Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis
Abstract
:1. Introduction
1.1. Conserved Functions of Transcription Factors across Mammals
1.2. Transcription Factors, Potential Targets for Vaccine Development
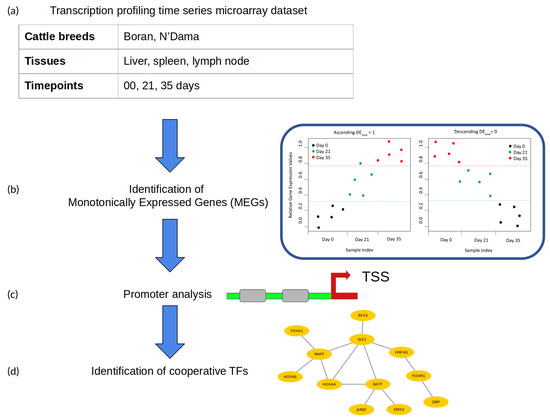
2. Materials and Methods
2.1. Microarray Dataset
2.2. Identification of Monotonically Expressed Genes
2.3. Identification of Transcription Factor Cooperation
- Promoter sequences: The promoter sequence (covering the −500 to 100 bp regions relative to a transcription start site) of each significant monotonically expressed gene (MEG) is extracted from the UCSC genome browser [43].
- Creation of the PWM library and TFBS detection: For the detection of TFBSs in the promoters of MEGs, we obtained PWMs from the TRANSFAC database (release 2018.1) [44].Until now, based on the functional analysis and comprehensive performance evaluation strategies, different studies have shown that the computational TFBS detection methods using PWMs are well established and highly applied. However, their prediction performance is prone to high rates of false positive predictions [30,45,46]. In order to eliminate the false predictions to some extent in our analysis, we manually created a specific PWM library following our previous study [47]. For this purpose, we first obtained all available cattle TFs from AnimalTFDB 2.0 [48] and identified the expression values (TPM values) of their corresponding TF genes in the gene expression dataset, under study. Second, the TFs were excluded from further analysis if the TPM values of their TF genes were zero. After that, the corresponding PWMs of the remaining TFs were obtained from the TRANSFAC database [44]. Finally, based on the Pearson correlation between these PWMs, we applied hierarchical clustering and used only the PWMs with the highest information content from each cluster as representative to create our final non-redundant vertebrate PWM library (see Supplementary Table S1).In addition, we applied the Match program [49] using these PWMs and their TRANSFAC specific profile parameter minSum to minimize the sum of false positive and false negative rate for the detection of putative TFBSs in promoter sequences.
- Pre-defined distances: For the identification of cooperative TFs based on the co-occurrence of their TFBSs, the PC-TraFF algorithm requires pre-defined minimum and maximum distance thresholds. In this study, the recommended distance values of and were used for the minimum and maximum distance, respectively.
3. Results and Discussion
3.1. Data Processing
3.2. Identification of MEGs
3.3. Identification of Cooperative TFs
3.4. Cooperative TFs in Liver Tissue
3.5. Cooperative TFs in Spleen Tissue
3.6. Cooperative TFs in Lymph Node Tissue
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biello, D. Deadly by the Dozen: 12 Diseases climate change may Worsen. Sci. Am. 2008, 8, 12–18. [Google Scholar]
- Radostits, O.M.; Gay, C.C.; Hinchcliff, K.W.; Constable, P.D. Veterinary Medicine E-Book: A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats; Elsevier Health Sciences: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Firesbhat, A.; Desalegn, C. Epidemiology and Impacts of Trypanosomiasis in Cattle. Eur. J. Appl. Sci. 2015, 7, 220–225. [Google Scholar]
- Donelson, J.E.; Hill, K.L.; El-Sayed, N.M. Multiple mechanisms of immune evasion by African trypanosomes. Mol. Biochem. Parasitol. 1998, 91, 51–66. [Google Scholar] [CrossRef]
- Losos, G.J.; Ikede, B. Review of pathology of diseases in domestic and laboratory animals caused by Trypanosoma congolense, T. vivax, T. brucei, T. rhodesiense and T. gambiense. Vet. Pathol. 1972, 9, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Morrison, L.J.; Vezza, L.; Rowan, T.; Hope, J.C. Animal African trypanosomiasis: Time to increase focus on clinically relevant parasite and host species. Trends Parasitol. 2016, 32, 599–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristjanson, P.M.; Swallow, B.M.; Rowlands, G.; Kruska, R.; De Leeuw, P. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric. Syst. 1999, 59, 79–98. [Google Scholar] [CrossRef]
- Stijlemans, B.; De Baetselier, P.; Magez, S.; Van Ginderachter, J.A.; De Trez, C. African trypanosomiasis-associated anemia: The contribution of the interplay between parasites and the mononuclear phagocyte system. Front. Immunol. 2018, 9, 218. [Google Scholar] [CrossRef]
