated DNA because the region was specific for cccDNA rather than partially double-stranded DNA found in viral particles. The primers used are (5′-CTATGCTGGGTCTTCCAAATT-3′) which anneals to near the codon for amino acid 80 in human HBc open reading frame (ORF) and (5′-TTTCTGTGTAAACAATATCTG-3′) which anneals to near the codon for amino acid 680 in HBV Pol ORF. Therefore, if cccDNA was present in the isolated DNA, an amplified 1 kb product could be obtained. In this test, DNA isolated from mock-infected PNHHs was used as the negative control. A PCR test was performed with RNA to confirm whether HBV transcripts were produced. Reverse transcription amplification was performed with primers specific for the epsilon regions: (5′-CAACTTTTTTTCACCTCTGCCTA-3′) which anneals to DR1 and the reverse primer (5′-GATCTCGTACTGAAAGGAAAGA-3′). In addition, to detect HBV genome, real-time PCR was also performed as previously described[12].
RNA was isolated using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. With the total isolated RNA, reverse transcription was performed with Cy5-labeled dUTP in the experimental sample and Cy3-labeled dUTP in the control. Fifty micrograms of RNA, 1.5 ng oligo dT primer, and 1 ng control RNA containing lambda DNA sequences with a poly A sequence at the 3′ ends for reverse transcription, were mixed and volume of the mixture was adjusted to 20 μL. The mixture was incubated at 70°C for 5 min. After incubation, the mixture was quickly cooled on ice. With this whole reaction mixture, the labeling reaction was performed under the following conditions: 1 × reverse transcription buffer, 0.6 mmol/L Cy3- or Cy5-dUTP, 40 U of RNase inhibitor (Roche), 50 U of AMV-RT (Roche) and a dNTP mix containing 1 mmol/L dATP, 1 mmol/L dGTP, 1 mmol/L dCTP, and 0.4 mmol/L dTTP at 42°C for 1 h. After 1 h, 50 U of AMV-RT was added to the reaction mixture and the mixture was further incubated for 1 h for complete reverse transcription. Reverse transcription was stopped by the addition of 5 μL 0.5 mol/L EDTA. The synthesized cDNA was purified using a chromaspin column (Clonetech) as instructed by the manufacturer and precipitated with ethanol. Both Cy3- and Cy5- labeled cDNAs were resuspended with 100 μL hybridization buffer, containing 6 × standard saline citrate (SSC), 2 g/L sodium dodecyl sulfate (SDS), 5 × Denhardt solution, and 1 mg/mL salmon sperm DNA. The labeled cDNA was used for hybridization to the cDNA microarray chip at 62°C. The chip was arrayed using a GMS417 arrayer (Genetic MicroSystems Inc., Woburn, MA) with 4224 cDNAs and internal standards such as tubulin and actin and external standards such as lambda DNA. After 16-18 h of hybridization, the hybridized array was washed twice at 58°C for 30 min with washing bufferIcontaining 2 × SSC and 2 g/L SDS and washed once with washing buffer II containing 0.05 × SSC at room temperature for 5 min.
For quantification of the signals, the chips were scanned using an array scanner generation III (Molecular Dynamics) followed by image analysis using ImaGene ver. 3.0 software (BioDiscovery Ltd., Swansea, UK). The signal intensity of each spot was adjusted to obtain more accurate data by subtracting the background signals from the immediate surroundings. In this analysis, a difference in the ratio of more than two folds was considered significant.
HepG2, Huh7, and Chang liver cells were maintained in minimum essential media (Sigma) supplemented with 100 mL/L fetal bovine serum. For reverse transcription-polymerase chain reaction and luciferase reporter assay, cells were seeded in 12- well plates at a density of 0.2 × 106 cells per well and transfected on the following day with the aperopriate DNA and fugene 6 (Roche) as described by the manufacturer. To normalize the total DNA, pUC119 and backbone DNA of pHBV1.2× were used. The transfection efficiency for HepG2 with fugene 6 was usually 10%-20%.
Cells were transfected with pUC119 and pHBV1.2×. After 48 h of transfection, total RNA was extracted with TRIzol reagent (Life Technologies) as described by the manufacturer. cDNA was produced by reverse transcription using the same procedure as cDNA microarray analysis. Following reverse transcription, the synthesized cDNA was amplified with 2.5 U Hot start Taq polymerase (Takara), GAPDH specific primer set, and appropriate primer set. The sequences of the primer set are as follows: TRAF2 specific forward(5′-AGGGGACCCTGAAAGAATAC-3′), TRAF2 specific reverse(5′-CAGGGCTTCAATCTTGTCTT-3′), NIK specific forward(5′-TACCTCCACTCACGAAGGAT-3′), NIK specific reverse(5′-CAAGGGAGGAGACTTGTTTG-3′), LT-α specific forward(5′-AGCTATCCACCCACACAGAT-3′),
LT-α specific reverse(5′-GTTTATTGGGCTTCATCGAG-3′), GAPDH specific forward(5′-ATCATCCCTGCCTCTACTGG-3′), and GAPDH specific reverse (5′-TGGGTGTCGCTGTTGAAGTC-3′). PCR amplification was performed using Gene mp PCR system 2400 (Perkin-Elmer) with 5 min initial denaturation at 95°C and 35 cycles of 50 s at 95°C, 50 s at 60°C, and 50 s at 72°C, followed by 7 min of extension at 72°C. To separate the PCR fragments, 15% agarose gel was used.
To confirm NIK and TRAF2 protein expression, we performed immunoblot analysis with anti-NIK rabbit polyclonal antibody (Santa Cruz) and anti-TRAF2 mouse monoclonal antibody (Santa Cruz). HepG2 cells were seeded in 6- well plates at a density of 0.4 × 106 cells per well. Cells were lysed with RIPA buffer containing 25 mmol/L Tris-HCl (pH 7.4), 150 mmol/L KCl, 5 mmol/L EDTA, 10 mL/L Nonidet P-40 (NP-40), 5 g/L sodium deoxycholate, and 1 g/L SDS and centrifuged at 12 000 g for 10 min. The supernatants were separated by 12% SDS-PAGE protein gel for immunoblot analysis.
