Unraveling the Nrf2-ARE Signaling Pathway in the DF-1 Chicken Fibroblast Cell Line: Insights into T-2 Toxin-Induced Oxidative Stress Regulation
Abstract
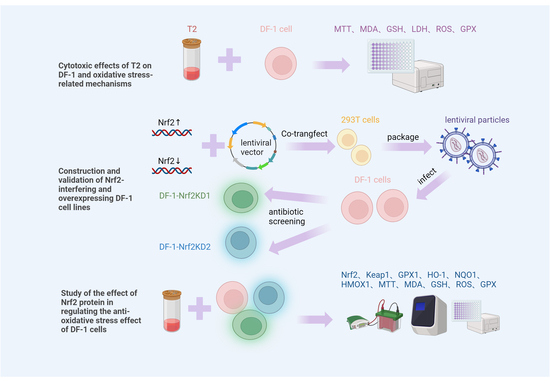
:1. Introduction
2. Results
2.1. Toxicity Effects of T2 in DF-1 Cells
2.2. The Effect of T2 on ROS Activity, MDA Content, GSH Content and GPX Activity in DF-1 Cells
2.3. The Impact of T2 on the mRNA Expression of Factors Relevant to Oxidative Stress in DF-1 Cells
2.4. Preparation and Formation of Recombinant Lentiviral Plasmids
2.5. Assessment of the Impact of Modulated Nrf2 Expression
2.6. The Effect of Nrf2 Expression on the Morphology and Viability of DF-1 Cells
2.7. The Effect of Nrf2 Expression on ROS, MDA, GSH Content and GPX Activity in DF-1 Cells
2.8. The Effect of Nrf2 Expression on the Expression of Oxidative Stress-Related Factor mRNA
2.9. The Effect of Nrf2 Expression on the Expression of Oxidative Stress-Related Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Kits
5.3. Cell Culture
5.4. The MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 Diphenyl Tetrazolium Bromide) Assay for Cell Activity and IC50 Value
5.5. Measurement of Intracellular GSH, LDH, ROS, MDA and GPX
5.6. Relative mRNA Levels of Oxidative Stress-Related Genes in DF-1 Cells
5.7. Preparation of Recombinant Lentiviral Plasmid
5.8. Construction of Nrf2 Knockdown and Overexpression in DF-1 Cell Line
5.9. Assessment of the Impact of Modulated Nrf2 Expression
5.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ueno, Y. Toxicological features of T-2 toxin and related trichothecenes. Fundam. Appl. Toxicol. 1984, 4, S124–S132. [Google Scholar] [CrossRef] [PubMed]
- Pier, A.C.; Richard, J.L.; Cysewski, S.J. Implications of mycotoxins in animal disease. J. Am. Vet. Med. Assoc. 1980, 176, 719–724. [Google Scholar] [PubMed]
- Abbas, H.K.; Mirocha, C.J.; Kommedahl, T.; Vesonder, R.F.; Golinski, P. Production of trichothecene and non-trichothecene mycotoxins by fusarium species isolated from maize in minnesota. Mycopathologia 1989, 108, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.Y.; Li, Q.; Meng, F.G.; Fu, Y.; Zhao, Z.J.; Wang, L.H. T-2 toxin contamination in grains and selenium concentration in drinking water and grains in kaschin-beck disease endemic areas of qinghai province. Biol. Trace Elem. Res. 2012, 150, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Beyer, M.; Ferse, I.; Mulac, D.; Würthwein, E.U.; Humpf, H.U. Structural elucidation of t-2 toxin thermal degradation products and investigations toward their occurrence in retail food. J. Agric. Food Chem. 2009, 57, 1867–1875. [Google Scholar] [CrossRef] [PubMed]
- Königs, M.; Mulac, D.; Schwerdt, G.; Gekle, M.; Humpf, H.U. Metabolism and cytotoxic effects of t-2 toxin and its metabolites on human cells in primary culture. Toxicology 2009, 258, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Tan, L.; Yi, S.; Wu, Y.; Hu, Y.; Wang, A.; Yang, L. Low-concentration t-2 toxin attenuates pseudorabies virus replication in porcine kidney 15 cells. Toxins 2022, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, L.; Sun, J.; Wang, L.; Guo, H.; Ye, Y.; Sun, X. Microbial detoxification of mycotoxins in food and feed. Crit. Rev. Food Sci. Nutr. 2022, 62, 4951–4969. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Beier, R.C.; Shen, J.; De Smet, D.; De Saeger, S.; Zhang, S. T-2 toxin, a trichothecene mycotoxin: Review of toxicity, metabolism, and analytical methods. J. Agric. Food Chem. 2011, 59, 3441–3453. [Google Scholar] [CrossRef]
