Published online Nov 15, 2004. doi: 10.3748/wjg.v10.i22.3299
Revised: February 4, 2004
Accepted: February 10, 2004
Published online: November 15, 2004
AIM: To investigate coxsackievirus B3 infection and its gene mutation in Keshan disease.
METHODS: The expression of Coxsackievirus B3 RNA was detected in autopsy specimens of acute (12 cases), sub-acute (27 cases) and chronic (15 cases) Keshan disease by in situ hybridization. In sub-acute Keshan disease specimens, 3 cases with positive result by in situ hybridization were selected RT-PCR analysis. The DNA segments were then sequenced.
RESULTS: Coxsackievirus B3 RNA was detected in the cytoplasm of myocardiocyte. The positive rate was 83% in acute, 67% in sub-acute and 80% in chronic Keshan disease. In the conservative region of Coxsackievirus B3 genome, there was a mutation in 234 (C-T) compared to the non-cardiovirulent strain, CVB3/0.
CONCLUSION: Coxsackievirus B3 RNA can survive and replicate in heart muscle of Keshan disease, which may play an important role in the occurrence of Keshan disease. The possible mechanism of occurrence of Keshan disease is associated with point a mutation in Coxsackievirus B3 genome.
- Citation: Ren LQ, Li XJ, Li GS, Zhao ZT, Sun B, Sun F. Coxsackievirus B3 infection and its mutation in Keshan disease. World J Gastroenterol 2004; 10(22): 3299-3302
- URL: https://www.wjgnet.com/1007-9327/full/v10/i22/3299.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i22.3299
Enteroviruses, particularly Coxsackie B viruses (CVB), are the most common cause of human viral myocarditis and are associated with dilated cardiomyopathy (DCM)[1-3]. In general, neonates and children suffer more severe clinical syndromes due to CVB infection, often severe or life threatening, because of inflammation and necrosis of heart muscle[4]. Many diseases are considered to have relationship with CVB3 infect, but the mechanism is still not well understood. Although the immune response of the host undoubtedly plays an important role in the pathogenesis of viral heart disease[5], direct viral cytotoxicity has been found to be crucial for organ pathology both during acute and persistent heart muscle infection. Coxsackievirus B3 (CVB3), one of six CVB serotypes, is a member of the genus enterovirus within the family Picornaviridae and its structure is well understood[6,7]. Some studies suggest that the 5'NTR of enteroviruses maybe play an important role in producing or sustaining the myocardial virulence[8-14].
Keshan disease is endemic exclusively in selenium-deficient rural areas of China, including 14 provinces and autonomous regions[15]. Its clinical features are low body selenium content and acute or chronic episodes of heart disorder characterized by cardiogenic shock, arrhythmia, ECG changes (for example, right branch block), and/or congestive heart failure, with an enlarged heart. Four types of the disease are seen: acute, subacute, chronic, and latent or compensated. The basic pathological change is multifocal myocardial necrosis and fibrous replacement throughout the myocardium, with various degrees of cellular infiltration and calcification, depending upon the type of disease[16,17]. Research suggests that not a single but several more factors may have a relationship with the occurrence of Keshan disease. The reasons are often considered as global, biological chemical factors (selenium deficiency), dietary nutritional factors (vitamin E deficiency) and infection (virus, especially enteroviruses). This hypothesis can explain both endemic and seasonal features of Keshan disease. Recent studies have also demonstrated a possible etiological role of enterovirus infection in a particular form of heart muscle disease, selenium deficiency-related endemic cardiomyopathy (Keshan disease), seen in China[18,19]. Presently, patients of Keshan disease are rare and there is no ideal animal model, research of viral etiology of Keshan disease is relatively few. In this study, we detected all the four types of Keshan disease using in situ hybridization and sequenced the 5’NTR of CVB3 from subacute Keshan disease in order to investigate the relationship between Keshan disease and CVB3 infection.