After seeded on 12-well plates, HepG2 cells were co-transfected with appropriate DNA (Figure 5), 0.1 μg pNF-κB-luciferase and 0.1 μg pCMV-β-galactosidase. After 48 h of transfection, cell extracts were prepared and luciferase reporter assay was performed using a luciferase assay system (Promega) as described by the manufacturer. The transfection efficiency was normalized by its galactosidase activity. The assay was triplicated and repeated at least twice.
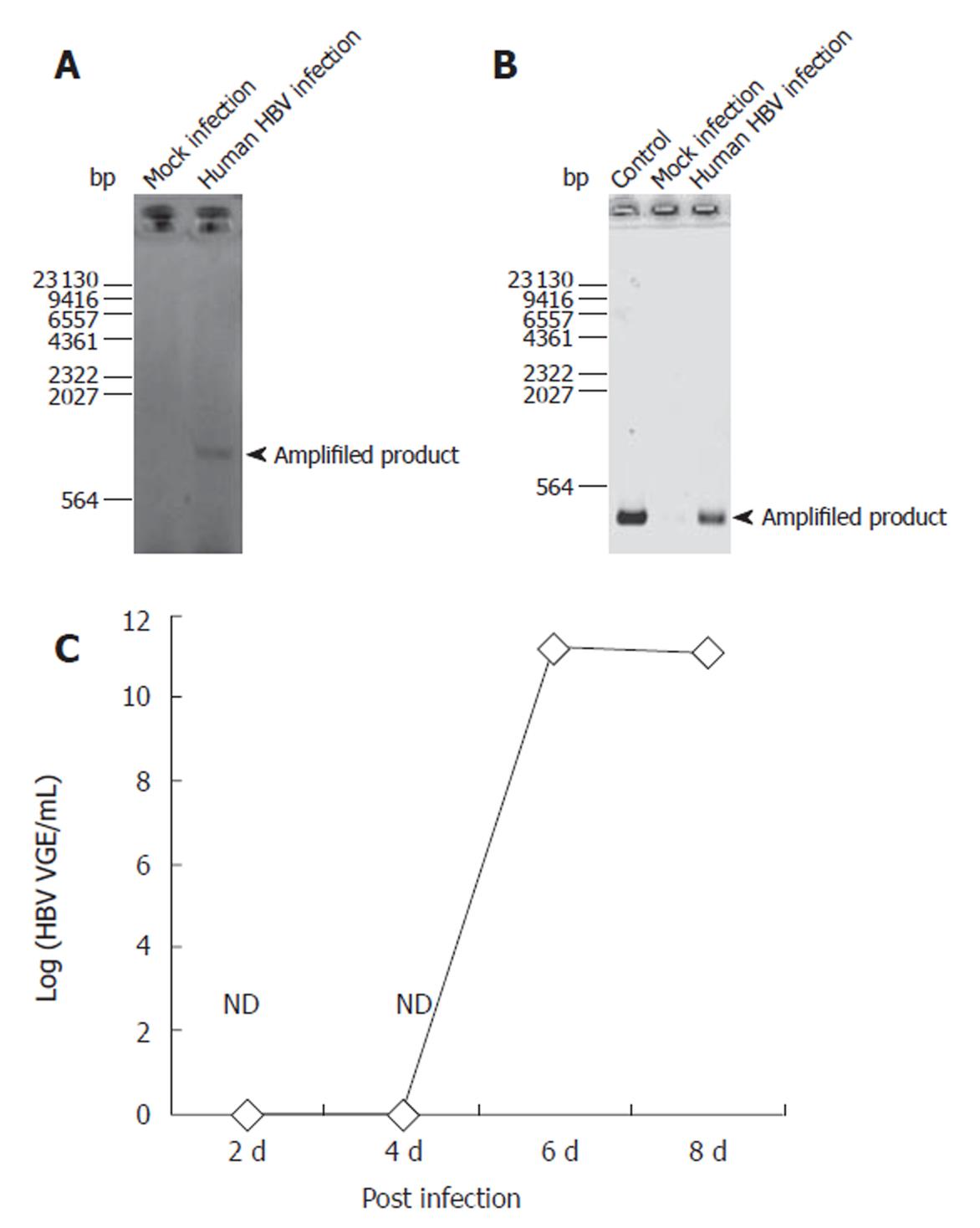
To confirm HBV infection to PNHHs, DNA was isolated during the RNA purification step with TRIzol reagent as instructed by the manufacturer. Infection was confirmed through PCR-based amplification specific only for cccDNA in nuclei of the infected cells. Figure 1 shows that the amplified product appeared in HBV- infected cells, indicating that HBV did infect PNHHs and that the nucleocapsid was transported into nuclei of the infected hepatocytes (Figure 1A). RT-PCR analysis of the HBV transcripts amplified with primers specific for epsilon and polymerase regions as described in Materials and Methods confirmed the presence of HBV RNA transcripts in the infected cells (Figure 1B). Real time PCR showed that HBV was not detected until 6 d after infection (Figure 1C).