- Yaro, M.; Munyard, K.; Stear, M.; Groth, D. Combatting African animal trypanosomiasis (AAT) in livestock: The potential role of trypanotolerance. Vet. Parasitol. 2016, 225, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Naessens, J. Bovine trypanotolerance: A natural ability to prevent severe anaemia and haemophagocytic syndrome? Int. J. Parasitol. 2006, 36, 521–528. [Google Scholar] [CrossRef]
- Murray, M.; D‘Ieteren, G.; Authie, E.; Wissocq, N. Trypanotolerance, an option for sustainable livestock production in areas at risk from trypanosomosis. OIE Rev. Sci. Tech. 1998, 17, 154–175. [Google Scholar]
- Starkey, P. N’Dama cattle-a productive trypanotolerant breed. FAO World Anim. Rev. 1984, 50, 2–15. [Google Scholar]
- Grootenhuis, J.; Dwinger, R.; Dolan, R.; Moloo, S.; Murray, M. Susceptibility of African buffalo and Boran cattle to Trypanosoma congolense transmitted by Glossina morsitans centralis. Vet. Parasitol. 1990, 35, 219–231. [Google Scholar] [CrossRef]
- Borst, P. Antigenic variation and allelic exclusion. Cell 2002, 109, 5–8. [Google Scholar] [CrossRef] [Green Version]
- Cross, G.A.; Wirtz, L.E.; Navarro, M. Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998, 91, 77–91. [Google Scholar] [CrossRef]
- Barry, J.D.; McCulloch, R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001, 49, 1–70. [Google Scholar]
- Cnops, J.; Magez, S.; De Trez, C. Escape mechanisms of African trypanosomes: Why trypanosomosis is keeping us awake. Parasitology 2015, 142, 417–427. [Google Scholar] [CrossRef]
- Anene, B.; Onah, D.; Nawa, Y. Drug resistance in pathogenic African trypanosomes: What hopes for the future? Vet. Parasitol. 2001, 96, 83–100. [Google Scholar] [CrossRef]
- Michel-Todó, L.; Bigey, P.; Reche, P.A.; Pinazo, M.J.; Gascón, J.; Alonso-Padilla, J. Design of an Epitope-Based Vaccine Ensemble for Animal Trypanosomiasis by Computational Methods. Vaccines 2020, 8, 130. [Google Scholar] [CrossRef] [Green Version]
- Noyes, H.; Brass, A.; Obara, I.; Anderson, S.; Archibald, A.L.; Bradley, D.G.; Fisher, P.; Freeman, A.; Gibson, J.; Gicheru, M.; et al. Genetic and expression analysis of cattle identifies candidate genes in pathways responding to Trypanosoma congolense infection. Proc. Natl. Acad. Sci. USA 2011, 108, 9304–9309. [Google Scholar] [CrossRef] [Green Version]
- Ruddle, N.H.; Akirav, E.M. Secondary lymphoid organs: Responding to genetic and environmental cues in ontogeny and the immune response. J. Immunol. 2009, 183, 2205–2212. [Google Scholar] [CrossRef]
- Clayton, C.E.; Selkirk, M.E.; Corsini, C.A.; Ogilvie, B.M.; Askonas, B.A. Murine trypanosomiasis: Cellular proliferation and functional depletion in the blood, peritoneum, and spleen related to changes in bone marrow stem cells. Infect. Immun. 1980, 28, 824–831. [Google Scholar] [PubMed]
- Kuriakose, S.M.; Singh, R.; Uzonna, J.E. Host intracellular signaling events and pro-inflammatory cytokine production in African trypanosomiasis. Front. Immunol. 2016, 7, 181. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Sun, H.J.; Chang, T.Y.; Lo, H.H.; Cheng, W.C.; Tseng, G.C.; Lin, C.T.; Chang, S.J.; Pal, N.R.; Chung, I.F. Discovering monotonic stemness marker genes from time-series stem cell microarray data. BMC Genom. 2015, 16, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S. Identification of monotonically differentially expressed genes for non-small cell lung cancer. BMC Bioinform. 2019, 20, 177. [Google Scholar] [CrossRef] [Green Version]
- Meckbach, C.; Tacke, R.; Hua, X.; Waack, S.; Wingender, E.; Gültas, M. PC-TraFF: Identification of potentially collaborating transcription factors using pointwise mutual information. BMC Bioinform. 2015, 16, 400. [Google Scholar] [CrossRef] [Green Version]
- Wlochowitz, D.; Haubrock, M.; Arackal, J.; Bleckmann, A.; Wolff, A.; Beißbarth, T.; Wingender, E.; Gültas, M. Computational identification of key regulators in two different colorectal cancer cell lines. Front. Genet. 2016, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Orozco-Solis, R.; Aguilar-Arnal, L. Circadian Regulation of Immunity Through Epigenetic Mechanisms. Front. Cell. Infect. Microbiol. 2020, 10, 96. [Google Scholar] [CrossRef] [Green Version]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, T.W.; Wang, J.; Collins, P.J.; Partridge, E.C.; Aldred, S.F.; Trinklein, N.D.; Myers, R.M.; Weng, Z. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012, 13, R50. [Google Scholar] [CrossRef] [Green Version]