All the samples were cardiac tissue isolated at post mortem and fixed in formaldehyde and embedded in paraffin wax and preserved in Institute of Keshan Disease, Jilin University from 1959-1985. The samples included 12 acute patients (7 males, 5 females, average age 16.3 years), 27 subacute patients (12 males, 15 females, average age 4.3 years) and 15 chronic patients (9 males, 6 females, average age 23.8 years). We used three non-Keshan disease patients as controls, including two normal and one perinatal period cardiomyopathy. The cultured human amniotic cell containing a cardiovirulent strain of CVB3 served as positive control and by RT-PCR.
An obligonucleotide probe was purchased from Shanghai Sangon Biological Engineering Company. This probe, labeled with biotin, was designed to detect coxsackievirus B3, whose sequence was 5’-GTCCGCAATGGCGGGCGGTGGTGGTGTCTC-3’.
All slides were treated by APES solution (1:50 acetone) in order to prevent the section slipping from the slide. The 5 μm thick section was baked 3-4 h at 80 °C and overnight at 60 °C.
The paraffin wax was removed by immersing the slide in xylene. The slides were then rehydrated, treated with 0.1 g/L Triton X-100 for 2 min, digested with proteinase K (0.25 mg/mL) in PBS for 5 min at room temperature, then sealed with histidine (2 mg/mL) for 2 min, and then dehydrated in graded concentrated ethyl alcohol. The tissue sections were denatured on heating block for 2 min at 80 °C and then cooled on ice immediately. They were pre-hybridized for 3-4 h at 42 °C by covering them with pre-hybridization liquid (6 × SSC, 5 × Denhardt, 5 g/L SDS, 100 μg/mL herring sperm DNA, 500 g/L deionized formamide) in a wet chamber. After removal of the pre-hybridization liquid, the sections were allowed to hybridize overnight under the same conditions as described above.
Following the hybridization, the slides were rinsed twice with 2 × SSC containing 500 g/L deionized formamide, twice with 2 × SSC, treated with 1 g/L Triton X-100 for 2 min and washed twice with buffer I (1 mol/L NaCl, 0.1 mol/L Tris, pH7.5). Buffer II (3 g bovine serum albumin dissolved in buffer I) was added to each section and incubated for 1 h. After pouring off buffer II, the SA-AP (streptavidin-alkaline phosphatase conjugation) was added onto the tissue sections, and allowed to react in this mixture for 20 min, rinsed with buffer I and then with buffer III (0.1 mol/L NaCl, 0.1 mol/L Tris-Cl, 50 mmol/L MgCl2, pH9.5). Finally, freshly prepared developer solution (BCIP/NBT) was applied to the slides and incubated for 30 min to 4 h. A brownish blue color reaction was produced. Following routine dehydration, the sections were cleared and mounted.
To assess the specificity of the hybridization signal, the culture cell infected by CVB3 were alternatively hybridized with the labeled obligonucleotide probe encoding CVB3 before and after digestion by Rnase, and hybridization solution without containing the labeled obligonucleotide probe encoding CVB3 was also used as negative control.
Total RNA was extracted from paraffin-embedded heart tissue of patients with Keshan disease according to the following procedures. Several section slices (5-10 μm) were put into a 1.5 mL Eppendorf tube in which approximately 1 mL xylene was added in order to exclude paraffin. The xylene was changed every 2 h and the last time it was kept overnight at room temperature. After being centrifuged at 5000 r/min for 5 min, the tissue was washed with non-water ethanol and then baked at 55 °C. The total RNA from the dried tissue was then extracted using Trizol regent (GIBCO). Positive controls were the cultured human amniotic cells containing CVB3. The RNA precipitate was dissolved in 10 μL of diethyl pyrocarbonate-treated H2O.
The enterovirus specific primers used for RT-PCR and sequencing of the 5'NTR were purchased from Sigma Genosys Australia Pty. Ltd. The sequence of the primers was 3P: 5’-TCAATTGTCACCATAAGCAGCAGCCA-3’ and 5P: 5’-CGGTACCTTTGTGCCCCTGTTTT-3’.
RT was carried out following the manufacturer’s instructions. A 2 μL of specific primer 3P and 1 μL of 10 mmol/L dNTPs were mixed with 8 μL of RNA template. The mixture was immediately put on ice after incubated at 65 °C for 5 min. Then the mixture of 4 μL of 5 × first strand buffer, 1 μL of Rnase inhibitor, 1 μL Superscript II Rnase H-free reverse transcriptase and 3 μL of diethyl pyrocarbonate-treated H2O was added to the reaction tube, followed by incubation at 42 °C for 5 min, and at 70 °C for 15 min in order to inactivate the reverse transcriptase.