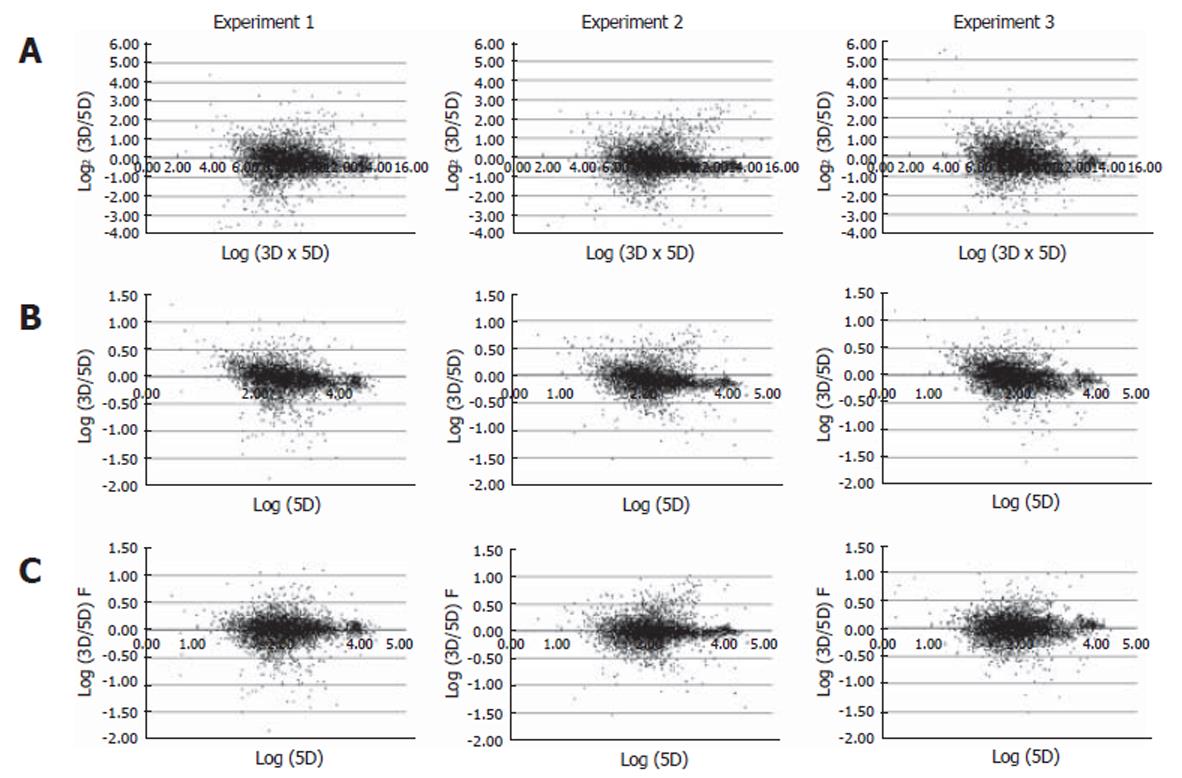
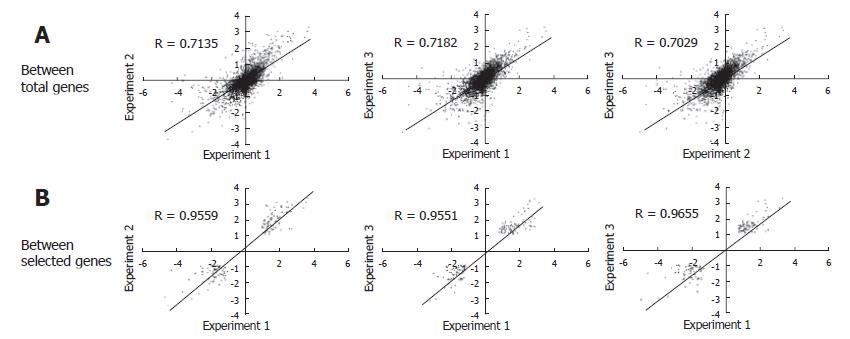
The experiments were carried out in triplicate at the infection step for more certain identification of genes differentially expressed by HBV infection. From each of the HBV infected cells for over 8 d, RNA was isolated and analyzed by microarray. As a result, three sets of cDNA array images were obtained. We analyzed the intensity of the raw image through scatterplot analyses. Figure 2A shows scatterplot analyses of log (Cy3 signal × Cy5 signal) vs log2 (Cy3 signal/Cy5 signal). This showed that each plot tended to divert from the general small curve (Figure 2A). But, each scatterplot analysis of Log (Cy3 signal/Cy5 signal) vs Log (Cy5 signal) showed a curve closer to the exponential decay (Figure 2B). Therefore, the data were fitted to an exponential decay curve for Cy3 per Cy5 channel correction (Figure 2C). Through these steps, we obtained a higher confidence ratio of the Cy3 signal compared to the Cy5 signal for each chip. With the ratios obtained, we analyzed the correlation coefficient between the data of the three chips. The correlation coefficient turned out to be more than 0.7 (Figure 3A), suggesting that the relationship between each chip was significant. The correlation coefficient for genes that were differentially expressed more than two folds was more than 0.95 (Figure 3B). Selected genes that were differentially expressed more than two folds, showed a high reproducibility among the triplicate microarray analyses.
Through a microarray analysis of PNHHs infected with HBV, we obtained the profile of 45 genes that were down regulated more than two folds compared to the control. The 45 down-regulated genes were analyzed classified by function (Table 1). Table 1 shows that many transcription factors related to RNA polymerase II, were down-regulated by HBV infection. In contrast, transcription factors such as C/EBP, which is used for transcription of HBV genes[19,20], were not differentially expressed. That is, the C/EBP expression level was changed less than two folds.
