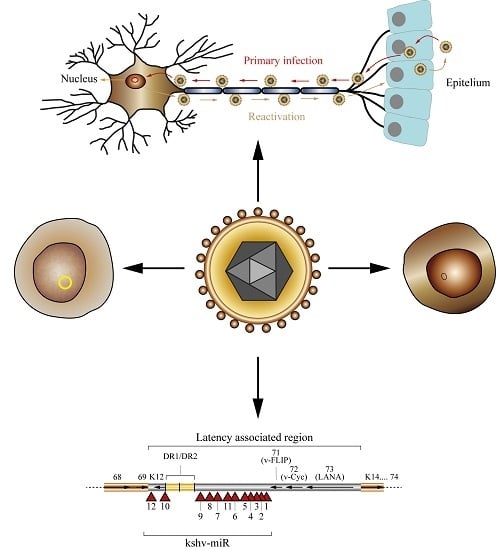
The Role of microRNAs in the Pathogenesis of Herpesvirus Infection
Abstract
:1. Introduction
2. Overview of miRNA Biogenesis
3. Alphaherpesvirus (HSV-1 and HSV-2) and microRNAs
3.1. Targeting Viral Transcripts to Maintain Latency
3.2. miRNAs Patterns of Accumulation in Alphaherpesvirus Support Viral miRNAs Predicted Functions
3.3. Cellular miRNAs Are Involved in Infection and Latency of Alphaherpesvirus
4. Human Cytomegalovirus microRNAs
4.1. HCMV miRNAs Target Cellular Genes to Evade Immune System and Control Cell Cycle as Well as Vesicle Trafficking
4.2. Viral miRNAs Target HCMV Transcripts
4.3. HCMV-Encoded miRNAs Are Expressed Differentially in Latent and Lytic Infection
4.4. HCMV Transcripts Alter Cellular miRNA Expression
4.5. Cellular miRNAs Target Viral Transcripts and Promote HCMV Latency
5. Gammaherpesviruses: Epstein–Barr Virus (EBV)-Encoded miRNAs
5.1. EBV-Encoded miRNAs Help Immune Evasion
5.2. EBV miRNAs Avoid Apoptosis by Targeting Cellular Pro-Apoptotic Genes
5.3. EBV-Encoded miRNA ebv-miR-BART6-5p Targets Dicer
5.4. Cellular miRNAs Regulate EBV Switch from Latent to Lytic Infection
5.5. EBV miRNAs Target Tumor Suppressor Genes
5.6. EBV miRNAs Play an Important Role in Viral-Induced Carcinomas and Lymphomas
5.7. EBV Is Able to Transfer Viral miRNAs through Exosomes
6. Human Herpesvirus 8/Kaposi’s Sarcoma-Associated Herpes Virus (HHV-8/KSHV) Encodes 25 Mature miRNAs
6.1. KSHV miRNAs Target Cellular mRNAs to Evade Immune Response and Modulate Cytokines Response
6.2. KSHV-Encoded miRNAs Regulate Cell Growth and Survival
6.3. KSHV miRNAs Facilitate Virus Entry
6.4. KSHV miRNAs Target Viral Transcripts to Regulate Latent and Lytic Infection
6.5. KSHV Encodes Orthologues of Cellular miRNAs
6.6. Cellular miRNAs Play a Key Role in KSHV Pathology
7. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| AIDS | Acquired immunodeficiency syndrome |
| APC | Adenomatous polyposis coli |
| ATP5B | Mitochondrial ATP synthase subunit beta |
| ATP6V0CP1 | ATPase, H+ transporting, lysosomal 16 kDa, V0 subunit C pseudogene 1 |
| BACH-1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| BAD | BCL2-associated agonist of cell death |
| BAX | BCL2-associated X Protein |
| BCL2 | B-cell CLL/lymphoma 2 |
| BCL6 | B-cell CLL/lymphoma 6 |
| BCLAF1 | BCL2-associated transcription factor 1 |
| BIM | BCL2 interacting mediator of cell death |
| BL | Burkitt’s lymphoma |
| BRCC3 | BRCA1/BRCA2-containing complex, subunit 3 |
| C/EBPβ | CCAAT/enhancer binding protein β |
| CCL5 | (C-C Motif) Ligand 5 |
| CCNE2 | Cyclin E2 |
| CD147 | Collagenase stimulatory factor |
| CDC42 | Cell division control protein 42 |
| CLASH | Cross-linking ligation and sequencing of hybrids |
| CXCL11 | C-X-C motif chemokine 11 |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 |
| DGCR8 | DiGeorge syndrome critical region gene-8 |
| DICE1 | Deleted in cancer 1 |
| DLBCL | Diffuse large B-cell lymphoma |
| DLL4 | Delta-Like 4 (Drosophila) |
| DNA | Deoxyribonucleic acid |
| EBNA | Epstein–Barr nuclear antigen |
| EBV | Epstein–Barr Virus |
| EID1 | EP300 interacting inhibitor of differentiation 1 |
| eIF4A1 | Eukaryotic initiation factor 4A1 |
| ERAP1 | Endoplasmic reticulum aminopeptidase 1 |
| ERC | Endocytic recycling compartment |
| FLICE | Fas-associated death domain-like interleukin-1 β-converting enzyme |
| GC | Gastric carcinoma |
| H3F3B | H3 histone, family 3B |
| HCMV | Human cytomegalovirus |
| HCV | Hepatitis C virus |
| HD | Hodgkin’s disease |
| HITS-CLIP | High-throughput sequencing of cross-linked immuno-precipitation |
| HIV-1 | Human Immunodeficiency Virus 1 |
| HSV-1 | Herpes Simplex Virus type 1 |
| HSV-2 | Herpes Simplex Virus type 2 |
| ICP0 | HSV-infected cell polypeptide 0 |
| ICP34.5 | HSV-infected cell polypeptide 34.5 |
| ICP4 | HSV-infected cell polypeptide 4 |
| IκBα | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| IKKε | IκB kinase epsilon |
| IL-18 | Interleukin 18 |
| IL-1α | Interleukin 1α |
| IL-1β | Interleukin 1β |
| IL-6 | Interleukin-6 |
| IRAK1 | Interleukin 1 receptor-associated kinase 1 |
| IRF1 | Interferon regulatory factor 1 |
| KLHL24 | Kelch-Like Family Member 24 |
| KS | Kaposi’s sarcoma |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| LANA | Latency-associated nuclear antigen |
| LAT | HSV latency-associated transcript |
| LMP | Latent membrane protein |
| MAF | v-Maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| MAPRE2 | Microtubule-associated protein, RP/EB family, member 2 |
| MCD | Multicentric Castleman’s disease |
| MCP-1 | Monocyte chemoattractant protein 1 |
| MHC-I | Major histocompatibility complex, class I |
| MICA | Major histocompatibility complex class-I related chain A |
| MICB | Major histocompatibility complex class-I related chain B |
| miRISC/RISC | miRNA-induced silencing complex |
| miRNA | microRNA |
| mRNA | Messenger RNA |
| MYD88 | Myeloid differentiation primary response 88 |
| NDRG1 | N-Myc Downstream-Regulated Gene 1 |
| NFIB | Nuclear factor I/B |
| NFκB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NKG2D | Killer cell lectin-like receptor subfamily K, member 1 |
| NKTL | NK/T-cell lymphoma |
| NLRP3 | NLR family, pyrin domain containing 3 |
| NPC | Nasopharyngeal carcinoma |
| ORF | Open reading frame |
| p21 | Cyclin-dependent kinase inhibitor 1A |
| PAR-CLIP | Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation |
| PEL | Primary effusion lymphoma |
| pri-miRNA | Primary-microRNA |
| PTEN | Phosphatase and tensin homolog |
| PTLD | Post-transplant lymphoma |
| PUMA | p53-Upregulated modulator of apoptosis |
| RAB11A | RAS-related GTP-binding protein 11A |
| RAB5C | RAS-related GTP-binding protein 5C |
| Rbl2 | Retinoblastoma-like protein 2 |