PCR was carried out with a 25 μL mixture containing 10 pmoL deoxynucleotide triphosphates, 15 pmoL forward primer (5P), 15 pmoL reverse primer (3P), 1 × reaction buffer (Dingguo Biocompany), and 3 μL of the RT reaction product. The PCR reaction mixture was pre-denatured at 95 °C for 2 min, and then amplified through 30 cycles, each cycle consisting of denaturation at 95 °C for 45 s, annealing at 55 °C for 45 s, and extension at 72 °C for 45 s. Finally, an additional extension was carried out at 72 °C for 10 min. A negative control containing 22 μL of the reaction mixture plus 3 μL of autoclaved H2O and a positive control containing 22 μL of the reaction mixture plus 3 μL of the RT reaction product from cells infected with the virulent laboratory strain of CVB3 were included in each set of PCR.
The amplification products of PCR were analyzed on 17 g/L agarose gel by electrophoresis and stained with ethidium bromide. To assess the specificity of RT-PCR, the extracted total RNA were digested by Rnase then used for PCR as template. The distilled water was also used as template in PCR reaction as negative control.
The sequencing reaction cycle was carried out using 4 μL of DNA polymerase FS-terminator mix (PE-ABI), 3.2 mmol/L primer, and 5 ng of PCR product per 100 bp length of template DNA with the volume made up to 10 μL with H2O. Reactions were run for 25 cycles as follows: 96 °C for 30 s, 50 °C for 15 s, and 60 °C for 4 min. DNA was precipitated, dissolved in loading buffer (167 g/L formamide, 733 g/L dextran), and separated on 41 g/L denaturing polyacrylamide gel on an ABI model 377 DNA sequencer. This procedure was performed by Shanghai Sangon Biological Engineering Company.
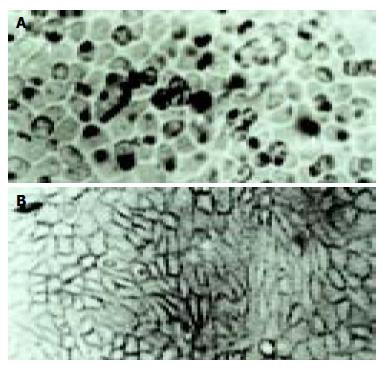
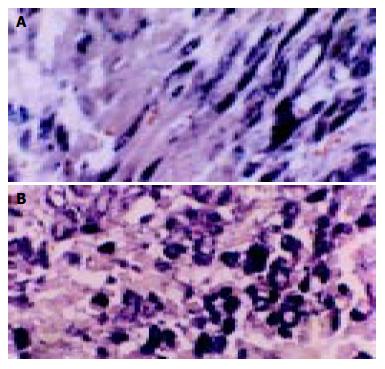
According to the feature of positive signal of the culture cell, the positive hybridization signals of the Coxsackievirus RNA appeared as deep brownish blue mass or granules (Figure 1). This criterion was applied to the tissue sample. In longitudinal sections, they were seen to adhere to myofibrils in one or several strings of beads (Figure 2A). They varied in size and, in cross section, were usually localized in the sarcoplasm in proximity to myocyte nuclei, but no signals were found in the nuclei themselves. The color of signal was the darkest in the myocardium of papillary muscles, trabeculae carneae and subendocardial muscle. It was lighter and less intense in the cells of the outer layers than those in the middle layers of ventricular walls. The myocardial cells surviving in fresh lesions and surrounding old necrotic foci often showed the hybridization signal. No signal was found in necrotic foci, and there were slight differences in the distribution and manifestation of positive hybrid signals in several types of Keshan disease. In most acute and subacute cases, the signal appeared predominantly in the myocyte surrounding foci and surviving in fresh foci as well as cells beneath the endocardium and of papillary muscles. The hybridization-positive granules were thick, and had clear border. The sarcoplasm appeared dark, and sometimes, homogeneously blue (Figure 2B). In most of the chronic cases, the granules were usually distributed in all parts of the myocardium, and were small and without clear borders, like dirt. The detection rate in subacute specimens was the highest, and the rate in acute specimens was higher than that in chronic specimens, but there were no significant differences among the three types of Keshan disease.
