Bacterial Type I Toxins: Folding and Membrane Interactions
Abstract
:1. Introduction
2. Overview of the Membrane-Associated Type I Toxins across the Bacterial Species
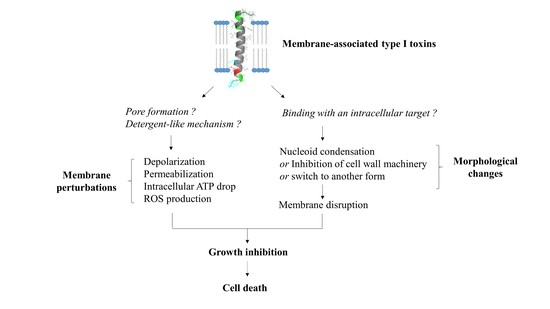
3. Membrane-Associated Type I Toxins Inducing Membrane Perturbations as a Primary Detected Effect
3.1. Membrane-Associated Type I Toxins Inducing Pore Formation
3.2. Membrane-Associated Type I Toxins Inducing Membrane Depolarization and/or Permeabilization
4. Membrane-Associated Type I Toxins Inducing Cell Morphology Changes as a Primary Detected Effect
5. Protein Folding of the S. aureus sprG1-Encoded Type I Toxins
6. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Melderen, L.; Saavedra De Bast, M. Bacterial Toxin–Antitoxin Systems: More Than Selfish Entities? PLoS Genet. 2009, 5, e1000437. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P. Toxin-Antitoxin Loci Are Highly Abundant in Free-Living but Lost from Host-Associated Prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yao, J.; Sun, Y.-C.; Wood, T.K. Type VII Toxin/Antitoxin Classification System for Antitoxins That Enzymatically Neutralize Toxins. Trends Microbiol. 2021, 29, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Kim, W.; Suk, S.; Park, H.; Bak, G.; Yoon, J.; Lee, Y. The Small RNA, SdsR, Acts as a Novel Type of Toxin in Escherichia Coli. RNA Biol. 2018, 15, 1319–1335. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef]
- Lawarée, E.; Jankevicius, G.; Cooper, C.; Ahel, I.; Uphoff, S.; Tang, C.M. DNA ADP-Ribosylation Stalls Replication and Is Reversed by RecF-Mediated Homologous Recombination and Nucleotide Excision Repair. Cell Rep. 2020, 30, 1373–1384.e4. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Lee, B.-J. Structure, Biology, and Therapeutic Application of Toxin–Antitoxin Systems in Pathogenic Bacteria. Toxins 2016, 8, 305. [Google Scholar] [CrossRef]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of Type I Toxin–Antitoxin Systems in Bacteria: Searches for New Candidates and Discovery of Novel Families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef]
- Gerdes, K.; Bech, F.W.; Jørgensen, S.T.; Løbner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of Postsegregational Killing by the Hok Gene Product of the ParB System of Plasmid R1 and Its Homology with the RelF Gene Product of the E. Coli RelB Operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [CrossRef]
- Pedersen, K.; Gerdes, K. Multiple Hok Genes on the Chromosome of Escherichia Coli. Mol. Microbiol. 1999, 32, 1090–1102. [Google Scholar] [CrossRef]
- Wen, J.; Fozo, E. SRNA Antitoxins: More than One Way to Repress a Toxin. Toxins 2014, 6, 2310–2335. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Masachis, S.; Darfeuille, F. Type I Toxin-Antitoxin Systems: Regulating Toxin Expression via Shine-Dalgarno Sequence Sequestration and Small RNA Binding. In Regulating with RNA in Bacteria and Archaea; Storz, G., Papenfort, K., Eds.; ASM Press: Washington, DC, USA, 2018; pp. 171–190. ISBN 978-1-68367-051-3. [Google Scholar]