- Blanchette, M.; Tompa, M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002, 12, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Blanchette, M.; Schwikowski, B.; Tompa, M. Algorithms for phylogenetic footprinting. J. Comput. Biol. 2002, 9, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Yan, J.; Whitington, T.; Toivonen, J.; Nitta, K.R.; Rastas, P.; Morgunova, E.; Enge, M.; Taipale, M.; Wei, G.; et al. DNA-binding specificities of human transcription factors. Cell 2013, 152, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitta, K.R.; Jolma, A.; Yin, Y.; Morgunova, E.; Kivioja, T.; Akhtar, J.; Hens, K.; Toivonen, J.; Deplancke, B.; Furlong, E.E.; et al. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 2015, 4, e04837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Franco-Zorrilla, J.M.; López-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Moussaif, M.; Choi, S.; Xu, L.; Sze, J.Y. RFX transcription factor DAF-19 regulates 5-HT and innate immune responses to pathogenic bacteria in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1003324. [Google Scholar] [CrossRef] [Green Version]
- Powell, N.; Canavan, J.; MacDonald, T.; Lord, G. Transcriptional regulation of the mucosal immune system mediated by T-bet. Mucosal Immunol. 2010, 3, 567–577. [Google Scholar] [CrossRef]
- Rothenberg, E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014, 32, 283–321. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; Papavassiliou, A.G. Transcription factor therapeutics: Long-shot or lodestone. Curr. Med. Chem. 2005, 12, 691–701. [Google Scholar] [CrossRef]
- Moustaqil, M.; Gambin, Y.; Sierecki, E. Biophysical Techniques for Target Validation and Drug Discovery in Transcription-Targeted Therapy. Int. J. Mol. Sci. 2020, 21, 2301. [Google Scholar] [CrossRef] [Green Version]
- Papavassiliou, K.A.; Papavassiliou, A.G. Transcription factor drug targets. J. Cell. Biochem. 2016, 117, 2693–2696. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Briefings Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stormo, G.D. DNA binding sites: Representation and discovery. Bioinformatics 2000, 16, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Tompa, M.; Li, N.; Bailey, T.L.; Church, G.M.; De Moor, B.; Eskin, E.; Favorov, A.V.; Frith, M.C.; Fu, Y.; Kent, W.J.; et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005, 23, 137. [Google Scholar] [CrossRef]
- Steuernagel, L.; Meckbach, C.; Heinrich, F.; Zeidler, S.; Schmitt, A.O.; Gültas, M. Computational identification of tissue-specific transcription factor cooperation in ten cattle tissues. PLoS ONE 2019, 14, e0216475. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.M.; Liu, T.; Liu, C.J.; Song, S.; Zhang, X.; Liu, W.; Jia, H.; Xue, Y.; Guo, A.Y. AnimalTFDB 2.0: A resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2014, 43, D76–D81. [Google Scholar] [CrossRef]
- Wingender, E.; Chen, X.; Fricke, E.; Geffers, R.; Hehl, R.; Liebich, I.; Krull, M.; Matys, V.; Michael, H.; Ohnhaeuser, R.; et al. Match-a tool for searching transcription factor binding sites in DNA sequences. Nucl. Acids Res. 2001, 29, 281–283. [Google Scholar] [CrossRef] [Green Version]
- Wingender, E.; Schoeps, T.; Dönitz, J. TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 2012, 41, D165–D170. [Google Scholar] [CrossRef]
- Fajas, L.; Landsberg, R.L.; Huss-Garcia, Y.; Sardet, C.; Lees, J.A.; Auwerx, J. E2Fs regulate adipocyte differentiation. Dev. Cell 2002, 3, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Denechaud, P.D.; Fajas, L.; Giralt, A. E2F1, a novel regulator of metabolism. Front. Endocrinol. 2017, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Haim, Y.; Blüher, M.; Slutsky, N.; Goldstein, N.; Klöting, N.; Harman-Boehm, I.; Kirshtein, B.; Ginsberg, D.; Gericke, M.; Guiu Jurado, E.; et al. Elevated autophagy gene expression in adipose tissue of obese humans: A potential non-cell-cycle-dependent function of E2F1. Autophagy 2015, 11, 2074–2088. [Google Scholar] [CrossRef] [PubMed]
- Bozek, K.; Relógio, A.; Kielbasa, S.M.; Heine, M.; Dame, C.; Kramer, A.; Herzel, H. Regulation of clock-controlled genes in mammals. PLoS ONE 2009, 4, e4882. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Stienstra, R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie 2017, 136, 75–84. [Google Scholar] [CrossRef]
- Roussigne, M.; Cayrol, C.; Clouaire, T.; Amalric, F.; Girard, J.P. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene 2003, 22, 2432. [Google Scholar] [CrossRef] [Green Version]