| RIP-CHIP | RNA-binding protein immunoprecipitation-microarray (Chip) |
| RNA | Ribonucleic acid |
| RNAi | RNA interference |
| RTA | R transactivator |
| siRNA | Small interference RNA |
| SMAD5 | SMAD family member 5 |
| SNAP23 | Synaptosomal-associated protein, 23 kDa |
| TGF-β | Transforming growth factor beta |
| TGFBR2 | Transforming growth factor, β-receptor II |
| TIMP3 | Tissue inhibitors of metalloprotease 3 |
| TLR2 | Toll-like receptor 2 |
| TNF-α | Tumor necrosis factor α |
| TOMM22 | Translocase of outer mitochondrial membrane 22 homolog (yeast) |
| TR | Terminal repeat |
| TWEAK | TNF-related weak inducer of apoptosis |
| TWEAKR | TWEAK receptor |
| UTR | Untranslated region |
| VAC | Virion assembly compartment |
| VAMP3 | Vesicle-associated membrane protein 3 |
| WIF1 | Wnt inhibitor factor 1 |
| xCT | Solute carrier family 7 (anionic amino acid transporter light chain, xC-system), member 11 |
| ZEB1/ZEB2 | Zinc finger E-box-binding homeobox (1 or 2) |
References
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell. Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, W.P.; Plasterk, R.H.A. The Diverse Functions of MicroRNAs in Animal Development and Disease. Dev. Cell 2006, 11, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell. Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Mendell, J.T. microRNAs in Vertebrate Physiology and Human Disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 215–239. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Stoffel, M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab. 2006, 4, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, L.; Szittya, G.; Silhavy, D.; Burgyán, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004, 23, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Vance, V.; Vaucheret, H. RNA silencing in plants--defense and counterdefense. Science 2001, 292, 2277–2280. [Google Scholar] [CrossRef] [PubMed]
- Katze, M.G.; He, Y.; Gale, M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002, 2, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. How Do Viruses Avoid Inhibition by Endogenous Cellular MicroRNAs? PLoS Pathog. 2013, 9, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Cullen, B.R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef] [PubMed]
- Bennasser, Y.; Yeung, M.L.; Jeang, K.-T. HIV-1 TAR RNA Subverts RNA Interference in Transfected Cells through Sequestration of TAR RNA-binding Protein, TRBP. J. Biol. Chem. 2006, 281, 27674–27678. [Google Scholar] [CrossRef] [PubMed]
- Harwig, A.; Das, A.T.; Berkhout, B. Retroviral microRNAs. Curr. Opin. Virol. 2014, 7, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Grundhoff, A.; Sullivan, C.S. Virus-encoded microRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef] [PubMed]
- Luna, J.M.; Scheel, T.K.H.; Rice, C.M.; Darnell, R.B. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell 2015, 160, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Lecellier, C.H.; Dunoyer, P.; Arar, K.; Lehmann-Che, J.; Eyquem, S.; Himber, C.; Saib, A.; Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science 2005, 308, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.T.; Nicot, C. miR-28-3p is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J. Biol. Chem. 2015, 290, 5381–5390. [Google Scholar] [CrossRef] [PubMed]
- Delorme-Axford, E.; Donker, R.B.; Mouillet, J.-F.; Chu, T.; Bayer, A.; Ouyang, Y.; Wang, T.; Stolz, D.B.; Sarkar, S.N.; Morelli, A.E.; et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12048–12053. [Google Scholar] [CrossRef] [PubMed]
- Frappier, L. Regulation of Herpesvirus Reactivation by Host MicroRNAs. J. Virol. 2015, 89, 2456–2458. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Herpesvirus microRNAs: Phenotypes and functions. Curr. Opin. Virol. 2011, 1, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, R.P.; Sullivan, C.S. Virus-Encoded microRNAs: An Overview and a Look to the Future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, Q. The miRNAs of herpes simplex virus (HSV). Virol. Sin. 2012, 27, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hook, L.; Hancock, M.; Landais, I.; Grabski, R.; Britt, W.; Nelson, J.A. Cytomegalovirus microRNAs. Curr. Opin. Virol. 2014, 7, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Meister, G.; Grässer, F.A. EBV-encoded miRNAs. Biochim. Biophys. Acta 2011, 1809, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E. Kaposi’s Sarcoma-Associated Herpesvirus microRNAs. Front. Microbiol. 2012, 3, 165. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Haecker, I.; Yang, Y.; Gao, S.-J.; Renne, R. γ-Herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Curr. Opin. Virol. 2013, 3, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Boss, I.W.; Renne, R. Viral miRNAs and immune evasion. Biochim. Biophys. Acta 2011, 1809, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Kuzembayeva, M.; Hayes, M.; Sugden, B. Multiple functions are mediated by the miRNAs of Epstein-Barr virus. Curr. Opin. Virol. 2014, 7, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Ziegelbauer, J.M. Functions of Kaposi’s sarcoma-associated herpesvirus microRNAs. Biochim. Biophys. Acta 2011, 1809, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Chekulaeva, M.; Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell. Biol. 2009, 21, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, 404–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, O.; Nadon, R. Re-analysis of genome wide data on mammalian microRNA-mediated suppression of gene expression. Translation 2013, 1, e24557/1–e24557/9. [Google Scholar] [CrossRef] [PubMed]