- Brantl, S.; Jahn, N. SRNAs in Bacterial Type I and Type III Toxin-Antitoxin Systems. FEMS Microbiol. Rev. 2015, 39, 413–427. [Google Scholar] [CrossRef]
- Brielle, R.; Pinel-Marie, M.-L.; Felden, B. Linking Bacterial Type I Toxins with Their Actions. Curr. Opin. Microbiol. 2016, 30, 114–121. [Google Scholar] [CrossRef]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a Small RNA) Form a Type I Toxin–Antitoxin System in Escherichia Coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An Antisense RNA Controls Synthesis of an SOS-Induced Toxin Evolved from an Antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Prock, S.; Reichert, J.; Wadhwani, P.; Zimpfer, B.; Bürck, J.; Berditsch, M.; Elstner, M.; Ulrich, A.S. Peptide-Lipid Interactions of the Stress-Response Peptide TisB That Induces Bacterial Persistence. Biophys. J. 2012, 103, 1460–1469. [Google Scholar] [CrossRef]
- Mok, W.W.K.; Patel, N.H.; Li, Y. Decoding Toxicity. J. Biol. Chem. 2010, 285, 41627–41636. [Google Scholar] [CrossRef]
- Göbl, C.; Dulle, M.; Hohlweg, W.; Grossauer, J.; Falsone, S.F.; Glatter, O.; Zangger, K. Influence of Phosphocholine Alkyl Chain Length on Peptide−Micelle Interactions and Micellar Size and Shape. J. Phys. Chem. B 2010, 114, 4717–4724. [Google Scholar] [CrossRef]
- Göbl, C.; Kosol, S.; Stockner, T.; Rückert, H.M.; Zangger, K. Solution Structure and Membrane Binding of the Toxin Fst of the Par Addiction Module. Biochemistry 2010, 49, 6567–6575. [Google Scholar] [CrossRef]
- Sayed, N.; Nonin-Lecomte, S.; Réty, S.; Felden, B. Functional and Structural Insights of a Staphylococcus Aureus Apoptotic-like Membrane Peptide from a Toxin-Antitoxin Module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef] [PubMed]
- Korkut, D.N.; Alves, I.D.; Vogel, A.; Chabas, S.; Sharma, C.M.; Martinez, D.; Loquet, A.; Salgado, G.F.; Darfeuille, F. Structural Insights into the AapA1 Toxin of Helicobacter Pylori. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129423. [Google Scholar] [CrossRef]
- Scheglmann, D.; Werner, K.; Eiselt, G.; Klinger, R. Role of Paired Basic Residues of Protein C-Termini in Phospholipid Binding. Protein Eng. Des. Sel. 2002, 15, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E.; Reddy, S.G.; Brinkman, C.L.; Patel, S.; Bayles, K.W.; Endres, J.L. Identification and Characterization of a Family of Toxin–Antitoxin Systems Related to the Enterococcus Faecalis Plasmid PAD1 Par Addiction Module. Microbiology 2009, 155, 2930–2940. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R J. 2015, 7, 4. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Unoson, C.; Wagner, E.G.H. A Small SOS-Induced Toxin Is Targeted against the Inner Membrane in Escherichia Coli: Mode of Action of TisB. Mol. Microbiol. 2008, 70, 258–270. [Google Scholar] [CrossRef]
- Thisted, T.; Sørensen, N.S.; Wagner, E.G.; Gerdes, K. Mechanism of Post-Segregational Killing: Sok Antisense RNA Interacts with Hok MRNA via Its 5′-End Single-Stranded Leader and Competes with the 3′-End of Hok MRNA for Binding to the Mok Translational Initiation Region. EMBO J. 1994, 13, 1960–1968. [Google Scholar] [CrossRef]