The positive rate of hybridization in the three type of Keshan disease was 83% (10/12) in acute, 67% (18/27) in sub-acute and 80% (12/15) in chronic Keshan disease. No positive signals were found in any negative controls.
Results of 3 myocardial control specimens investigated with in situ hybridization were negative.
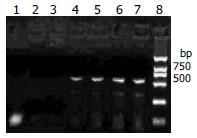
RT-PCR revealed that the RNA extracted from the myocardial tissue with positive control of in situ hybridization amplified a DNA fragment about 541 bp, while the three negative controls did not amplify (Figure 3). The RNA extracted from cultured cell infected by CVB3 were also amplified the DNA fragment about 541 bp.
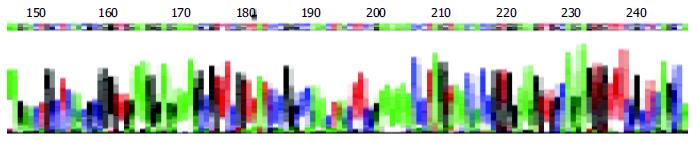
The DNA segments from RT-PCR were sequenced and compared to the non-cardiovirulent control, CVB3/0, and the cardiovirulent strain, CVB3/Nancy and CVB3/20. We found that the sequence of the amplified segment had one nucleotide different compared to that of CVB3/0. The 234th nucleotide in the genome of CVB3/0 was C while ours was T. But the sequence of our segment was similar that of CVB3/Nancy, CVB3/20 and the positive control (Figure 4).
Keshan disease has been studied for over 60 years in China. One of the key issues is the complex etiology of this endemic disease, with a focus on environmental nutritional factors and infectious agents. Selenium deficiency in the food chain has been recognized as a major but not exclusive environmental-nutritional factor, and increasing evidence supports an etiological role of enteroviruses in Keshan disease. Recently, Peng et al[20] determined the sequences of the 5’NTR and the 3’ end of the VP1 coding region from six enteroviruses isolate from cases of Keshan disease or outbreak myocarditis. From their results, they drew a conclusion that the sequence data confirm that coxsackievirus group B serotypes are predominant in the region where Keshan disease is endemic, and may be the etiological factors in outbreaks of myocarditis, and VP1 genotyping of enteroviruses is accurate and reliable. Animal experiments indicate that isolates may differ in pathogenicity. Virus infection combined with selenium deficiency in environments and vitamin E deficiency in dietary can illustrate both endemic and seasonal features of Keshan disease. The hypothesis of virus infection was accepted once it appeared, but the methods used in virology limited the development of a virus infection hypothesis of Keshan disease. The rapid development in molecular biology brings vitality to virology.
In situ hybridization is a method to detect the nucleic acids in a tissue or cell using a labeled obligonucleotide chain as probe. This technique has already been widely used in biology and medicine because of its high specificity, sensitivity and location precision. Using a cloned cDNA probe obtained from CVB4 infected cells and labeled by biotin-11-dUTP, Easton et al[21] found that 46% (6/13) patients who had been diagnosed coxsackievirus myocarditis can be found enterovirus infection in their myocardial tissue. Archard et al[22] examined enteroviruses infection in human myocardial tissue using an enterovirus specific probe, and found that the rate of enterovirus infection was 45% (21/47) in myocarditis patients and 43% (35/82) in cured myocarditis and DMC patients. In our research, the specific obligonucleotide probe of CVB3 was labeled by biotin on 5’ end, and the specimen was paraffin-embedded myocardial tissue from autopsy patients of Keshan disease. The object was to detect the coxsackievirus B3 RNA by in situ hybridization. Among the three types of Keshan disease patients, we found the positive rate was 83%, 67%, and 80% in acute, sub-acute and chronic Keshan disease patients, respectively. Our results provide a new support to the hypothesis that enterovirus, especially CVB3, might play an important role in pathogenesis of Keshan disease. Moreover, the area, range and the distribution of the positive signal of CVB3 were associated with the development of Keshan disease. The positive signals in acute and sub-acute Keshan disease were located in the surviving myocardiocyte in or around the necrosis foci while they were dispersed in almost all the myocardiocyte in chronic Keshan disease. The results of in situ hybridization suggested that enterovirus could lead to heart muscle lesion when they involved in the myocardial tissue, and they can dispersed to the normal heart muscle with increase of time.