- Gervais, V.; Campagne, S.; Durand, J.; Muller, I.; Milon, A. NMR studies of a new family of DNA binding proteins: The THAP proteins. J. Biomol. NMR 2013, 56, 3–15. [Google Scholar] [CrossRef]
- Scott, C.C.; Vossio, S.; Rougemont, J.; Gruenberg, J. TFAP2 transcription factors are regulators of lipid droplet biogenesis. Elife 2018, 7, e36330. [Google Scholar] [CrossRef]
- Melo, R.C.; Weller, P.F. Lipid droplets in leukocytes: Organelles linked to inflammatory responses. Exp. Cell Res. 2016, 340, 193–197. [Google Scholar] [CrossRef] [Green Version]
- Konige, M.; Wang, H.; Sztalryd, C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Cross, J.C.; Flannery, M.L.; Blanar, M.A.; Steingrimsson, E.; Jenkins, N.A.; Copeland, N.; Rutter, W.; Werb, Z. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 1995, 121, 2513–2523. [Google Scholar]
- Hollenberg, S.M.; Sternglanz, R.; Cheng, P.F.; Weintraub, H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 1995, 15, 3813–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cserjesi, P.; Brown, D.; Lyons, G.E.; Olson, E.N. Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev. Biol. 1995, 170, 664–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, D.; Cserjesi, P.; Olson, E.N. A subclass of bHLH proteins required for cardiac morphogenesis. Science 1995, 270, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.G.; Barbosa, A.C.; Richardson, J.A.; Schneider, M.D.; Srivastava, D.; Olson, E.N. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 2005, 132, 189–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meckbach, C.; Wingender, E.; Gültas, M. Removing background co-occurrences of transcription factor binding sites greatly improves the prediction of specific transcription factor cooperations. Front. Genet. 2018, 9, 189. [Google Scholar] [CrossRef]
- Zeidler, S.; Meckbach, C.; Tacke, R.; Raad, F.S.; Roa, A.; Uchida, S.; Zimmermann, W.H.; Wingender, E.; Gültas, M. Computational detection of stage-specific transcription factor clusters during heart development. Front. Genet. 2016, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Molina, L.; Conquet, F.; Dubois-Dauphin, M.; Schibler, U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997, 16, 6762–6771. [Google Scholar] [CrossRef] [Green Version]
- Olds, W. Sleep, Circadian Rhythms, and Metabolism: The Rhythm of Life; Apple Academic Press: Palm Bay, FL, USA, 2014. [Google Scholar]
- Huang, W.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Investig. 2011, 121, 2133–2141. [Google Scholar] [CrossRef]
- Laposky, A.D.; Bass, J.; Kohsaka, A.; Turek, F.W. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett. 2008, 582, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Lundkvist, G.B.; Kristensson, K.; Bentivoglio, M. Why trypanosomes cause sleeping sickness. Physiology 2004, 19, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676. [Google Scholar] [CrossRef] [PubMed]
- Borgs, L.; Beukelaers, P.; Vandenbosch, R.; Belachew, S.; Nguyen, L.; Malgrange, B. Cell “circadian” cycle: New role for mammalian core clock genes. Cell Cycle 2009, 8, 832–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balsalobre, A. Clock genes in mammalian peripheral tissues. Cell Tissue Res. 2002, 309, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Wuarin, J.; Schibler, U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 1990, 63, 1257–1266. [Google Scholar] [CrossRef]
- Reinke, H.; Asher, G. Circadian clock control of liver metabolic functions. Gastroenterology 2016, 150, 574–580. [Google Scholar] [CrossRef] [Green Version]
- Asher, G.; Schibler, U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011, 13, 125–137. [Google Scholar] [CrossRef] [Green Version]
- Cretenet, G.; Le Clech, M.; Gachon, F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 2010, 11, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H.; Alberta, J.A.; Gonzalez, F.J.; Waxman, D.J. Multiple, functional DBP sites on the promoter of the cholesterol 7 alpha-hydroxylase P450 gene, CYP7. Proposed role in diurnal regulation of liver gene expression. J. Biol. Chem. 1994, 269, 14681–14689. [Google Scholar]
- Zannini, M.; Francis-Lang, H.; Plachov, D.; Di Lauro, R. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol. Cell. Biol. 1992, 12, 4230–4241. [Google Scholar] [CrossRef]
- Plachov, D.; Chowdhury, K.; Walther, C.; Simon, D.; Guenet, J.L.; Gruss, P. Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development 1990, 110, 643–651. [Google Scholar]