| Categoty | UniGene | Gene nam | Symbol | Locus | Function | Control/HBV infection | P-value |
| Transcription/ RNA Pol II | Hs.442675 | Thyroid hormone receptor interactor 8 | TRIP8 | 10 | Transcription co-activator of Pol II promoter | 3.469 | 0.030 |
| Transcription | Hs.57475 | Sex comb on midleg homolog 1 | SCMH1 | 1p34 | Pol II transcription | 3.315 | 0.019 |
| Transcription | Hs.119014 | Zinc finger protein 175 | ZNF175 | 19q13.4 | C2H2 zinc-finger protein 175 | 2.942 | 0.017 |
| Transcription/ RNA Pol II | Hs.437905 | Spi-B transcription factor (Spi-1/PU.1 related) | SPIB | 19q13.3-q13.4 | RNA polymerase II transcription factor | 2.743 | 0.017 |
| Transcription/ RNA Pol II | Hs.148427 | LIM homeobox protein 3 | LHX3 | 9q34.3 | RNA Pol 2 transcription factor and activate pituitary hormone genes | 2.492 | 0.006 |
| Signal | Hs.17154 | Dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 4 | DYRK4 | 12p13.32 | Dual-specificity protein kinase 4 | 3.938 | 0.005 |
| Signal | Hs.262886 | Inositol polyphosphate-5- phosphatase, 145kD | INPP5D | 2q36-q37 | Modulating cytokine signaling within the hemopoietic system | 3.587 | 0.009 |
| Signal | Hs.75249 | ADP-ribosylation factor-like 6 interacting protein | ARL6IP | 16p12-p11.2 | Activator of phospholipase D (PLD) | 2.801 | 0.010 |
| Tumor/Suppress | Hs.77793 | c-src tyrosine kinase | CSK | 15q23-q25 | Downregulate the tyrosine kinase activity of the c-src oncoprotein | 3.377 | 0.021 |
| Tumor/Induce | Hs.89839 | EphA1 | EPHA1 | 7q34 | Overexpression of EPH mRNA was found in a hepatoma | 3.039 | 0.022 |
| Tumor/Induce | Hs.79070 | V-myc avian myelocytomatosis viral oncogene homolog | MYC | 8q24.12-q24.13 | Promotes cell proliferation and transformation | 2.357 | 0.011 |
| Immune response | Hs.118354 | Human MHC Class I region proline rich protein mRNA | CAT56 | 6p21.32 | Immune response | 2.588 | 0.024 |
| Miscellaneous | Hs.180610 | Splicing factor proline/ glutamine rich | SFPQ | 1p34.3 | Pre-mRNA splicing factor required for pre-mRNA splicing | 10.471 | 0.006 |
| Miscellaneous/ Cytoskeleton | Hs.75064 | Tubulin-specific chaperone c | TBCC | 6pter-p12.1 | Cofactor in the folding pathway of beta-tubulin | 10.2 | 0.020 |
| Miscellaneous | Hs.438683 | BCM-like membrane protein precursor | SBB142 | 1q23.1 | BCM-like membrane protein precursor | 3.751 | 0.004 |
| Miscellaneous | Hs.8203 | Endomembrane protein emp70 precursor isolog | LOC56889 | 10q24.2 | Low similarity to human endosomal protein P76 | 3.502 | 0.038 |
| Miscellaneous | Hs.311609 | Nuclear RNA helicase, DECD variant of DEAD box family | DDXL | 19p13.13 | Member of the DEAD/H box ATP- dependent RNA helicase family | 2.868 | 0.026 |
| Miscellaneous/ Energy | Hs.150922 | BCS1 (yeast homolog)-like | BCS1L | 2q33 | Function in the assembly of complex III of the respiratory chain | 2.682 | 0.007 |
| Miscellaneous | Hs.6679 | hHDC for homolog of Drosophila headcase | LOC51696 | 6q23-q24 | hHDC for homolog of Drosophila headcase | 2.611 | 0.020 |
| Miscellaneous | Hs.5300 | Bladder cancer associated protein | BLCAP | 20q11.2-q12 | Appears to be down-regulated during bladder cancer progression | 2.459 | 0.028 |
| Miscellaneous | Hs.179526 | Upregulated by 1, 25-dihydroxyvitamin D-3 | VDUP1 | 1q21.2 | Upregulated by 1, 25-dihydroxyvitamin D-3 | 2.394 | 0.043 |
| Miscellaneous | Hs.440961 | Calpastatin | CAST | 5q15-q21 | Inhibitor of the cysteine (thiol) protease calpain | 2.276 | 0.000 |
| Miscellaneous | Hs.275775 | Selenoprotein P, plasma, 1 | SEPP1 | 5q31 | An oxidant defense in the extracellular space | 2.186 | 0.007 |
| EST | Hs.371233 | ESTs | Xp22.3 | Moderately similar to T08795 hypothetical protein DKFZp586J1822.1 | 7.826 | 0.025 | |
| EST | Hs.229338 | ESTs | X | 4.687 | 0.007 | ||
| EST | Hs.212957 | ESTs | 3q26.1 | Moderately similar to ZN91_HUMAN ZINC FINGER PROTEIN 91 | 4.611 | 0.019 | |
| EST | Hs.211823 | ESTs | 2q37.1 | 4.519 | 0.030 | ||
| EST | Hs.57836 | ESTs | 17 | 3.323 | 0.029 | ||
| EST | Hs.87912 | ESTs | 14q24.1 | 3.314 | 0.013 | ||
| EST | Hs.12429 | ESTs | FLJ22479 | 4q26-q27 | Hypothetical protein FLJ22479 | 3.217 | 0.041 |
| EST | Hs.213586 | ESTs | 7 | 2.759 | 0.044 | ||
| EST | Hs.2755711 | ESTs | 22 | Weakly similar to T20379 hypothetical protein | 2.723 | 0.019 | |
| EST | Hs.191435 | ESTs | 8p23.1-p22 | Weakly similar to S65657 alpha-1C- adrenergic receptor splice form 2 | 2.638 | 0.023 | |
| EST | Hs.31293 | ESTs | 9p13.1 | 2.286 | 0.035 | ||
| Predicted protein | Hs.414464 | Hypothetical protein | HSD3.1 | 14q31.3 | 7.314 | 0.008 | |