- Verstraeten, N.; Knapen, W.J.; Kint, C.I.; Liebens, V.; Van den Bergh, B.; Dewachter, L.; Michiels, J.E.; Fu, Q.; David, C.C.; Fierro, A.C.; et al. Obg and Membrane Depolarization Are Part of a Microbial Bet-Hedging Strategy That Leads to Antibiotic Tolerance. Mol. Cell 2015, 59, 9–21. [Google Scholar] [CrossRef]
- Verstraeten, N.; Gkekas, S.; Kint, C.I.; Deckers, B.; Van den Bergh, B.; Herpels, P.; Louwagie, E.; Knapen, W.; Wilmaerts, D.; Dewachter, L.; et al. Biochemical Determinants of ObgE-mediated Persistence. Mol. Microbiol. 2019, 112, 1593–1608. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Bayoumi, M.; Dewachter, L.; Knapen, W.; Mika, J.T.; Hofkens, J.; Dedecker, P.; Maglia, G.; Verstraeten, N.; Michiels, J. The Persistence-Inducing Toxin HokB Forms Dynamic Pores That Cause ATP Leakage. mBio 2018, 9, e00744-18. [Google Scholar] [CrossRef]
- Edelmann, D.; Berghoff, B.A. Type I Toxin-Dependent Generation of Superoxide Affects the Persister Life Cycle of Escherichia Coli. Sci. Rep. 2019, 9, 14256. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Dewachter, L.; De Loose, P.-J.; Bollen, C.; Verstraeten, N.; Michiels, J. HokB Monomerization and Membrane Repolarization Control Persister Awakening. Mol. Cell 2019, 75, 1031–1042.e4. [Google Scholar] [CrossRef]
- Argaman, L.; Hershberg, R.; Vogel, J.; Bejerano, G.; Wagner, E.G.H.; Margalit, H.; Altuvia, S. Novel Small RNA-Encoding Genes in the Intergenic Regions of Escherichia Coli. Curr. Biol. 2001, 11, 941–950. [Google Scholar] [CrossRef]
- Wassarman, K.M. Identification of Novel Small RNAs Using Comparative Genomics and Microarrays. Genes Dev. 2001, 15, 1637–1651. [Google Scholar] [CrossRef]
- Vogel, J.; Argaman, L.; Wagner, E.G.H.; Altuvia, S. The Small RNA IstR Inhibits Synthesis of an SOS-Induced Toxic Peptide. Curr. Biol. 2004, 14, 2271–2276. [Google Scholar] [CrossRef]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS System: A Complex and Tightly Regulated Response to DNA Damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef]
- Darfeuille, F.; Unoson, C.; Vogel, J.; Wagner, E.G.H. An Antisense RNA Inhibits Translation by Competing with Standby Ribosomes. Mol. Cell 2007, 26, 381–392. [Google Scholar] [CrossRef]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin Causes Persister Formation by Inducing the TisB Toxin in Escherichia Coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, B.A.; Hoekzema, M.; Aulbach, L.; Wagner, E.G.H. Two Regulatory RNA Elements Affect TisB-Dependent Depolarization and Persister Formation: RNA-Based Regulation of Depolarization and Persistence. Mol. Microbiol. 2017, 103, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, D.; Oberpaul, M.; Schäberle, T.F.; Berghoff, B.A. Post-transcriptional Deregulation of the tisB/istR-1 Toxin–Antitoxin System Promotes SOS -independent Persister Formation in Escherichia coli. Environ. Microbiol. Rep. 2021, 13, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Gurnev, P.A.; Ortenberg, R.; Dörr, T.; Lewis, K.; Bezrukov, S.M. Persister-Promoting Bacterial Toxin TisB Produces Anion-Selective Pores in Planar Lipid Bilayers. FEBS Lett. 2012, 586, 2529–2534. [Google Scholar] [CrossRef]