Though there was a high enterovirus infection rate in myocardial tissue of Keshan disease patients[23,24], it is well known that the enterovirus could also be detected in many patients who had no heart muscle lesion. The change of viral cardiovirulence can undoubtedly play an important role. Some research has demonstrated that under some conditions, such as selenium deficiency, vitamin deficiency, the phenotype of enterovirus can be changed, that is, a non-cardiovirulent strain can change into a cardiovirulent strain, or the cardiovirulence can be increased in these conditions[25-27]. Beck et al[28] sequenced the mutated strain, CVB3/0se-, which was extracted from the heart muscle of selenium deficient mice, and found that there were six mutation points in CVB3/0se- and compared to the two-cardiovirulent strains, CVB3/20, CVB3/M1, all mutated points were turned into that of a cardiovirulent strain. Can the same episodes also happen in Keshan disease patients? In our research, we found an about 541 bp DNA segment by RT-PCR. Compared to CVB3/0, a non-cardiovirulent strain, the sequence of our segment had only one nucleotide different, which was 234 T. But the segment had the same sequence as that of CVB3/Nancy and CVB3/20, cardiovirulent strain. This result can give us some indications that the mutation of virus genome might play an important role in the pathogeny of Keshan disease. But this needs to further investigation.
As we know that RNA is degraded easily, so its detection is difficult, especially in paraffin-embedded tissue[29,30]. But the paraffin-embedded myocardial tissue we select have been kept in our institute for a long time (10-50 years), and are generally thought not to be optimal for RNA research, but our results displayed that results of both in situ hybridization and RT-PCR were satisfactory.
Edited by Kumar M Proofread by Xu FM
| 1. | Woodruff JF. Viral myocarditis. A review. Am J Pathol. 1980;101:425-484. [PubMed] [Cited in This Article: ] |
| 2. | Bowles NE, Richardson PJ, Olsen EG, Archard LC. Detection of Coxsackie-B-virus-specific RNA sequences in myocardial biopsy samples from patients with myocarditis and dilated cardiomyopathy. Lancet. 1986;1:1120-1123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 450] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 3. | Martino TA, Liu P, Sole MJ. Viral infection and the pathogenesis of dilated cardiomyopathy. Circ Res. 1994;74:182-188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 166] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Kaplan MH, Klein SW, McPhee J, Harper RG. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev Infect Dis. 1983;5:1019-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 182] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Knowlton KU, Badorff C. The immune system in viral myocarditis: maintaining the balance. Circ Res. 1999;85:559-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Muckelbauer JK, Kremer M, Minor I, Diana G, Dutko FJ, Groarke J, Pevear DC, Rossmann MG. The structure of coxsackievirus B3 at 3.5 A resolution. Structure. 1995;3:653-667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 198] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 458] [Cited by in F6Publishing: 434] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 8. | Agol VI. The 5'-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 87] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | Minor PD. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065-3077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 164] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Rohll JB, Percy N, Ley R, Evans DJ, Almond JW, Barclay WS. The 5'-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384-4391. [PubMed] [Cited in This Article: ] |
| 11. | Le SY, Siddiqui A, Maizel JV. A common structural core in the internal ribosome entry sites of picornavirus, hepatitis C virus, and pestivirus. Virus Genes. 1996;12:135-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Rinehart JE, Gómez RM, Roos RP. Molecular determinants for virulence in coxsackievirus B1 infection. J Virol. 1997;71:3986-3991. [PubMed] [Cited in This Article: ] |