| Predicted protein | Hs.100914 | Hypothetical protein FLJ10352 | FLJ10352 | 18p11.21 | 6.239 | 0.006 | |
| Predicted protein | Hs.181112 | HSPC126 protein | HSPC126 | 13q14.12 | 3.322 | 0.045 | |
| Predicted protein | Hs.306711 | KIAA1081 protein | ELKS | 12p13.3 | 3.122 | 0.036 | |
| Predicted protein | Hs.101891 | KIAA1193 protein | KIAA1193 | 19p13.3 | Weakly similar to RPB1_HUMAN DNA-directed RNA Pol II largest subunit | 3.029 | 0.030 |
| Predicted protein | Hs.272759 | KIAA1457 protein | KIAA1457 | 12q24.31 | 2.98 | 0.020 | |
| Predicted protein | Hs.172089 | Homo sapiens mRNA; cDNA DKFZp586I2022 | 11q22.1 | 2.784 | 0.036 | ||
| Predicted protein | Hs.7049 | Hypothetical protein FLJ11305 | FLJ11305 | 13q34 | 2.65 | 0.028 | |
| Predicted protein | Hs.445255 | KIAA0368 protein | KIAA0368 | 9q32 | 2.423 | 0.016 | |
| Predicted protein | Hs.192190 | KIAA0782 protein | KIAA0782 | 11q13.3 | 2.332 | 0.009 | |
| Predicted protein | Hs.169910 | KIAA0173 gene product | KIAA0173 | 2p24.3-p24.1 | Similar to S72482 hypothetical protein | 2.171 | 0.014 |
From the analysis by cDNA microarray, 53 up-regulated genes were identified by an increase of more than two folds in their differential expression. Table 2 shows that growth- and tumor-related molecules comprised a proportion of the up-regulated genes. The positive effector genes for tumor and proliferation have found to be GDF11[21] and NOL1[22] and the negative effector genes EXTL3[23] and RAD50. The most interesting genes have found to be the TNF signaling pathway- related genes. LT-α is an inflammatory cytokine and induces the TNF signaling pathway as a ligand for TNF receptor (TNFR). LT-α binds to TNFR and recruits TRAF2. MAP3K14 (NF-κB inducing kinase, NIK) binds to TRAF2 and activates NF-κB[24]. LT-α, TRAF2, and NIK were also up-regulated in the experiment (Table 2). This means that HBV activates NF-κB through up-regulation of LT-α, TRAF2, and NIK.
| Categoty | UniGene | Gene name | Symbol | Locus | Function | Control/HBVinfection | P-value |
| Signal | Hs.82887 | Protein phosphatase 1, regulatory (inhibitor) subunit 11 | PPP1R11 | 6p21.3 | Soluble protein phosphatase inhibitor(reppresor) | 3.623 | 0.026 |
| Signal | Hs.437575 | TNF receptor-associated factor 2 | TRAF2 | 9q34 | Required for activation of NFkappaB | 3.23 | 0.026 |
| Signal | Hs.6527 | G protein-coupled receptor 56 | GPR56 | 16q13 | Member of the G protein-coupled receptor family | 2.817 | 0.009 |
| Signal | Hs.29203 | Homo sapiens G protein beta subunit mRNA, partial cds | GBL | 16p13.3 | G protein-linked receptor protein for signalling pathway | 2.76 | 0.009 |
| Signal/ Cytoskeleton | Hs.2157 | Wiskott-Aldrich syndrome | WAS | Xp11.4-p11.21 | Involved in transduction of signals from receptors on the cell surface to the actin cytoskeleton | 2.446 | 0.018 |
| Signal | Hs.440315 | Mitogen-activated protein kinase kinase kinase 14 | MAP3K14 | 17q21 | Binds to TRAF2 and stimulates NF-kappaB activity | 2.185 | 0.043 |
| Tumor/ Induce | Hs.15243 | Nucleolar protein 1 (120kD) | NOL1 | 12p13.3 | Transforms NIH3T3 cells when overexpressed | 5.273 | 0.004 |
| Tumor/ Supress | Hs.9018 | Exostoses (multiple)-like 3 | EXTL3 | 8p21 | Tumor suppressor, glycosyltransferase activity | 5.07 | 0.093 |
| Tumor/ Supress | Hs.41587 | RAD50 (S. cerevisiae) homolog | RAD50 | 5q31 | Associates with MRE11, nibrin (NBS1) and the tumor suppressor BRAC1 | 2.443 | 0.013 |
| Growth/ Positive | Hs.511740 | Growth differentiation factor 11 | GDF11 | 12q13.13 | Regulators of cell growth and differentiation in both embryonic and adult tissues | 2.969 | 0.023 |
| Cell cycle/ Negative | Hs.76364 | Allograft inflammatory factor 1 | AIF1 | 6p21.3 | Involved in negative regulation of growth of vascular smooth muscle cells | 3.573 | 0.031 |
| Cell cycle/ Positive | Hs.25313 | Microspherule protein 1 | MCRS1 | 12q13.12 | Involved in cell-cycle-dependent stabilization of ICP22 in HSV1- infected cells | 3.273 | 0.044 |
| Cell cycle/ Positive | Hs.371833 | Nuclear receptor binding factor-2 | NRBF-2 | 10 | A possible gene activator protein interacting with nuclear hormone receptors | 2.568 | 0.018 |
| Cell cycle/ Positive | Hs.440606 | Centrosomal protein 2 | CEP2 | 20q11.22-q12 | Regulate centriole-centriole cohesion during the cell cycle | 2.422 | 0.022 |
| Enzyme/ Glycosylation | Hs.4814 | Mannosidase, alpha, class 1B, member 1 | MAN1B1 | 9q34 | N-linked glycosylation | 3.097 | 0.008 |
| Enzyme/ lysophospholipase | Hs.889 | Charot-Leyden crystal protein | CLC | 19q13.1 | Phospholipid metabolism and anti-pathogen | 3.065 | 0.035 |