- Djuranovic, S.; Nahvi, A.; Green, R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012, 336, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Béthune, J.; Artus-Revel, C.G.; Filipowicz, W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012, 13, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Mathonnet, G.; Sundermeier, T.; Mathys, H.; Zipprich, J.T.; Svitkin, Y.V.; Rivas, F.; Jinek, M.; Wohlschlegel, J.; Doudna, J.A.; et al. Mammalian miRNA RISC Recruits CAF1 and PABP to Affect PABP-Dependent Deadenylation. Mol. Cell 2009, 35, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, T.; Han, Z.; Zhou, G.; Roizman, B. Patterns of accumulation of miRNAs encoded by herpes simplex virus during productive infection, latency, and on reactivation. Proc. Natl. Acad. Sci. USA 2015, 112, E49–E55. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.K.; Devi-Rao, G.; Feldman, L.T.; Dobson, A.T.; Zhang, Y.F.; Flanagan, W.M.; Stevens, J.G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J. Virol. 1988, 62, 1194–1202. [Google Scholar] [PubMed]
- Randall, G.; Lagunoff, M.; Roizman, B. Herpes simplex virus 1 open reading frames O and P are not necessary for establishment of latent infection in mice. J. Virol. 2000, 74, 9019–9027. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Griffiths, A.; Li, G.; Silva, L.M.; Kramer, M.F.; Gaasterland, T.; Wang, X.-J.; Coen, D.M. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 2006, 80, 5499–5508. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Patel, A.; Krause, P.R. Novel Less-Abundant Viral MicroRNAs Encoded by Herpes Simplex Virus 2 Latency-Associated Transcript and Their Roles in Regulating ICP34.5 and ICP0 mRNAs. J. Virol. 2009, 83, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Bertke, A.S.; Patel, A.; Wang, K.; Cohen, J.I.; Krause, P.R. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. USA 2008, 105, 10931–10936. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Nagel, M.A.; Cohrs, R.J.; Gilden, D.H.; Cullen, B.R. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 2009, 83, 10677–10683. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Wang, K.; Tang, S.; Krause, P.R.; Mont, E.K.; Cohen, J.I.; Cullen, B.R. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 2010, 84, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Kramer, M.F.; Mellor, J.C.; van Lint, A.L.; Roth, F.P.; Knipe, D.M.; Coen, D.M. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 2010, 84, 4659–4672. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Guo, Z.; Zhang, X.; Guo, L.; Liu, L.; Liao, Y.; Wang, J.; Wang, L.; Li, Q. A microRNA encoded by HSV-1 inhibits a cellular transcriptional repressor of viral immediate early and early genes. Sci. China Life Sci. 2013, 56, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Munson, D.J.; Burch, A.D. A novel miRNA produced during lytic HSV-1 infection is important for efficient replication in tissue culture. Arch. Virol. 2012, 157, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Liao, J.; Huang, Q.; Nie, Y.; Wu, K. HSV-1 miR-H6 Inhibits HSV-1 Replication and IL-6 Expression in Human Corneal Epithelial Cells In Vitro. Clin. Dev. Immunol. 2012, 2012, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Bertke, A.S.; Patel, A.; Margolis, T.P.; Krause, P.R. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J. Virol. 2011, 85, 4501–4509. [Google Scholar] [CrossRef] [PubMed]
- Samaniego, L.A.; Wu, N.; DeLuca, N.A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 1997, 71, 4614–4625. [Google Scholar] [PubMed]
- Zheng, S.; Li, Y.; Zhang, Y.; Li, X.; Tang, H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antivir. Res. 2011, 89, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Sun, H.; Fan, H.; Wang, C.; Li, Y.; Liu, M.; Tang, H. MiR-23a facilitates the replication of HSV-1 through the suppression of interferon regulatory factor 1. PLoS ONE 2014, 9, e114021. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Zhao, Y.; Clement, C.; Neumann, D.M.; Lukiw, W.J. HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. NeuroReport 2009, 20, 1500–1505. [Google Scholar] [CrossRef] [PubMed]
- Varani, S.; Landini, M. Cytomegalovirus-induced immunopathology and its clinical consequences. Herpesviridae 2011, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.R.; Li, J.Y.; Gleadle, J.M. Human cytomegalovirus encoded microRNAs: Hitting targets. Expert Rev. Anti-Infect. Ther. 2015, 13, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grässer, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Antoniewicz, A.; Allen, E.; Saugstad, J.; McShea, A.; Carrington, J.C.; Nelson, J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 2005, 79, 12095–12099. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.; Trang, P.; Zhong, Q.; Yang, E.; van Belle, C.; Liu, F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell. Microbiol. 2005, 7, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Stark, T.J.; Arnold, J.D.; Spector, D.H.; Yeo, G.W. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J. Virol. 2012, 86, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Elefant, N.; Zimmermann, A.; Wolf, D.G.; Saleh, N.; Biton, M.; Horwitz, E.; Prokocimer, Z.; Prichard, M.; Hahn, G.; et al. Host immune system gene targeting by a viral miRNA. Science 2007, 317, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Nachmani, D.; Lankry, D.; Wolf, D.G.; Mandelboim, O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010, 11, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Qi, Y.; Ma, Y.; He, R.; Ji, Y.; Sun, Z.; Ruan, Q. The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112-1. J. Virol. 2013, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Esteso, G.; Luzon, E.; Sarmiento, E.; Gomez-Caro, R.; Steinle, A.; Murphy, G.; Carbone, J.; Vales-Gomez, M.; Reyburn, H.T. Altered MicroRNA Expression after Infection with Human Cytomegalovirus Leads to TIMP3 Downregulation and Increased Shedding of Metalloprotease Substrates, Including MICA. J. Immunol. 2014, 193, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, S.; Kim, S.; Kim, D.; Ahn, J.-H.; Ahn, K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012, 8, e1002577. [Google Scholar] [CrossRef] [PubMed]