- Sayed, N.; Jousselin, A.; Felden, B. A Cis-Antisense RNA Acts in Trans in Staphylococcus Aureus to Control Translation of a Human Cytolytic Peptide. Nat. Struct. Mol. Biol. 2012, 19, 105–112. [Google Scholar] [CrossRef]
- Pichon, C.; Felden, B. From The Cover: Small RNA Genes Expressed from Staphylococcus Aureusgenomic and Pathogenicity Islands with Specific Expression among Pathogenic Strains. Proc. Natl. Acad. Sci. USA 2005, 102, 14249–14254. [Google Scholar] [CrossRef]
- Solecki, O.; Mosbah, A.; Baudy Floc’h, M.; Felden, B. Converting a Staphylococcus Aureus Toxin into Effective Cyclic Pseudopeptide Antibiotics. Chem. Biol. 2015, 22, 329–335. [Google Scholar] [CrossRef]
- Weaver, K. The Fst/Ldr Family of Type I TA System Toxins: Potential Roles in Stress Response, Metabolism and Pathogenesis. Toxins 2020, 12, 474. [Google Scholar] [CrossRef]
- Folli, C.; Levante, A.; Percudani, R.; Amidani, D.; Bottazzi, S.; Ferrari, A.; Rivetti, C.; Neviani, E.; Lazzi, C. Toward the Identification of a Type I Toxin-Antitoxin System in the Plasmid DNA of Dairy Lactobacillus Rhamnosus. Sci. Rep. 2017, 7, 12051. [Google Scholar] [CrossRef]
- Maggi, S.; Yabre, K.; Ferrari, A.; Lazzi, C.; Kawano, M.; Rivetti, C.; Folli, C. Functional Characterization of the Type I Toxin Lpt from Lactobacillus Rhamnosus by Fluorescence and Atomic Force Microscopy. Sci. Rep. 2019, 9, 15208. [Google Scholar] [CrossRef]
- Weaver, K.E. The Par Toxin-Antitoxin System from Enterococcus Faecalis Plasmid PAD1 and Its Chromosomal Homologs. RNA Biol. 2012, 9, 1498–1503. [Google Scholar] [CrossRef]
- Wen, J.; Harp, J.R.; Fozo, E.M. The 5’ UTR of the Type I Toxin ZorO Can Both Inhibit and Enhance Translation. Nucleic Acids Res. 2017, 45, 4006–4020. [Google Scholar] [CrossRef]
- Otsuka; Ishikawa; Takahashi; Masuda A Short Peptide Derived from the ZorO Toxin Functions as an Effective Antimicrobial. Toxins 2019, 11, 392. [CrossRef]
- Rudd, K.E. Novel Intergenic Repeats of Escherichia Coli K-12. Res. Microbiol. 1999, 150, 653–664. [Google Scholar] [CrossRef]
- Fozo, E.M.; Kawano, M.; Fontaine, F.; Kaya, Y.; Mendieta, K.S.; Jones, K.L.; Ocampo, A.; Rudd, K.E.; Storz, G. Repression of Small Toxic Protein Synthesis by the Sib and OhsC Small RNAs. Mol. Microbiol. 2008, 70, 1076–1093. [Google Scholar] [CrossRef]
- Han, K.; Kim, K.; Bak, G.; Park, H.; Lee, Y. Recognition and Discrimination of Target MRNAs by Sib RNAs, a Cis-Encoded SRNA Family. Nucleic Acids Res. 2010, 38, 5851–5866. [Google Scholar] [CrossRef]
- Manganelli, R.; Gennaro, M.L. Protecting from Envelope Stress: Variations on the Phage-Shock-Protein Theme. Trends Microbiol. 2017, 25, 205–216. [Google Scholar] [CrossRef]
- Fozo, E.M. New Type I Toxin-Antitoxin Families from “Wild” and Laboratory Strains of E. Coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012, 9, 1504–1512. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Tokunaga, N.; Inouye, M.; Phadtare, S. Characterization of LdrA (Long Direct Repeat A) Protein of Escherichia Coli. J. Mol. Microbiol. Biotechnol. 2014, 24, 91–97. [Google Scholar] [CrossRef]