- Macchia, P.E.; Lapi, P.; Krude, H.; Pirro, M.T.; Missero, C.; Chiovato, L.; Souabni, A.; Baserga, M.; Tassi, V.; Pinchera, A.; et al. PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. Nat. Genet. 1998, 19, 83. [Google Scholar] [CrossRef] [PubMed]
- Laury, A.R.; Perets, R.; Piao, H.; Krane, J.F.; Barletta, J.A.; French, C.; Chirieac, L.R.; Lis, R.; Loda, M.; Hornick, J.L.; et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am. J. Surg. Pathol. 2011, 35, 816–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, Y.; Alford, F.; Burger, H. The 24-hour plasma thyrotrophin profile. Clin. Sci. 1972, 43, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutayoba, B.; Gombe, S. Effect of African trypanosomiasis on plasma cortisol and thyroxine concentration in goats. Res. Vet. Sci. 1989, 47, 315–318. [Google Scholar] [CrossRef]
- Abebe, G. The Integrity of Hypothalamic-Pituitary-Adrenal Axis in Boran (Bos Indicus) Cattle Infected with Trypanosoma Congolense. Ph.D. Thesis, Brunel University, Uxbridge, UK, 1991. [Google Scholar]
- Mukhopadhyay, D.; Plateroti, M.; Anant, S.; Nassir, F.; Samarut, J.; Davidson, N.O. Thyroid hormone regulates hepatic triglyceride mobilization and apolipoprotein B messenger ribonucleic acid editing in a murine model of congenital hypothyroidism. Endocrinology 2003, 144, 711–719. [Google Scholar] [CrossRef] [Green Version]
- McAninch, E.A.; Bianco, A.C. Thyroid hormone signaling in energy homeostasis and energy metabolism. Ann. N. Y. Acad. Sci. 2014, 1311, 77. [Google Scholar] [CrossRef] [Green Version]
- Mathe, D.; Chevallier, F. Effects of the thyroid state on cholesterol metabolism in the rat. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1976, 441, 155–164. [Google Scholar] [CrossRef]
- Ness, G.C.; Dugan, R.E.; Lakshmanan, M.; Nepokroeff, C.M.; Porter, J.W. Stimulation of hepatic β-hydroxy-β-methylglutaryl coenzyme A reductase activity in hypophysectomized rats by L-triiodothyronine. Proc. Natl. Acad. Sci. USA 1973, 70, 3839–3842. [Google Scholar] [CrossRef] [Green Version]
- Ness, G.C.; Pendleton, L.C.; Li, Y.C.; Chiang, J.Y. Effect of thyroid hormone on hepatic cholesterol 7α hydroxylase, LDL receptor, HMG-CoA reductase, farnesyl pyrophosphate synthetase and apolipoprotein AI mRNA levels in hypophysectomized rats. Biochem. Biophys. Res. Commun. 1990, 172, 1150–1156. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Clouaire, T.; Roussigne, M.; Ecochard, V.; Mathe, C.; Amalric, F.; Girard, J.P. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. USA 2005, 102, 6907–6912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roussigne, M.; Kossida, S.; Lavigne, A.C.; Clouaire, T.; Ecochard, V.; Glories, A.; Amalric, F.; Girard, J.P. The THAP domain: A novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 2003, 28, 66–69. [Google Scholar] [CrossRef]
- Zakirova, Z.; Fanutza, T.; Bonet, J.; Readhead, B.; Zhang, W.; Yi, Z.; Beauvais, G.; Zwaka, T.P.; Ozelius, L.J.; Blitzer, R.D.; et al. Mutations in THAP1/DYT6 reveal that diverse dystonia genes disrupt similar neuronal pathways and functions. PLoS Genet. 2018, 14, e1007169. [Google Scholar] [CrossRef] [PubMed]
- Valli, V.; Forsberg, C.; Robinson, G. The pathogenesis of Trypanosoma congolense infection in calves. I. Clinical observations and gross pathological changes. Vet. Pathol. 1978, 15, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva Pereira, S.; Trindade, S.; De Niz, M.; Figueiredo, L.M. Tissue tropism in parasitic diseases. Open Biol. 2019, 9, 190036. [Google Scholar] [CrossRef] [Green Version]
- Kierstein, S.; Noyes, H.; Naessens, J.; Nakamura, Y.; Pritchard, C.; Gibson, J.; Kemp, S.; Brass, A. Gene expression profiling in a mouse model for African trypanosomiasis. Genes Immun. 2006, 7, 667–679. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Xie, X.; Liao, W.; Zhang, H.; Cao, H.; Fei, R.; Wang, X.; Wei, L.; Shao, Q.; Chen, H. The transcription factor RFX5 is a transcriptional activator of the TPP1 gene in hepatocellular carcinoma. Oncol. Rep. 2017, 37, 289–296. [Google Scholar] [CrossRef]
- Aftab, S.; Semenec, L.; Chu, J.S.C.; Chen, N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol. Biol. 2008, 8, 226. [Google Scholar] [CrossRef] [Green Version]