- Maghazachi, A.A.; Al-Aoukaty, A.; Schall, T.J. CC chemokines induce the generation of killer cells from CD56+ cells. Eur. J. Immunol. 1996, 26, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Landais, I.; Pelton, C.; Streblow, D.; DeFilippis, V.; McWeeney, S.; Nelson, J.A. Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway. PLoS Pathog. 2015, 11, e1004881. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Shin, J.; Kim, Y.; Evnouchidou, I.; Kim, D.; Kim, Y.-K.; Kim, Y.-E.; Ahn, J.-H.; Riddell, S.R.; et al. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8+ T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 2011, 12, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Noriega, V.; Redmann, V.; Gardner, T.; Tortorella, D. Diverse immune evasion strategies by human cytomegalovirus. Immunol. Res. 2012, 54, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Meyers, H.; White, E.A.; Spector, D.H.; Nelson, J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007, 3, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Tirabassi, R.; Meyers, H.; Wu, G.; McWeeney, S.; Hook, L.; Nelson, J.A. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 2010, 6, e1000967. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Qi, Y.; Ma, Y.; He, R.; Ji, Y.; Sun, Z.; Ruan, Q. Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett. 2013, 587, 2266–2271. [Google Scholar] [CrossRef] [PubMed]
- Pavelin, J.; Reynolds, N.; Chiweshe, S.; Wu, G.; Tiribassi, R.; Grey, F. Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication. PLoS Pathog. 2013, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hook, L.M.; Grey, F.; Grabski, R.; Tirabassi, R.; Doyle, T.; Hancock, M.; Landais, I.; Jeng, S.; McWeeney, S.; Britt, W.; et al. Cytomegalovirus miRNAs Target Secretory Pathway Genes to Facilitate Formation of the Virion Assembly Compartment and Reduce Cytokine Secretion. Cell Host Microbe 2014, 15, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.; Vanicek, J.; Robins, H.; Shenk, T.; Levine, A.J. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc. Natl. Acad. Sci. USA 2008, 105, 5453–5458. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Saleh, N.; Goldberg, M.D.; Prichard, M.; Wolf, D.G.; Mandelboim, O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J. Virol. 2009, 83, 10684–10693. [Google Scholar] [CrossRef] [PubMed]
- Tirabassi, R.; Hook, L.; Landais, I.; Grey, F.; Meyers, H.; Hewitt, H.; Nelson, J. Human Cytomegalovirus US7 Is Regulated Synergistically by Two Virally Encoded MicroRNAs and by Two Distinct Mechanisms. J. Virol. 2011, 85, 11938–11944. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Qi, Y.; Ma, Y.; He, R.; Ji, Y.; Sun, Z.; Ruan, Q. Down-regulation of human cytomegalovirus UL138, a novel latency-associated determinant, by hcmv-miR-UL36. J. Biosci. 2013, 38, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Meshesha, M.K.; Bentwich, Z.; Solomon, S.A.; Avni, Y.S. In vivo expression of human cytomegalovirus (HCMV) microRNAs during latency. Gene 2016, 575, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gao, Y.; Zhou, Q.; Zhang, Q.; Peng, Y.; Tian, K.; Wang, J.; Zheng, X. Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA. Gene 2014, 536, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-R.; Liu, X.-J.; Li, X.-J.; Shen, Z.-Z.; Yang, B.; Wu, C.-C.; Li, J.-F.; Miao, L.-F.; Ye, H.-Q.; Qiao, G.-H.; et al. miR-21 Attenuates Human Cytomegalovirus Replication in Neural Cells by Targeting Cdc25a. J. Virol. 2014, 89, 1070–1082. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.H.; Hannemann, H.; Kulkarni, A.S.; Schwartz, P.H.; O’Dowd, J.M.; Fortunato, E.A. Human Cytomegalovirus Infection Causes Premature and Abnormal Differentiation of Human Neural Progenitor Cells. J. Virol. 2010, 84, 3528–3541. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Weber, F.; Croce, C.; Liu, C.; Liao, X.; Pellett, P.E. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 2008, 82, 9065–9074. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Vanicek, J.; Murphy, E.A. Host miRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol. 2014, 88, 5524–5532. [Google Scholar]
- Lisboa, L.F.; Egli, A.; O’Shea, D.; Åsberg, A.; Hartmann, A.; Rollag, H.; Pang, X.L.; Tyrrell, D.L.; Kumar, D.; Humar, A. Hcmv-miR-UL22A-5p: A Biomarker in Transplantation With Broad Impact on Host Gene Expression and Potential Immunological Implications. Am. J. Transpl. 2015, 15, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Thi, E.P.; Mire, C.E.; Lee, A.C.H.; Geisbert, J.B.; Zhou, J.Z.; Agans, K.N.; Snead, N.M.; Deer, D.J.; Barnard, T.R.; Fenton, K.A.; et al. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature 2015, 521, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.A.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Thorley-Lawson, D.A.; Hawkins, J.B.; Tracy, S.I.; Shapiro, M. The pathogenesis of Epstein-Barr virus persistent infection. Curr. Opin. Virol. 2013, 3, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Delecluse, H.-J.; Feederle, R.; O’Sullivan, B.; Taniere, P. Epstein Barr virus-associated tumours: An update for the attention of the working pathologist. J. Clin. Pathol. 2007, 60, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Sato, Y.; Kimura, H. Modes of infection and oncogenesis by the Epstein-Barr virus. Rev. Med. Virol. 2014, 24, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Pratt, Z.L.; Kuzembayeva, M.; Sengupta, S.; Sugden, B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology 2009, 386, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Schäfer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed]