- Chi, M.S.; Mirocha, C.J.; Kurtz, H.J.; Weaver, G.; Bates, F.; Shimoda, W.; Burmeister, H.R. Acute toxicity of t-2 toxin in broiler chicks and laying hens. Poult. Sci. 1977, 56, 103–116. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Velkov, T.; Tang, S.; Dai, C. T-2 toxin-induced toxicity in neuroblastoma-2a cells involves the generation of reactive oxygen, mitochondrial dysfunction and inhibition of nrf2/ho-1 pathway. Food Chem. Toxicol. 2018, 114, 88–97. [Google Scholar] [CrossRef] [PubMed]
- He, S.J.; Hou, J.F.; Dai, Y.Y.; Zhou, Z.L.; Deng, Y.F. N-acetyl-cysteine protects chicken growth plate chondrocytes from t-2 toxin-induced oxidative stress. J. Appl. Toxicol. JAT 2012, 32, 980–985. [Google Scholar] [CrossRef] [PubMed]
- Mackei, M.; Orbán, K.; Molnár, A.; Pál, L.; Dublecz, K.; Husvéth, F.; Neogrády, Z.; Mátis, G. Cellular effects of t-2 toxin on primary hepatic cell culture models of chickens. Toxins 2020, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Q.; Li, M.; Tang, S.; Dai, C. T-2 toxin induces apoptotic cell death and protective autophagy in mouse microglia bv2 cells. J. Fungi 2022, 8, 761. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, V.I.; Anderson, P. Trichothecene mycotoxins trigger a ribotoxic stress response that activates c-jun n-terminal kinase and p38 mitogen-activated protein kinase and induces apoptosis. J. Biol. Chem. 1999, 274, 13985–13992. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Han, S.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q. T-2 toxin induces oxidative stress, apoptosis and cytoprotective autophagy in chicken hepatocytes. Toxins 2020, 12, 90. [Google Scholar] [CrossRef]
- Young, J.C.; Zhou, T.; Yu, H.; Zhu, H.; Gong, J. Degradation of trichothecene mycotoxins by chicken intestinal microbes. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2007, 45, 136–143. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of nf-e2-related factor 2 (nrf2), a nf-e2-like basic leucine zipper transcriptional activator that binds to the tandem nf-e2/ap1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef]
- Cho, H.Y.; Kleeberger, S.R. Nrf2 protects against airway disorders. Toxicol. Appl. Pharmacol. 2010, 244, 43–56. [Google Scholar] [CrossRef]
- Sykiotis, G.P.; Habeos, I.G.; Samuelson, A.V.; Bohmann, D. The role of the antioxidant and longevity-promoting nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 41–48. [Google Scholar] [CrossRef]
- Lacher, S.E.; Levings, D.C.; Freeman, S.; Slattery, M. Identification of a functional antioxidant response element at the hif1a locus. Redox Biol. 2018, 19, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Stępkowski, T.M.; Kruszewski, M.K. Molecular cross-talk between the nrf2/keap1 signaling pathway, autophagy, and apoptosis. Free Radic. Biol. Med. 2011, 50, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, Y.; Huang, T.; Liu, Y.; Chen, F.; Chen, Y.; Jiang, Y.; Zhang, C.; Yang, X. T-2 toxin metabolism and its hepatotoxicity: New insights on the molecular mechanism and detoxification. Environ. Pollut. 2023, 330, 121784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Geng, X.; Dong, Q.; Li, X.; Ye, P.; Lin, M.; Xu, B.; Jiang, H. Crosstalk between autophagy and the keap1-nrf2-are pathway regulates realgar-induced neurotoxicity. J. Ethnopharmacol. 2023, 301, 115776. [Google Scholar] [CrossRef] [PubMed]
- Weidner, M.; Lenczyk, M.; Schwerdt, G.; Gekle, M.; Humpf, H.U. Neurotoxic potential and cellular uptake of t-2 toxin in human astrocytes in primary culture. Chem. Res. Toxicol. 2013, 26, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Zhang, G.; Xue, B.; Ding, Q.; Han, L.; Huang, J.C.; Wu, F.; Li, C.; Yang, C. Toxicity and detoxification of t-2 toxin in poultry. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2022, 169, 113392. [Google Scholar] [CrossRef] [PubMed]