- Villard, J.; Peretti, M.; Masternak, K.; Barras, E.; Caretti, G.; Mantovani, R.; Reith, W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell. Biol. 2000, 20, 3364–3376. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr. Thymic selection: Two pathways to life and two to death. Immunity 1994, 1, 3–6. [Google Scholar] [CrossRef]
- Lazzaro, D.; Price, M.; de Felice, M.; Di Lauro, R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 1991, 113, 1093–1104. [Google Scholar] [PubMed]
- Ikeda, K.; Clark, J.C.; Shaw-White, J.R.; Stahlman, M.T.; Boutell, C.J.; Whitsett, J.A. Gene structure and expression of human thyroid transcription factor-1 in respiratory epithelial cells. J. Biol. Chem. 1995, 270, 8108–8114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.C.; Hsu, Y.C.; Chai, C.Y. Role of TTF-1, CK20, and CK7 immunohistochemistry for diagnosis of primary and secondary lung adenocarcinoma. Kaohsiung J. Med Sci. 2006, 22, 14–19. [Google Scholar] [CrossRef] [Green Version]
- Belalcazar, A.; Krishnamurti, U.; Shaib, W. TTF-1 expression in rectal adenocarcinoma: A case report and review of the literature. Case Rep. Oncol. 2016, 9, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Aversa, S.; Bellan, C. TTF1 Expression in Pulmonary Metastatic Rectal Adenocarcinoma. Case Rep. Gastrointest. Med. 2018, 2018, 6405125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbro, D.; Di Loreto, C.; Beltrami, C.A.; Belfiore, A.; Di Lauro, R.; Damante, G. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res. 1994, 54, 4744–4749. [Google Scholar]
- Ros, P.; Rossi, D.L.; Acebrón, A.; Santisteban, P. Thyroid-specific gene expression in the multi-step process of thyroid carcinogenesis. Biochimie 1999, 81, 389–396. [Google Scholar] [CrossRef]
- Zhang, P.; Zuo, H.; Nakamura, Y.; Nakamura, M.; Wakasa, T.; Kakudo, K. Immunohistochemical analysis of thyroid-specific transcription factors in thyroid tumors. Pathol. Int. 2006, 56, 240–245. [Google Scholar] [CrossRef]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef]
- Daynes, R.A.; Jones, D.C. Emerging roles of PPARs in inflammation and immunity. Nat. Rev. Immunol. 2002, 2, 748. [Google Scholar] [CrossRef]
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001, 108, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Waki, H.; Murakami, K.; Motojima, K.; Komeda, K.; Ide, T.; Kubota, N.; Terauchi, Y.; Tobe, K.; et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. J. Biol. Chem. 2001, 276, 41245–41254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spohrer, S.; Groß, R.; Nalbach, L.; Schwind, L.; Stumpf, H.; Menger, M.D.; Ampofo, E.; Montenarh, M.; Götz, C. Functional interplay between the transcription factors USF1 and PDX-1 and protein kinase CK2 in pancreatic β-cells. Sci. Rep. 2017, 7, 16367. [Google Scholar] [CrossRef] [Green Version]
- Sirito, M.; Lin, Q.; Deng, J.M.; Behringer, R.R.; Sawadogo, M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. USA 1998, 95, 3758–3763. [Google Scholar] [CrossRef] [Green Version]
- Corre, S.; Galibert, M.D. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005, 18, 337–348. [Google Scholar] [CrossRef]
- Tan, Y.; Chen, Y.; Du, M.; Peng, Z.; Xie, P. USF2 inhibits the transcriptional activity of Smurf1 and Smurf2 to promote breast cancer tumorigenesis. Cell. Signal. 2019, 53, 49–58. [Google Scholar] [CrossRef]
- Lam, E.W.F.; Gomes, A.R. Forkhead box transcription factors in cancer initiation, progression and chemotherapeutic drug response. Front. Oncol. 2014, 4, 305. [Google Scholar] [CrossRef] [Green Version]
- Liao, G.B.; Li, X.Z.; Zeng, S.; Liu, C.; Yang, S.M.; Yang, L.; Hu, C.J.; Bai, J.Y. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal. 2018, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zona, S.; Bella, L.; Burton, M.J.; de Moraes, G.N.; Lam, E.W.F. FOXM1: An emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 1316–1322. [Google Scholar] [CrossRef] [Green Version]
- Nandi, D.; Cheema, P.S.; Jaiswal, N.; Nag, A. FoxM1: Repurposing an oncogene as a biomarker. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 52, pp. 74–84. [Google Scholar]
- Han, Z.; Yi, P.; Li, X.; Olson, E.N. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006, 133, 1175–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.M.; Dai, X.Y.; Qiu, X.B.; Yuan, F.; Li, R.G.; Xu, Y.J.; Qu, X.K.; Huang, R.T.; Xue, S.; Yang, Y.Q. HAND1 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin. Chem. Lab. Med. (CCLM) 2016, 54, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Abramovich, C.; Humphries, R.K. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 2005, 12, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S. Homeobox genes in normal hematopoiesis and leukemogenesis. Int. J. Hematol. 1998, 68, 343–353. [Google Scholar] [CrossRef]