- Cosmopoulos, K.; Pegtel, M.; Hawkins, J.; Moffett, H.; Novina, C.; Middeldorp, J.; Thorley-Lawson, D.A. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 2009, 83, 2357–2367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundhoff, A. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 2006, 12, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Lung, R.W.-M.; Tong, J.H.-M.; Sung, Y.-M.; Leung, P.-S.; Ng, D.C.-H.; Chau, S.-L.; Chan, A.W.-H.; Ng, E.K.-O.; Lo, K.-W.; To, K.-F. Modulation of LMP2A Expression by a Newly Identified Epstein-Barr Virus-Encoded MicroRNA miR-BART22. Neoplasia 2009, 11, 1174–IN17. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Pfuhl, T.; Motsch, N.; Barth, S.; Nicholls, J.; Grasser, F.; Meister, G. Identification of Novel Epstein-Barr Virus MicroRNA Genes from Nasopharyngeal Carcinomas. J. Virol. 2009, 83, 3333–3341. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.F.; To, K.F.; Lo, K.W.; Lung, R.W.M.; Hui, J.W.Y.; Liao, G.; Hayward, S.D. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. USA 2007, 104, 16164–16169. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; O’Hara, A.; Araujo, I.; Barreto, J.; Carvalho, E.; Sapucaia, J.B.; Ramos, J.C.; Luz, E.; Pedroso, C.; Manrique, M.; et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008, 68, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Nam, C.S.; Raab-Traub, N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 2011, 412, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Vereide, D.T.; Seto, E.; Chiu, Y.-F.; Hayes, M.; Tagawa, T.; Grundhoff, A.; Hammerschmidt, W.; Sugden, B. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 2014, 33, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Yuen, K.-S.; Xu, R.; Tsao, S.W.; Chen, H.; Li, M.; Kok, K.-H.; Jin, D.-Y. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int. J. Cancer 2013, 133, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.Y.-W.; Siu, K.-L.; Kok, K.-H.; Lung, R.W.-M.; Tsang, C.M.; To, K.-F.; Kwong, D.L.-W.; Tsao, S.W.; Jin, D.-Y. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008, 205, 2551–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godshalk, S.; Bhaduri-McIntosh, S.; Slack, F.J. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle 2008, 7, 3595–3600. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, H.; Wulff, B.-E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.M.G.; Kong, K.L.; Tsang, J.W.H.; Kwong, D.L.W.; Guan, X.-Y. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 2012, 118, 698–710. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.-A.; Pich, D.; McInnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.J.; Masters, S.L. Cutting Edge: miR-223 and EBV miR-BART15 Regulate the NLRP3 Inflammasome and IL-1 Production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, H.; Lee, S.K. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett. 2015, 356, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, S.; Pan, Q.; Blencowe, B.J.; Claycomb, J.M.; Frappier, L. Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. J. Virol. 2014, 88, 11166–11177. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Connell, A.L.; Iempridee, T.; Xu, I.; Mertz, J.E. Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication. J. Virol. 2010, 84, 10329–10343. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, X.; Fewell, C.; Cameron, J.; Yin, Q.; Flemington, E.K. Differential expression of the miR-200 family microRNAs in epithelial and B cells and regulation of Epstein-Barr virus reactivation by the miR-200 family member miR-429. J. Virol. 2010, 84, 7892–7897. [Google Scholar] [CrossRef] [PubMed]
- Klinke, O.; Feederle, R.; Delecluse, H.J. Genetics of Epstein-Barr virus microRNAs. Semin. Cancer Biol. 2014, 26, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Kraus, R.J.; Perrigoue, J.G.; Mertz, J.E. ZEB negatively regulates the lytic-switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 2003, 77, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A.L.; Wang, Z.; Yu, X.; Mertz, J.E. Either ZEB1 or ZEB2/SIP1 can play a central role in regulating the Epstein-Barr virus latent-lytic switch in a cell-type-specific manner. J. Virol. 2010, 84, 6139–6152. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.; Rossi, A.; Rossi, D.; Kroening, S.; Bonatti, S.; Mallardo, M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-κB pathway. Nucleic Acids Res. 2008, 36, 6608–6619. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.E.; Fewell, C.; Yin, Q.; McBride, J.; Wang, X.; Lin, Z.; Flemington, E.K. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 2008, 382, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Linnstaedt, S.D.; Gottwein, E.; Skalsky, R.L.; Luftig, M.A.; Cullen, B.R. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 2010, 84, 11670–11678. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-J.; Chen, G.-H.; Chen, Y.-H.; Liu, C.-Y.; Chang, K.-P.; Chang, Y.-S.; Chen, H.-C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS ONE 2010, 5, e12745. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.N.; Chae, H.-S.; Oh, S.T.; Kang, J.-H.; Park, C.H.; Park, W.S.; Takada, K.; Lee, J.M.; Lee, W.-K.; Lee, S.K. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 2007, 81, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Chugh, P.E.; Dittmer, D.P.; Raab-Traub, N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J. Virol. 2014, 88, 1389–1393. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.; Motsch, N.; Zhu, J.Y.; Barth, S.; Okoniewski, M.; Reineke, T.; Tinguely, M.; Faggioni, A.; Trivedi, P.; Meister, G.; et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011, 39, 1880–1893. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, R.; Fitzsimmons, L.; Thomas, W.A.; Kelly, G.L.; Rowe, M.; Bell, A.I. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J. Virol. 2011, 85, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Motsch, N.; Alles, J.; Imig, J.; Zhu, J.; Barth, S.; Reineke, T.; Tinguely, M.; Cogliatti, S.; Dueck, A.; Meister, G.; et al. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS ONE 2012, 7, e42193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.-M.; Lyu, X.-M.; Luo, W.-R.; Cui, X.-F.; Ye, Y.-F.; Yuan, C.-C.; Peng, Q.-X.; Wu, D.-H.; Liu, T.-F.; Wang, E.; et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015, 34, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ye, Y.; Jiang, Q.; Chen, Y.; Lyu, X.; Li, J.; Wang, S.; Liu, T.; Cai, H.; Yao, K.; et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2015, 6, 7353. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Miyata, M.; Kano, M.; Kondo, S.; Yoshizaki, T.; Iizasa, H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J. Virol. 2015, 89, 2684–2697. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Wurdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G.; Shair, K.H.Y.; Marquitz, A.R.; Kung, C.-P.; Edwards, R.H.; Raab-Traub, N. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 20370–20375. [Google Scholar] [CrossRef] [PubMed]
- Gourzones, C.; Gelin, A.; Bombik, I.; Klibi, J.; Vérillaud, B.; Guigay, J.; Lang, P.; Témam, S.; Schneider, V.; Amiel, C.; et al. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. J. Virol. 2010, 7, 271. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; Munger, K.L. Epstein–Barr Virus Infection and Multiple Sclerosis: A Review. J. Neuroimmune Pharmacol. 2010, 5, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Handunnetthi, L.; Giovannoni, G.; Ramagopalan, S.V. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE 2010, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Levin, L.I.; Munger, K.L.; O’Reilly, E.J.; Falk, K.I.; Ascherio, A. Primary infection with the Epstein-Barr virus and risk of multiple sclerosis. Ann. Neurol. 2010, 67, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Pohl, D. Epstein-Barr virus and multiple sclerosis. J. Neurol. Sci. 2009, 286, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Thacker, E.L.; Mirzaei, F.; Ascherio, A. Infectious mononucleosis and risk for multiple sclerosis: A meta-analysis. Ann. Neurol. 2006, 59, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Jakymiw, A.; Findlay, V.; Parsons, C. KSHV-Encoded MicroRNAs: Lessons for Viral Cancer Pathogenesis and Emerging Concepts. Int. J. Cell Biol. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lu, S.; Zhang, Z.; Gonzalez, C.M.; Damania, B.; Cullen, B.R. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5570–5575. [Google Scholar] [CrossRef] [PubMed]
- Samols, M.A.; Hu, J.; Skalsky, R.L.; Renne, R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2005, 79, 9301–9305. [Google Scholar] [CrossRef] [PubMed]
- Marshall, V.; Parks, T.; Bagni, R.; Wang, C.D.; Samols, M.A.; Hu, J.; Wyvil, K.M.; Aleman, K.; Little, R.F.; Yarchoan, R.; et al. Conservation of virally encoded microRNAs in Kaposi sarcoma--associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J. Infect. Dis 2007, 195, 645–659. [Google Scholar] [PubMed]
- Umbach, J.L.; Cullen, B.R. In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 2010, 84, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Marshall, V.; Martró, E.; Labo, N.; Ray, A.; Wang, D.; Mbisa, G.; Bagni, R.K.; Volfovsky, N.; Casabona, J.; Whitby, D. Kaposi sarcoma (KS)-associated herpesvirus microRNA sequence analysis and KS risk in a European AIDS-KS case control study. J. Infect. Dis 2010, 202, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Cullen, B.R. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 2010, 84, 5229–5237. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Bai, Z.; Ye, F.; Xie, J.; Kim, C.-G.; Huang, Y.; Gao, S.-J. Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat. Cell. Biol. 2010, 12, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Zhu, Y.; Jones, T.; Bai, Z.; Huang, Y.; Gao, S.-J. A Kaposi’s sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival. J. Virol. 2012, 86, 11698–11711. [Google Scholar] [CrossRef] [PubMed]
- Suffert, G.; Malterer, G.; Hausser, J.; Viiliäinen, J.; Fender, A.; Contrant, M.; Ivacevic, T.; Benes, V.; Gros, F.; Voinnet, O.; et al. Kaposi’s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 2011, 7, e1002405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Freitas, E.; Sullivan, R.; Mohan, S.; Bacelieri, R.; Branch, D.; Romano, M.; Kearney, P.; Oates, J.; Plaisance, K.; et al. Upregulation of xCT by KSHV-encoded microRNAs facilitates KSHV dissemination and persistence in an environment of oxidative stress. PLoS Pathog. 2010, 6, e1000742. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.; Henderson, S.; Lagos, D.; Nikitenko, L.; Coulter, E.; Roberts, S.; Gratrix, F.; Plaisance, K.; Renne, R.; Bower, M.; et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010, 24, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Samols, M.A.; Plaisance, K.B.; Boss, I.W.; Riva, A.; Lopez, M.C.; Baker, H.V.; Renne, R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007, 81, 12836–12845. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Mukherjee, N.; Sachse, C.; Frenzel, C.; Majoros, W.H.; Chi, J.-T.A.; Braich, R.; Manoharan, M.; Soutschek, J.; Ohler, U.; et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007, 450, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-C.; Li, Z.; Chu, C.