| 13. | Dunn JJ, Chapman NM, Tracy S, Romero JR. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: localization to the 5' nontranslated region. J Virol. 2000;74:4787-4794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 84] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Tu Z, Chapman NM, Hufnagel G, Tracy S, Romero JR, Barry WH, Zhao L, Currey K, Shapiro B. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5' nontranslated region. J Virol. 1995;69:4607-4618. [PubMed] [Cited in This Article: ] |
| 15. | Ge K, Yang G. The epidemiology of selenium deficiency in the etiological study of endemic diseases in China. Am J Clin Nutr. 1993;57:259S-263S. [PubMed] [Cited in This Article: ] |
| 16. | Li GS, Wang F, Kang D, Li C. Keshan disease: an endemic cardiomyopathy in China. Hum Pathol. 1985;16:602-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 95] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Gu BQ. Pathology of Keshan disease. A comprehensive review. Chin Med J (Engl). 1983;96:251-261. [PubMed] [Cited in This Article: ] |
| 18. | Li Y, Peng T, Yang Y, Niu C, Archard LC, Zhang H. High prevalence of enteroviral genomic sequences in myocardium from cases of endemic cardiomyopathy (Keshan disease) in China. Heart. 2000;83:696-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Li Y, Yang Y, Chen H. [Detection of enteroviral RNA in paraffin-embedded myocardial tissue from patients with Keshan by nested PCR]. Zhonghua Yixue Zazhi. 1995;75:344-345, 382. [PubMed] [Cited in This Article: ] |
| 20. | Peng T, Li Y, Yang Y, Niu C, Morgan-Capner P, Archard LC, Zhang H. Characterization of enterovirus isolates from patients with heart muscle disease in a selenium-deficient area of China. J Clin Microbiol. 2000;38:3538-3543. [PubMed] [Cited in This Article: ] |
| 21. | Easton AJ, Eglin RP. The detection of coxsackievirus RNA in cardiac tissue by in situ hybridization. J Gen Virol. 1988;69:285-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Archard LC, Bowles NE, Cunningham L, Freeke CA, Olsen EG, Rose ML, Meany B, Why HJ, Richardson PJ. Molecular probes for detection of persisting enterovirus infection of human heart and their prognostic value. Eur Heart J. 1991;12 Suppl D:56-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 69] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Huang Z, Xia Y, Jin Q, Wu H. [Coxsackievirus B3 infection and Keshan disease]. Wei Sheng Yan Jiu. 2002;31:261-263. [PubMed] [Cited in This Article: ] |
| 24. | Guanqing H. On the etiology of Keshan disease: two hypotheses. Chin Med J (Engl). 1979;92:416-422. [PubMed] [Cited in This Article: ] |
| 25. | Levander OA, Beck MA. Interacting nutritional and infectious etiologies of Keshan disease. Insights from coxsackie virus B-induced myocarditis in mice deficient in selenium or vitamin E. Biol Trace Elem Res. 1997;56:5-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Beck MA, Kolbeck PC, Rohr LH, Shi Q, Morris VC, Levander OA. Benign human enterovirus becomes virulent in selenium-deficient mice. J Med Virol. 1994;43:166-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 106] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 27. | Beck MA, Kolbeck PC, Rohr LH, Shi Q, Morris VC, Levander OA. Vitamin E deficiency intensifies the myocardial injury of coxsackievirus B3 infection of mice. J Nutr. 1994;124:345-358. [PubMed] [Cited in This Article: ] |
| 28. | Beck MA, Shi Q, Morris VC, Levander OA. Rapid genomic evolution of a non-virulent coxsackievirus B3 in selenium-deficient mice results in selection of identical virulent isolates. Nat Med. 1995;1:433-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 240] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 29. | Jackson DP, Quirke P, Lewis F, Boylston AW, Sloan JM, Robertson D, Taylor GR. Detection of measles virus RNA in paraffin-embedded tissue. Lancet. 1989;1:1391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Sariol CA, Pelegrino JL, Martinez A, Arteaga E, Kouri G, Guzman MG. Detection and genetic relationship of dengue virus sequences in seventeen-year-old paraffin-embedded samples from Cuba. Am J Trop Med Hyg. 1999;61:994-1000. [PubMed] [Cited in This Article: ] |