- Cillo, C.; Cantile, M.; Faiella, A.; Boncinelli, E. Homeobox genes in normal and malignant cells. J. Cell. Physiol. 2001, 188, 161–169. [Google Scholar] [CrossRef]
- Grier, D.; Thompson, A.; Kwasniewska, A.; McGonigle, G.; Halliday, H.; Lappin, T. The pathophysiology of HOX genes and their role in cancer. J. Pathol. J. Pathol. Soc. G. B. Irel. 2005, 205, 154–171. [Google Scholar]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef] [Green Version]
- Collins, E.M.; Thompson, A. HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 2020, 62, 847–856. [Google Scholar] [CrossRef]
- Elias, M.H.; Baba, A.A.; Husin, A.; Sulong, S.; Hassan, R.; Sim, G.A.; Wahid, A.; Fadilah, S.; Ankathil, R. HOXA4 gene promoter hypermethylation as an epigenetic mechanism mediating resistance to imatinib mesylate in chronic myeloid leukemia patients. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Dickson, G.J.; Kwasniewska, A.; Mills, K.I.; Lappin, T.R.; Thompson, A. Hoxa6 potentiates short-term hemopoietic cell proliferation and extended self-renewal. Exp. Hematol. 2009, 37, 322–333. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, C.; Zhang, J. HMBOX1 negatively regulates NK cell functions by suppressing the NKG2D/DAP10 signaling pathway. Cell. Mol. Immunol. 2011, 8, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schraml, B.U.; Hildner, K.; Ise, W.; Lee, W.L.; Smith, W.A.E.; Solomon, B.; Sahota, G.; Sim, J.; Mukasa, R.; Cemerski, S.; et al. The AP-1 transcription factor Batf controls T H 17 differentiation. Nature 2009, 460, 405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, J. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell. Mol. Life Sci. 2009, 66, 565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klesert, T.R.; Cho, D.H.; Clark, J.I.; Maylie, J.; Adelman, J.; Snider, L.; Yuen, E.C.; Soriano, P.; Tapscott, S.J. Mice deficient in Six5 develop cataracts: Implications for myotonic dystrophy. Nat. Genet. 2000, 25, 105. [Google Scholar] [CrossRef] [PubMed]
- Meola, G.; Cardani, R. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 594–606. [Google Scholar] [CrossRef] [Green Version]
- Zeng, R.M.; Lu, X.H.; Lin, J.; Hu, J.; Rong, Z.J.; Xu, W.C.; Liu, Z.W.; Zeng, W.T. Knockdown of FOXM1 attenuates inflammatory response in human osteoarthritis chondrocytes. Int. Immunopharmacol. 2019, 68, 74–80. [Google Scholar] [CrossRef]
- Vallochi, A.L.; Teixeira, L.; Oliveira, K.d.S.; Maya-Monteiro, C.M.; Bozza, P.T. Lipid droplet, a key player in host-parasite interactions. Front. Immunol. 2018, 9, 1022. [Google Scholar] [CrossRef]
- Martínez-Hernández, A.; Gutierrez-Malacatt, H.; Carrillo-Sánchez, K.; Saldaña-Alvarez, Y.; Rojas-Ochoa, A.; Crespo-Solis, E.; Aguayo-González, A.; Rosas-López, A.; Ayala-Sanchez, J.M.; Aquino-Ortega, X.; et al. Small MAF genes variants and chronic myeloid leukemia. Eur. J. Haematol. 2014, 92, 35–41. [Google Scholar] [CrossRef]
- Mukiihi, J.; Nyirenda, C.; Paul, B.; Aduwuyi, J.; Mzula, E.; Malata, H. Chronic myeloid leukaemia in central Africans. East Afr. Med. J. 2003, 80, 470–475. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Zhang, G.; Wan, Y.Y. SKI and SMAD4 are essential for IL-21-induced Th17 differentiation. Mol. Immunol. 2019, 114, 260–268. [Google Scholar] [CrossRef]
- Rhee, J.; Park, S.H.; Kim, S.K.; Kim, J.H.; Ha, C.W.; Chun, C.H.; Chun, J.S. Inhibition of BATF/JUN transcriptional activity protects against osteoarthritic cartilage destruction. Ann. Rheum. Dis. 2017, 76, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, S.F.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Manolescu, A.; Sainz, J.; Helgason, A.; Stefansson, H.; Emilsson, V.; Helgadottir, A.; et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006, 38, 320. [Google Scholar] [CrossRef] [PubMed]
- Ledent, V.; Paquet, O.; Vervoort, M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002, 3, research0030-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Pontual, L.; Mathieu, Y.; Golzio, C.; Rio, M.; Malan, V.; Boddaert, N.; Soufflet, C.; Picard, C.; Durandy, A.; Dobbie, A.; et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum. Mutat. 2009, 30, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Hrckulak, D.; Janeckova, L.; Lanikova, L.; Kriz, V.; Horazna, M.; Babosova, O.; Vojtechova, M.; Galuskova, K.; Sloncova, E.; Korinek, V. Wnt effector tcf4 is dispensable for wnt signaling in human cancer cells. Genes 2018, 9, 439. [Google Scholar] [CrossRef] [Green Version]