-Y.; Feng, J.; Feng, J.; Sun, R.; Rana, T.M. MicroRNAs encoded by Kaposi’s sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010, 11, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Stedman, W.; Yousef, M.; Renne, R.; Lieberman, P.M. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 2010, 84, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Ziegelbauer, J.M.; Sullivan, C.S.; Ganem, D. Tandem array–based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 2009, 41, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Abend, J.R.; Ramalingam, D.; Kieffer-Kwon, P.; Uldrick, T.S.; Yarchoan, R.; Ziegelbauer, J.M. Kaposi’s sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J. Virol. 2012, 86, 11663–11674. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liang, D.; He, Z.; Deng, Q.; Robertson, E.S.; Lan, K. miR-K12-7-5p encoded by Kaposi’s sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS ONE 2011, 6, e16224. [Google Scholar] [CrossRef] [PubMed]
- Bellare, P.; Ganem, D. Regulation of KSHV Lytic Switch Protein Expression by a Virus-Encoded MicroRNA: An Evolutionary Adaptation that Fine-Tunes Lytic Reactivation. Cell Host Microbe 2009, 6, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Abend, J.R.; Uldrick, T.; Ziegelbauer, J.M. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi’s sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol. 2010, 84, 12139–12151. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Gao, Y.; Lin, X.; He, Z.; Zhao, Q.; Deng, Q.; Lan, K. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKε. Cell Res. 2011, 21, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Boss, I.W.; Nadeau, P.E.; Abbott, J.R.; Yang, Y.; Mergia, A.; Renne, R. A Kaposi’s sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rγnull mice. J. Virol. 2011, 85, 9877–9886. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, R.; Lin, X.; Liang, D.; Deng, Q.; Lan, K. Kaposi’s sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J. Virol. 2012, 86, 1372–1381. [Google Scholar] [CrossRef] [PubMed]
- Moody, R.; Zhu, Y.; Huang, Y.; Cui, X.; Jones, T.; Bedolla, R.; Lei, X.; Bai, Z.; Gao, S.-J. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 2013, 9, e1003857. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral MicroRNA Targetome of KSHV-Infected Primary Effusion Lymphoma Cell Lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Shamulailatpam, P.; Raja, A.N.; Gottwein, E. Kaposi’s sarcoma-associated herpesvirus encodes a mimic of cellular miR-23. J. Virol. 2013, 87, 11821–11830. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Peruzzi, F.; Reiss, K.; Dai, L. Role of host microRNAs in Kaposi’s sarcoma-associated herpesvirus pathogenesis. Viruses 2014, 6, 4571–4580. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-H.; Wu, M.-F.; Wu, Y.-H.; Chang, S.-J.; Lin, S.-F.; Sharp, T.V.; Wang, H.-W. The M type K15 protein of Kaposi’s sarcoma-associated herpesvirus regulates microRNA expression via its SH2-binding motif to induce cell migration and invasion. J. Virol. 2009, 83, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-H.; Hu, T.-F.; Chen, Y.-C.; Tsai, Y.-N.; Tsai, Y.-H.; Cheng, C.-C.; Wang, H.-W. The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood 2011, 118, 2896–2905. [Google Scholar] [CrossRef] [PubMed]
- Bridge, G.; Monteiro, R.; Henderson, S.; Emuss, V.; Lagos, D.; Georgopoulou, D.; Patient, R.; Boshoff, C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 2012, 120, 5063–5072. [Google Scholar] [CrossRef] [PubMed]
- Punj, V.; Matta, H.; Schamus, S.; Tamewitz, A.; Anyang, B.; Chaudhary, P.M. Kaposi’s sarcoma-associated herpesvirus-encoded viral FLICE inhibitory protein (vFLIP) K13 suppresses CXCR4 expression by upregulating miR-146a. Oncogene 2010, 29, 1835–1844. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-G.; Majerciak, V.; Uldrick, T.S.; Wang, X.; Kruhlak, M.; Yarchoan, R.; Zheng, Z.-M. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol. 2011, 225, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-G.; Pripuzova, N.; Majerciak, V.; Kruhlak, M.; Le, S.-Y.; Zheng, Z.-M. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol. 2011, 85, 2620–2630. [Google Scholar] [CrossRef] [PubMed]
- Lagos, D.; Pollara, G.; Henderson, S.; Gratrix, F.; Fabani, M.; Milne, R.S.B.; Gotch, F.; Boshoff, C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 2010, 12, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Ma, X.; Shen, C.; Cao, X.; Feng, N.; Qin, D.; Zeng, Y.; Zhu, J.; Gao, S.-J.; Lu, C. Inhibition of Kaposi’s sarcoma-associated herpesvirus lytic replication by HIV-1 Nef and cellular microRNA hsa-miR-1258. J. Virol. 2014, 88, 4987–5000. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Li, W.; Tang, Q.; Yao, S.; Lv, Z.; Feng, N.; Ma, X.; Bai, Z.; Zeng, Y.; Qin, D.; et al. Cellular microRNAs 498 and 320d regulate herpes simplex virus 1 induction of Kaposi’s sarcoma-associated herpesvirus lytic replication by targeting RTA. PLoS ONE 2013, 8, e55832. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Nam, J.-W.; Farh, K.K.-H.; Chiang, H.R.; Shkumatava, A.; Bartel, D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 2010, 38, 789–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Dölken, L.; Malterer, G.; Erhard, F.; Kothe, S.; Friedel, C.C.; Suffert, G.; Marcinowski, L.; Motsch, N.; Barth, S.; Beitzinger, M.; et al. Systematic Analysis of Viral and Cellular MicroRNA Targets in Cells Latently Infected with Human γ-Herpesviruses by RISC Immunoprecipitation Assay. Cell Host Microbe 2010, 7, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Haecker, I.; Gay, L.A.; Yang, Y.; Hu, J.; Morse, A.M.; McIntyre, L.M.; Renne, R. Ago HITS-CLIP Expands Understanding of Kaposi’s Sarcoma-associated Herpesvirus miRNA Function in Primary Effusion Lymphomas. PLoS Pathog. 