| Enzyme/Protease | Hs.75890 | Site-1 protease | MBTPS1 | 16q24 | A sterol-regulated subtilisin-like serine protease | 2.873 | 0.023 |
| Immune response | Hs.2014 | T cell receptor delta locus | TRD@ | 14q11.2 | T-cell antigen receptor, delta polypeptide | 3.69 | 0.023 |
| Immune response | Hs.465511 | Granzyme M | GZMM | 19p13.3 | Serine protease for anti-pathogen response | 3.201 | 0.023 |
| Transcription | Hs.436871 | Zinc finger protein 173 | ZNF173 | 6p21.3 | DNA/protein binding, transcriptional protein | 3.411 | 0.002 |
| Transcription | Hs.108139 | Zinc finger protein 212 | ZNF212 | 7q36.1 | DNA/protein binding, transcriptional protein | 2.341 | 0.045 |
| Apoptosis | Hs.36 | Lymphotoxin alpha | LTA | 6p21.3 | A member of the tumor necrosis factor family | 14.912 | 0.019 |
| Miscellaneous | Hs.434384 | Titin | TTN | 2q31 | Large myofilament protein | 4.087 | 0.012 |
| Miscellaneous | Hs.58927 | Nuclear VCP-like | NVL | 1q41-q42.2 | Member of the AAA family of ATPases | 4.005 | 0.035 |
| Miscellaneous | Hs.122552 | G-2 and S-phase expressed 1 | GTSE1 | 22q13.2-q13.3 | Accumulates in late S/G2 phase, is phosphorylated in mitosis, and disappears in G1 phase | 3.491 | 0.007 |
| Miscellaneous/Glycosylation | Hs.82921 | Solute carrier family 35 (CMP- sialic acid transporter), member 1 | SLC35A1 | 6q15 | Important for normal sialylation of glycoproteins and glycolipids | 3.352 | 0.013 |
| Miscellaneous | Hs.410455 | Unc119 (C.elegans) homolog | UNC119 | 17q11.2 | May function in photoreceptor neurotransmission | 3.328 | 0.030 |
| Miscellaneous | Hs.55041 | CGI-22 protein | MRPL2 | 6p21.3 | Unknown | 3.052 | 0.01 |
| Miscellaneous/Cytoskeleton | Hs.74088 | Bridging integrator-3 | BIN3 | 8q21.2 | Related to actin assembly- competent state | 2.677 | 0.018 |
| Miscellaneous | Hs.25237 | Mesenchymal stem cell protein DSCD75 | LOC51337 | 8q24.3 | Moderately similar to uncharacterized Drosophila CG4666 | 2.615 | 0.042 |
| EST | Hs.95867 | Homo sapiens EST00098 gene, last exon | EST00098 | 9q34.1 | 8.781 | 0.027 | |
| EST | Hs.98785 | ESTs | KSP37 | 4p16 | 3.749 | 0.011 | |
| EST | Hs.136912 | ESTs | MGC10796 | 3q13.13 | 3.435 | 0.003 | |
| EST | Hs.101774 | ESTs | FLJ23045 | 20p11.23 | 3.387 | 0.032 | |
| EST | Hs.420262 | ESTs | 13 | 3.355 | 0.022 | ||
| EST | Hs.124840 | ESTs | 11q13.1 | 3.114 | 0.021 | ||
| EST | Hs.272299 | ESTs | RP4-622L5 | 1p36.11-p34.2 | 3.008 | 0.034 | |
| EST | Hs.415048 | ESTs | 5 | 2.891 | 0.025 | ||
| EST | Hs.531268 | ESTs | 16 | 2.889 | 0.013 | ||
| EST | Hs.273830 | ESTs | FLJ12742 | 1 | 2.739 | 0.020 | |
| EST | Hs.190162 | ESTs | 1p32.3 | 2.708 | 0.021 | ||
| EST | Hs.303172 | ESTs | 18 | 2.578 | 0.04 | ||
| EST | Hs.59203 | ESTs | 7 | 2.437 | 0.022 | ||
| EST | Hs.231444 | ESTs | 1 | 2.343 | 0.005 | ||
| Unknown sequence | Hs.284265 | Homo sapiens pRGR1 mRNA, partial cds | 6q27 | 2.966 | 0.043 | ||
| Unknown sequence | Hs.291385 | Homo sapiens clone 23664 and 23905 mRNA sequence | 4p14-p12 | 2.439 | 0.041 | ||
| Predicted protein | Hs.31718 | Homo sapiens cDNA FLJ11034 fis, clone PLACE1004258 | VRL | 17.359 | 0.000 | ||
| Predicted protein | Hs.61960 | Hypothetical protein | FLJ20040 | 16p13.3 | 9.166 | 0.002 | |
| Predicted protein | Hs.274552 | Homo sapiens cDNA FLJ10720 fis, clone NT2RP3001116 | FLJ10720 | 5 | 4.751 | 0.015 | |
| Predicted protein | Hs.279761 | HSPC134 protein | HSPC134 | 14q11.2 | 3.501 | 0.038 | |
| Predicted protein | Hs.283716 | Hypothetical protein PRO1584 | PRO1584 | 8p21.2 | 3.493 | 0.033 | |
| Predicted protein | Hs.464526 | Homo sapiens clone 23649 and 23755 unknown mRNA, partial cds | 18q11.2 | 3.198 | 0.032 | ||
| Predicted protein | Hs.274412 | Homo sapiens cDNA FLJ10207 fis, clone HEMBA1005475 | UPF3A | 17p11.2 | 3.076 | 0.012 |
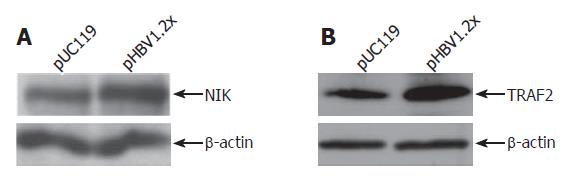
According to the cDNA microarray data, three genes related to the TNF signaling pathway, LT-α, TRAF2, and NIK, were up-regulated. Upregulation of these genes was confirmed by RT-PCR. For RT-PCR analysis, primer sets specific to LT-α, TRAF2, and NIK, were used and experiments were performed in hepatoma-derived cell lines, including HepG2, Huh7, and Chang liver cells. As a result, the mRNA levels of these three genes in each cell line were increased by pHBV1.2× transfection (Figure 4). In addition to RT-PCR, the expression of NIK and TRAF2 was confirmed at the protein level (Figure 5). The expression of LT-α, was confirmed by immunofluorescence staining analysis (data not shown).