- Schoof, M.; Hellwig, M.; Harrison, L.; Holdhof, D.; Lauffer, M.C.; Niesen, J.; Virdi, S.; Indenbirken, D.; Schüller, U. The basic helix-loop-helix transcription factor TCF4 impacts brain architecture as well as neuronal morphology and differentiation. Eur. J. Neurosci. 2020. [Google Scholar] [CrossRef]
- Gilliet, M.; Cao, W.; Liu, Y.J. Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008, 8, 594. [Google Scholar] [CrossRef]
- Reizis, B.; Bunin, A.; Ghosh, H.S.; Lewis, K.L.; Sisirak, V. Plasmacytoid dendritic cells: Recent progress and open questions. Annu. Rev. Immunol. 2011, 29, 163–183. [Google Scholar] [CrossRef] [Green Version]
- Sisirak, V.; Ganguly, D.; Lewis, K.L.; Couillault, C.; Tanaka, L.; Bolland, S.; D’Agati, V.; Elkon, K.B.; Reizis, B. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 2014, 211, 1969–1976. [Google Scholar] [CrossRef] [Green Version]
- Han, S.M.; Na, H.Y.; Ham, O.; Choi, W.; Sohn, M.; Ryu, S.H.; In, H.; Hwang, K.C.; Park, C.G. TCF4-targeting miR-124 is differentially expressed amongst dendritic cell subsets. Immune Netw. 2016, 16, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Jain, N.; Hartert, K.; Tadros, S.; Fiskus, W.; Havranek, O.; Ma, M.C.J.; Bouska, A.; Heavican, T.; Kumar, D.; Deng, Q.; et al. Targetable genetic alterations of TCF4 (E2-2) drive immunoglobulin expression in diffuse large B cell lymphoma. Sci. Transl. Med. 2019, 11, eaav5599. [Google Scholar] [CrossRef] [PubMed]
- Takano, K.; Lyons, M.; Moyes, C.; Jones, J.; Schwartz, C. Two percent of patients suspected of having Angelman syndrome have TCF4 mutations. Clin. Genet. 2010, 78, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Torshizi, A.D.; Armoskus, C.; Zhang, H.; Forrest, M.P.; Zhang, S.; Souaiaia, T.; Evgrafov, O.V.; Knowles, J.A.; Duan, J.; Wang, K. Deconvolution of Transcriptional Networks Identifies TCF4 as a Master Regulator in Schizophrenia. Sci. Adv. 2019, 5, eaau4139. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhu, Y.; Morozov, Y.M.; Chen, X.; Page, S.C.; Rannals, M.D.; Maher, B.J.; Rakic, P. Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatry 2019, 24, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Santpere, G.; Kawasawa, Y.I.; Evgrafov, O.V.; Gulden, F.O.; Pochareddy, S.; Sunkin, S.M.; Li, Z.; Shin, Y.; Zhu, Y.; et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 2018, 362, eaat7615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steverding, D. The history of African trypanosomiasis. Parasites Vectors 2008, 1, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Boran | N’Dama | |||
|---|---|---|---|---|
| Ascending | Descending | Ascending | Descending | |
| Liver | 741 | 308 | 757 | 124 |
| Spleen | 669 | 126 | 13 | 139 |
| Lymph node | 87 | 5 | 119 | 114 |
| Cooperative Transcription Factor Pairs | |||
|---|---|---|---|
| Breed | Liver | Spleen | Lymph Node |
| Boran | PPARG–RFX5 HAND1E47–THAP1 HAND1E47–TFAP2A THAP1–E2F1 HOXA6–BATF TTF1–RFX5 DBP–PAX8 E2F1–PPARA E2F1–E2F1 | EMX2–BATF FOXM1–JUND HOXA4–HOXB7 PPARA–E2F1 E2F1–E2F1 E2F1–TFAP2A | JUND–BATF HOXA6–MAFF SIX3–MAFF HAND1E47–E2F1 HOXA4–BATF FOXM1–HNF4G DBP–FOXM1 SIX3–RFX5 HAND1E47–FOSL1 FOXA1–MAFF HOXA4–MAFF SIX3–BATF SIX3–HNF4G HOXA4–SIX3 THAP1–PPARA E2F1–PPARA EMX2–BATF THAP1–THAP1 |
| N’Dama | PPARG–SIX5 FOXM1–DLX3 HAND1E47–E2F1 HAND1E47–USF2 SIX5–PPARA SIX3–THAP1 THAP1–THAP1 PPARA–DBP PPARA–TFAP2A E2F1–E2F1 | HMBOX–BATF HOXB7–BATF SIX5–THAP1 HOXA4–HMBOX1 HAND1E47–DBP HOXA6–BATF SIX5–E2F1 HMBOX1–RFX5 E2F1–TFAP2A | SMAD4–E2F1 HOXA6–BATF HOXA4–HOXB7 TCF4–HNF4G E2F1–TFAP2A THAP1–HNF4G DBP–TFAP2A HAND1E47–RFX5 THAP1–PPARA E2F1–PPARA EMX2–BATF HOXA4–SIX3 THAP1–THAP1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajavel, A.; Heinrich, F.; Schmitt, A.O.; Gültas, M. Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis. Vaccines 2020, 8, 246. https://doi.org/10.3390/vaccines8020246
Rajavel A, Heinrich F, Schmitt AO, Gültas M. Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis. Vaccines. 2020; 8(2):246. https://doi.org/10.3390/vaccines8020246
Chicago/Turabian StyleRajavel, Abirami, Felix Heinrich, Armin Otto Schmitt, and Mehmet Gültas. 2020. "Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis" Vaccines 8, no. 2: 246. https://doi.org/10.3390/vaccines8020246