2012, 8, e1002884. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.J.; Rabinowitz, G.S.; Yario, T.A.; Luna, J.M.; Darnell, R.B.; Steitz, J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012, 31, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, A.M.; Das, S.; Xiao, Z.; Andresson, T.; Kieffer-Kwon, P.; Happel, C.; Ziegelbauer, J. Proteomic Screening of Human Targets of Viral microRNAs Reveals Functions Associated with Immune Evasion and Angiogenesis. PLoS Pathog. 2013, 9, e1002484. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kalejta, R.F.; Kerry, J.; Semmes, O.J.; O’Connor, C.M.; Khan, Z.; Garcia, B.A.; Shenk, T.; Murphy, E. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2012, 109, 9575–9580. [Google Scholar] [CrossRef] [PubMed]
- Grosswendt, S.; Filipchyk, A.; Manzano, M.; Klironomos, F.; Schilling, M.; Herzog, M.; Gottwein, E.; Rajewsky, N. Unambiguous Identification of miRNA: Target site interactions by different types of ligation reactions. Mol. Cell 2014, 54, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Tollervey, D. Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nat. Protoc. 2014, 9, 711–728. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.R.; Kara, M.; Coleman, C.B.; Grau, K.R.; Oko, L.M.; Krueger, B.J.; Renne, R.; van Dyk, L.F.; Tibbetts, S.A. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio 2014, 5, e00981-14. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.H.; Teng, M.; Sun, A.J.; Yu, L.L.; Hu, B.; Qu, L.H.; Ding, K.; Cheng, X.C.; Liu, J.X.; Cui, Z.Z.; et al. Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek’s disease virus. Virology 2014, 448, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.F.; Jurak, I.; Pesola, J.M.; Boissel, S.; Knipe, D.M.; Coen, D.M. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 2011, 417, 239–247. [Google Scholar] [CrossRef] [PubMed]




| miRNA | Target * | Predicted Role | References | |
|---|---|---|---|---|
| EBV-encoded (ebv-miR-) | BHRF1-3 | CXCL11 (c) | Immune evasion | [121] |
| BART1 | BIM (c) | Inhibits apoptosis | [122] | |
| BART1-3p | Caspase-3 (c) | Inhibits apoptosis | [123] | |
| BART1-5p | LMP1 (v) | Immune evasion | [120] | |
| BART2-5p | MICB (c) | Immune evasion | [124] | |
| BART3 | BIM (c) | Inhibits apoptosis | [122] | |
| BART3-5p | DICE1 (c) | Cell transformation and proliferation | [125] | |
| BART5-5p | PUMA (c) | Inhibits apoptosis | [126] | |
| BART6-5p | Dicer (c) | Modulates biogenesis of microRNAs | [127,128] | |
| BART7 | APC (c) | Cell transformation and proliferation | [129] | |
| BART9 | BIM (c) | Inhibits apoptosis | [122] | |
| BART11 | BIM (c) | Inhibits apoptosis | [122] | |
| BART12 | BIM (c) | Inhibits apoptosis | [122] | |
| BART15 | NLRP3 (c) | Immune evasion | [130] | |
| BART16 | LMP1 (v) | Immune evasion | [130] | |
| TOMM22 (c) | Inhibits apoptosis | [122] | ||
| Caspase-3 (c) | Inhibits apoptosis | [123] | ||
| BART17-5p | LMP1 (v) | Immune evasion | [120] | |
| BART19-3p | WIF1 (c) | Cell transformation and proliferation | [129] | |
| APC (c) | Cell transformation and proliferation | [129] | ||
| BART20-5p | BAD (c) | Inhibits apoptosis | [131] | |
| Cell-encoded (hsa-miR-) | let-7a | Dicer (c) | Modulates biogenesis of microRNAs | [132] |
| 200b | ZEB1, ZEB2 (c) | Modulates latent/lytic infection | [133,134] | |
| 429 | ZEB1, ZEB2 (c) | Modulates latent/lytic infection | [133,134] | |
| KSHV-Encoded miRNA (kshv-miR-) | Target * | Predicted Role | References |
|---|---|---|---|
| K12-1 | MICB | Immune evasion | [124] |
| p21 | Oncogenesis | [164] | |
| IκBα | Cell survival | [165] | |
| Modulates latent/lytic infection | [166] | ||
| Caspase 3 | Cell survival | [167] | |
| xCT expression | Facilitates viral entry | [168,169,170,171] | |
| K12-3 | C/EBPβ | Immune evasion | [168,158] |
| Cell survival | [168,158] | ||
| Oncogenesis | [168,158] | ||
| Caspase 3 | Cell survival | [167] | |
| NFIB | Modulates latent/lytic infection | [172] | |
| K12-4 | RBL2 | Modulates latent/lytic infection | [173] |
| Caspase 3 | Cell survival | [167] | |
| K12-5 | BCLAF1 | Modulates latent/lytic infection | [174] |
| MYD88 | Immune evasion | [175] | |
| K12-6 | xCT expression | Facilitates viral entry | [168,169,170,171] |
| K12-7 | C/EBPβ | Immune evasion | [168] |
| Cell survival | [168] | ||
| Oncogenesis | [168] | ||
| KSHV ORF50 | Modulates latent/lytic infection | [176,177] | |
| K12-9 | IRAK1 | Immune evasion | [175] |
| xCT expression | Facilitates viral entry | [168,169,170,171] | |
| KSHV ORF50 | Modulates latent/lytic infection | [176,177] | |
| BCLAF1 | Modulates latent/lytic infection | [174] | |
| K12-10 | TWEAKR | Immune evasion | [178] |
| Cell survival | [178] | ||
| TGFBR2 | Cell survival | [166] | |
| Oncogenesis | [166] | ||
| K12-11 | IKKε | Immune evasion | [179] |
| Modulates latent/lytic infection | [179] | ||
| C/EBPβ | Immune evasion | [180] | |
| Cell survival | [180] | ||
| Oncogenesis | [180] | ||
| SMAD5 | Cell survival | [181] | |
| Oncogenesis | [181] | ||
| xCT expression | Facilitates viral entry | [168,169,170,171] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piedade, D.; Azevedo-Pereira, J.M. The Role of microRNAs in the Pathogenesis of Herpesvirus Infection. Viruses 2016, 8, 156. https://doi.org/10.3390/v8060156
Piedade D, Azevedo-Pereira JM. The Role of microRNAs in the Pathogenesis of Herpesvirus Infection. Viruses. 2016; 8(6):156. https://doi.org/10.3390/v8060156
Chicago/Turabian StylePiedade, Diogo, and José Miguel Azevedo-Pereira. 2016. "The Role of microRNAs in the Pathogenesis of Herpesvirus Infection" Viruses 8, no. 6: 156. https://doi.org/10.3390/v8060156