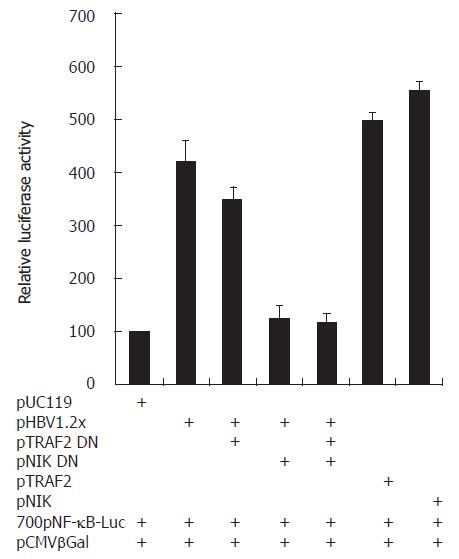
According to cDNA microarray and RT-PCR analysis, mRNA expression of LT-α, TRAF2, and NIK was up-regulated by HBV. Since their expression was related to NF-κB activation. HBV-mediated NF-κB activation might be involved in the up regulation of these genes. To determine whether these genes actually are involved in HBV-mediated NF-κB activation, we performed a luciferase assay with a pNF-κB-luciferase vector as a reporter plasmid. To elucidate whether HBV-mediated NF-κB activation is dependent on TRAF2 and NIK of three genes, we cotransfected pTRAF2 DN or pNIK DN, the dominant negative form of pTRAF2 or pNIK, with pNF-κB-luciferase, pCMV β-galactosidase, and pHBV1.2× (Figure 6). The experiment for LT-α was performed with anti- LT-α, to neutralize LT-α. The pHBV1.2× produced about a 4.2 folds greater increase in NF-κB luciferase activity than pUC119. However, pHBV1.2× cotransfection with TRAF2 DN or NIK DN produced about a 3.5 or 1.2 fold relative increase (Figure 6) and treatment with anti- LT-α decreased the ratio to less than 4.2 folds (data not shown). These findings led us to think that HBV could activate NF-κB and that this HBV-mediated NF-κB activation might require LT-α, TRAF2, and NIK. DC.
In this report, we focused on the interaction between HBV and hepatocytes during the initial stage of infection. To mimic hepatocyte infection with HBV under in vivo conditions, we isolated PNHHs and infected them with HBV. We chose this method because cultured cell lines such as HepG2 are seldom infected with HBV[25,26], and transformed cultured cells have many physiological properties that are altered in the original state of hepatocytes[27,28]. In this experiment, the same hepatocytes were used as a control. Since they are produced under identical conditions, a pair of samples of the same genetic background could be obtained. With these samples, we were able to analyze differentially expressed genes. As a result, we obtained gene expression profiles and 98 consistently differentially expressed genes were identified by gene expression profiles. Of these genes, 53 were up-regulated and 45 down-regulated. It was reported that there are no genes uniformly correlated with HBV DNA profile during the initial host response to HBV infection[29]. However, because this study was performed on chimpanzees, there are some considerations in making a comparision between this study with our report. Our report analyzed the effect of HBV on PNHHs at cellular level without any other cell types, including immunocytes. So the influence of immunocytes was not included in this analysis. In addition, the difference in human beings and chimpanzees needs to be taken into consideration.
The results of our study showed that a proportion of the down-regulated genes was transcription factor-related genes and a proportion of the up-regulated genes was TNF signaling pathway-related genes. Down regulation of transcription factors may be helpful for the transcription of the HBV gene because the transcripts of the host cell can be repressed and the transcriptional machinery can be efficiently used for viral transcription. C/EBP, which is involved in viral genome transcription[19,20], had no substantial differential expression in this experiment. In addition to down regulation of transcription factor for virus transcription, up regulation of cell proliferation-related genes may help viral replication. Of the up-regulated genes, LT-α, TRAF2, and NIK may induce cell proliferation via NF-κB activation.
In fact, LT-α is mainly related to the signal cascade for apoptosis and generally involves the host defense system[30]. However LT-α is also related to cell proliferation. Usually, TNF signaling including LT-α, can induce apoptosis and proliferation[31]. TNF signaling by LT-α has a signal cascade from TNFR to TRADD. In the case of apoptosis, TRADD-FADD interaction is needed to activate caspase 8[31]. In the case of proliferation, TRADD-TRAF2 interaction induces activation of NF-κB, a proliferation-inducing transcription factor[31]. After TRAF2 binds to TRADD, NIK binds to TRAF2 and activates NF-κB through IKK activation and IκB-α degradation[24,32-34]. In cDNA microarray data, among genes related to the two opposite effects initiated by LT-α, proliferation-related genes are up-regulated. FADD is not differentially altered by more than two folds. Therefore, HBV infection may strengthen the TNF signaling pathway to cell proliferation through the induction of gene expression.
In conclusion, HBV induces NF-κB activation by upregulating LT-α, TRAF2, and NIK, and cell proliferation by activating NF-κB.