- Karacaoğlu, E.; Selmanoğlu, G. T-2 toxin induces cytotoxicity and disrupts tight junction barrier in serw3 cells. Environ. Toxicol. Pharmacol. 2017, 56, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, P.; Zhang, X.; Zhang, J.; Cui, Y.; Song, M.; Li, Y. T-2 toxin causes dysfunction of sertoli cells by inducing oxidative stress. Ecotoxicol. Environ. Saf. 2021, 225, 112702. [Google Scholar] [CrossRef]
- Chaudhari, M.; Jayaraj, R.; Bhaskar, A.S.; Lakshmana Rao, P.V. Oxidative stress induction by t-2 toxin causes DNA damage and triggers apoptosis via caspase pathway in human cervical cancer cells. Toxicology 2009, 262, 153–161. [Google Scholar] [CrossRef]
- Tsutsui, H.; Kinugawa, S.; Matsushima, S. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc. Res. 2009, 81, 449–456. [Google Scholar] [CrossRef]
- Spitz, D.R.; Adams, D.T.; Sherman, C.M.; Roberts, R.J. Mechanisms of cellular resistance to hydrogen peroxide, hyperoxia, and 4-hydroxy-2-nonenal toxicity: The significance of increased catalase activity in H2O2-resistant fibroblasts. Arch. Biochem. Biophys. 1992, 292, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.K.; Surh, Y.J. Nrf2-keap1 signaling as a potential target for chemoprevention of inflammation-associated carcinogenesis. Pharm. Res. 2010, 27, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Itoh, K.; Takahashi, S.; Sato, H.; Yanagawa, T.; Katoh, Y.; Bannai, S.; Yamamoto, M. Transcription factor nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000, 275, 16023–16029. [Google Scholar] [CrossRef] [PubMed]
- Sena-Esteves, M.; Gao, G. Production of high-titer retrovirus and lentivirus vectors. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Tam, K.; Ma, F.; Tam, C.; Aleshe, B.; Wang, X.; Quintos, J.P.; Morselli, M.; Pellegrini, M.; Hollis, R.P.; et al. Β-globin lentiviral vectors have reduced titers due to incomplete vector rna genomes and lowered virion production. Stem Cell Rep. 2021, 16, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Acharya, R. The recent progresses in shrna-nanoparticle conjugate as a therapeutic approach. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 104, 109928. [Google Scholar] [CrossRef] [PubMed]
- Keleku-Lukwete, N.; Suzuki, M.; Yamamoto, M. An overview of the advantages of keap1-nrf2 system activation during inflammatory disease treatment. Antioxid. Redox Signal. 2018, 29, 1746–1755. [Google Scholar] [CrossRef]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Nrf2/are-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013, 65, 645–657. [Google Scholar] [CrossRef]
- Domijan, A.M.; Marjanović Čermak, A.M.; Vulić, A.; Tartaro Bujak, I.; Pavičić, I.; Pleadin, J.; Markov, K.; Mihaljević, B. Cytotoxicity of gamma irradiated aflatoxin b(1) and ochratoxin a. J. Environ. Sci. Health. Part. B Pestic. Food Contam. Agric. Wastes 2019, 54, 155–162. [Google Scholar] [CrossRef]
- Agyin, J.K.; Santhamma, B.; Nair, H.B.; Roy, S.S.; Tekmal, R.R. Bu-32: A novel proteasome inhibitor for breast cancer. Breast Cancer Res. BCR 2009, 11, R74. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the mtt assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, G.W.; Yan, Z.; Lang, W.; Masucci, J.A. The ic(50) concept revisited. Curr. Top. Med. Chem. 2012, 12, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Qingtao, H.; Yuetao, S.; Yipeng, T.; Chunmei, L. Determination and application of leakage rate of lactate dehydrogenase in the cultured medium of cells. Chin. J. Cell Biol. 2004, 26, 89–92. [Google Scholar]
- Owen, J.B.; Butterfield, D.A. Measurement of oxidized/reduced glutathione ratio. Methods Mol. Biol. 2010, 648, 269–277. [Google Scholar] [PubMed]