The authors thank Dr. SY Lee for providing pTRAF2 and pTRAF2 DN vector.
S- Editor Wang J L- Editor Wang XL E- Editor Ma WH
| 1. | Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129-1133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1838] [Cited by in F6Publishing: 1721] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 2. | Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 715] [Cited by in F6Publishing: 723] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 3. | Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651-693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 805] [Cited by in F6Publishing: 873] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 4. | Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 184] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 5. | Mason WS, Halpern MS, England JM, Seal G, Egan J, Coates L, Aldrich C, Summers J. Experimental transmission of duck hepatitis B virus. Virology. 1983;131:375-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 134] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 127] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 7. | Tagawa M, Omata M, Okuda K. Appearance of viral RNA transcripts in the early stage of duck hepatitis B virus infection. Virology. 1986;152:477-482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Honda M, Kaneko S, Kawai H, Shirota Y, Kobayashi K. Differential gene expression between chronic hepatitis B and C hepatic lesion. Gastroenterology. 2001;120:955-966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 141] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 9. | Delpuech O, Trabut JB, Carnot F, Feuillard J, Brechot C, Kremsdorf D. Identification, using cDNA macroarray analysis, of distinct gene expression profiles associated with pathological and virological features of hepatocellular carcinoma. Oncogene. 2002;21:2926-2937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 72] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Hashimoto K, Tangoku A, Hamada K. Differential gene expression in distinct virologic types of hepatocellular carcinoma: association with liver cirrhosis. Oncogene. 2003;22:3007-3014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol. 2004;10:509-513. [PubMed] [Cited in This Article: ] |
| 12. | Park SG, Lee SM, Jung G. Antisense oligodeoxynucleotides targeted against molecular chaperonin Hsp60 block human hepatitis B virus replication. J Biol Chem. 2003;278:39851-39857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158-6169. [PubMed] [Cited in This Article: ] |
| 14. | Song HY, Régnier CH, Kirschning CJ, Goeddel DV, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-kappaB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci USA. 1997;94:9792-9796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 470] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 15. | Chung YM, Park KJ, Choi SY, Hwang SB, Lee SY. Hepatitis C virus core protein potentiates TNF-alpha-induced NF-kappaB activation through TRAF2-IKKbeta-dependent pathway. Biochem Biophys Res Commun. 2001;284:15-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Schulze-Bergkamen H, Untergasser A, Dax A, Vogel H, Büchler P, Klar E, Lehnert T, Friess H, Büchler MW, Kirschfink M. Primary human hepatocytes--a valuable tool for investigation of apoptosis and hepatitis B virus infection. J Hepatol. 2003;38:736-744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Baccarani U, Sanna A, Cariani A, Sainz-Barriga M, Adani GL, Zambito AM, Piccolo G, Risaliti A, Nanni-Costa A, Ridolfi L. Isolation of human hepatocytes from livers rejected for liver transplantation on a national basis: results of a 2-year experience. Liver Transpl. 2003;9:506-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Guguen-Guillouzo C, Campion JP, Brissot P, Glaise D, Launois B, Bourel M, Guillouzo A. High yield preparation of isolated human adult hepatocytes by enzymatic perfusion of the liver. Cell Biol Int Rep. 1982;6:625-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 160] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Choi BH, Park GT, Rho HM. Interaction of hepatitis B viral X protein and CCAAT/ enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J Biol Chem. 1999;274:2858-2865. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 78] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Ott M, Thyagarajan SP, Gupta S. Phyllanthus amarus suppresses hepatitis B virus by interrupting interactions between HBV enhancer I and cellular transcription factors. Eur J Clin Invest. 1997;27:908-915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Nakashima M, Toyono T, Akamine A, Joyner A. Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech Dev. 1999;80:185-189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 187] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Fonagy A, Swiderski C, Wilson A, Bolton W, Kenyon N, Freeman JW. Cell cycle regulated expression of nucleolar antigen P120 in normal and transformed human fibroblasts. J Cell Physiol. 1993;154:16-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Van Hul W, Wuyts W, Hendrickx J, Speleman F, Wauters J, De Boulle K, Van Roy N, Bossuyt P, Willems PJ. Identification of a third EXT-like gene (EXTL3) belonging to the EXT gene family. Genomics. 1998;47:230-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 102] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Wang Q, Dziarski R, Kirschning CJ, Muzio M, Gupta D. Micrococci and peptidoglycan activate TLR2-->MyD88-->IRAK-->TRAF-->NIK-->IKK-->NF-kappaB signal transduction pathway that induces transcription of interleukin-8. Infect Immun. 2001;69:2270-2276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 140] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 25. | Galle PR, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology. 1994;106:664-673. [PubMed] [Cited in This Article: ] |
| 26. | Gripon P, Diot C, Thézé N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol. 1988;62:4136-4143. [PubMed] [Cited in This Article: ] |
| 27. | Otsuka M, Aizaki H, Kato N, Suzuki T, Miyamura T, Omata M, Seki N. Differential cellular gene expression induced by hepatitis B and C viruses. Biochem Biophys Res Commun. 2003;300:443-447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129-2137. [PubMed] [Cited in This Article: ] |
| 29. | Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669-6674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 520] [Cited by in F6Publishing: 514] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 30. | VanArsdale TL, VanArsdale SL, Force WR, Walter BN, Mosialos G, Kieff E, Reed JC, Ware CF. Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor kappaB. Proc Natl Acad Sci USA. 1997;94:2460-2465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 145] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1522] [Cited by in F6Publishing: 1496] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 32. | Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. 1997;385:540-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1057] [Cited by in F6Publishing: 1052] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 33. | Wajant H, Scheurich P. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int J Biochem Cell Biol. 2001;33:19-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 134] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276:36530-36534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 244] [Article Influence: 10.6] [Reference Citation Analysis (0)] |