- Xiao, H.; Li, P.; Zhang, W.; Zhang, W.; Tang, B. Recent advances in mitochondria-targetable organic molecule fluorescent probes for imaging of ros. SCIENTIA SINICA Chim. 2017, 47, 955–972. [Google Scholar]
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research progress of glutathione peroxidase family (gpx) in redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (mda) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Rao, M.Z.; Chen, J.; Zhang, J.; Yuan, L.; Liao, M.; Cao, W. A cell line resistant to avian leukosis virus subgroup b infection. Poult. Sci. 2019, 98, 6026–6033. [Google Scholar] [CrossRef]
- He, X.; He, Q.; Yu, W.; Huang, J.; Yang, M.; Chen, W.; Han, W. Optimized protocol for high-titer lentivirus production and transduction of primary fibroblasts. J. Basic Microbiol. 2021, 61, 430–442. [Google Scholar] [CrossRef]
- Zhang, T.L.; Wan, X.R.; Wu, R.; Wang, C. Effects of prp(c) on df-1 cells’ biological processes and rna-seq-based analysis of differential genes. J. Cell. Physiol. 2018, 233, 6671–6682. [Google Scholar] [CrossRef]
- Wan, Z.; Lu, Y.; Rui, L.; Yu, X.; Li, Z. Prdm1 overexpression induce g0/g1 arrest in df-1 cell line. Gene 2016, 592, 119–127. [Google Scholar] [CrossRef]










| Primer/Sequence Name | Primer Sequence (5′→3′) | GenBank Accession No. |
|---|---|---|
| Nrf2-F | CATAGAGCAAGTTTGGGAAGAG | MN416129.1 |
| Nrf2-R | GTTTCAGGGCTCGTGATTGT | MN416129.1 |
| Keap1-F | ACTTCGCTGAGGTCTCCAAG | MN416132.1 |
| Keap1-R | CAGTCGTACTGCACCCAGTT | MN416132.1 |
| GPX1-F | CTGTTCGCCTTCCTGAGAGA | NM_001277853.1 |
| GPX1-R | TGCAGTTTGATGGTCTCGAA | NM_001277853.1 |
| HO-1-F | AGCGCAGCGCTTCACGTCCC | NM_205344.2 |
| HO-1-R | ATAAAAGTCAATGTAAAGCG | NM_205344.2 |
| NQO1-F | AAGCCCTGCCCCGAGCGAAG | NM_001277621.1 |
| NQO1-R | GCAACAATAAATCGAGGTCT | NM_001277621.1 |
| GAPDH-F | GAACATCATCCCAGCGTCCA | NM_204305.1 |
| GAPDH-R | CGGCAGGTCAGGTCAACAAC | NM_204305.1 |
| Nrf2-MYC-F | CCGGAATTCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGAACTTGATTGACATC | MN416129.1 |
| Nrf2-Ha-R | CGCACCGGTTCAAGCGTAATCTGGAACATCGTATGGGTACAGTTTAGTCTCTGCCTT | MN416129.1 |
| Nrf2-shRNA-F-1 | CCGGGGAGACAGGTGAATTTGTTCCTCAAGAGGGAACAAATTCACCTGTCTCCTTTTT | MN416129.1 |
| Nrf2-shRNA-R-1 | AATTAAAAAGGAGACAGGTGAATTTGTTCCCTCTTGAGGAACAAATTCACCTGTCTCC | MN416129.1 |
| Nrf2-shRNA-F-2 | CCGGGCAAGTTTGGGAAGAGTTATTTCAAGAGAATAACTCTTCCCAAACTTGCTTTTT | MN416129.1 |
| Nrf2-shRNA-R-2 | AATTAAAAAGCAAGTTTGGGAAGAGTTATTCTCTTGAAATAACTCTTCCCAAACTTGC | MN416129.1 |
| Reagent | Dosage |
|---|---|
| cDNA | 1 μL |
| SYBR Green Mix | 5 μL |
| Upstream primers | 0.2 μL |
| Downstream primers | 0.2 μL |
| RNase free H2O | 3.6 μL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, S.; Wang, K.; Xiong, K.; Xiao, S.; Wu, C.; Zhou, M.; Li, L.; Yuan, G.; Jiang, L.; Xiong, Q.; et al. Unraveling the Nrf2-ARE Signaling Pathway in the DF-1 Chicken Fibroblast Cell Line: Insights into T-2 Toxin-Induced Oxidative Stress Regulation. Toxins 2023, 15, 627. https://doi.org/10.3390/toxins15110627
Gao S, Wang K, Xiong K, Xiao S, Wu C, Zhou M, Li L, Yuan G, Jiang L, Xiong Q, et al. Unraveling the Nrf2-ARE Signaling Pathway in the DF-1 Chicken Fibroblast Cell Line: Insights into T-2 Toxin-Induced Oxidative Stress Regulation. Toxins. 2023; 15(11):627. https://doi.org/10.3390/toxins15110627
Chicago/Turabian StyleGao, Suisui, Kaixin Wang, Kuankuan Xiong, Shuai Xiao, Chujian Wu, Mingxia Zhou, Linfeng Li, Guoxiang Yuan, Lihuang Jiang, Qianbo Xiong, and et al. 2023. "Unraveling the Nrf2-ARE Signaling Pathway in the DF-1 Chicken Fibroblast Cell Line: Insights into T-2 Toxin-Induced Oxidative Stress Regulation" Toxins 15, no. 11: 627. https://doi.org/10.3390/toxins15110627