- Fernández de Henestrosa, A.R.; Ogi, T.; Aoyagi, S.; Chafin, D.; Hayes, J.J.; Ohmori, H.; Woodgate, R. Identification of Additional Genes Belonging to the LexA Regulon in Escherichia Coli: Novel LexA-Regulated Genes in E. Coli. Mol. Microbiol. 2002, 35, 1560–1572. [Google Scholar] [CrossRef]
- Weel-Sneve, R.; Kristiansen, K.I.; Odsbu, I.; Dalhus, B.; Booth, J.; Rognes, T.; Skarstad, K.; Bjørås, M. Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities. PLoS Genet. 2013, 9, e1003260. [Google Scholar] [CrossRef]
- Kristiansen, K.I.; Weel-Sneve, R.; Booth, J.A.; Bjørås, M. Mutually Exclusive RNA Secondary Structures Regulate Translation Initiation of DinQ in Escherichia Coli. RNA 2016, 22, 1739–1749. [Google Scholar] [CrossRef]
- Berghoff, B.A.; Wagner, E.G.H. RNA-Based Regulation in Type I Toxin–Antitoxin Systems and Its Implication for Bacterial Persistence. Curr. Genet. 2017, 63, 1011–1016. [Google Scholar] [CrossRef]
- Weaver, K.E.; Walz, K.D.; Heine, M.S. Isolation of a Derivative OfEscherichia Coli–Enterococcus FaecalisShuttle Vector PAM401 Temperature Sensitive for Maintenance InE. Faecalisand Its Use in Evaluating the Mechanism of PAD1par-Dependent Plasmid Stabilization. Plasmid 1998, 40, 225–232. [Google Scholar] [CrossRef]
- Greenfield, T.J.; Ehli, E.; Kirshenmann, T.; Franch, T.; Gerdes, K.; Weaver, K.E. The Antisense RNA of the Par Locus of PAD1 Regulates the Expression of a 33-Amino-Acid Toxic Peptide by an Unusual Mechanism: PAD1 Par Stability Function. Mol. Microbiol. 2002, 37, 652–660. [Google Scholar] [CrossRef]
- Greenfield, T.J.; Franch, T.; Gerdes, K.; Weaver, K.E. Antisense RNA Regulation of the Par Post-Segregational Killing System: Structural Analysis and Mechanism of Binding of the Antisense RNA, RNAII and Its Target, RNAI. Mol. Microbiol. 2001, 42, 527–537. [Google Scholar] [CrossRef]
- Shokeen, S.; Patel, S.; Greenfield, T.J.; Brinkman, C.; Weaver, K.E. Translational Regulation by an Intramolecular Stem-Loop Is Required for Intermolecular RNA Regulation of the Par Addiction Module. JB 2008, 190, 6076–6083. [Google Scholar] [CrossRef]
- Shokeen, S.; Greenfield, T.J.; Ehli, E.A.; Rasmussen, J.; Perrault, B.E.; Weaver, K.E. An Intramolecular Upstream Helix Ensures the Stability of a Toxin-Encoding RNA in Enterococcus Faecalis. JB 2009, 191, 1528–1536. [Google Scholar] [CrossRef]
- Patel, S.; Weaver, K.E. Addiction Toxin Fst Has Unique Effects on Chromosome Segregation and Cell Division in Enterococcus Faecalis and Bacillussubtilis. JB 2006, 188, 5374–5384. [Google Scholar] [CrossRef]
- Weaver, K.E.; Weaver, D.M.; Wells, C.L.; Waters, C.M.; Gardner, M.E.; Ehli, E.A. Enterococcus Faecalis Plasmid PAD1-Encoded Fst Toxin Affects Membrane Permeability and Alters Cellular Responses to Lantibiotics. JB 2003, 185, 2169–2177. [Google Scholar] [CrossRef]
- Brinkman, C.L.; Bumgarner, R.; Kittichotirat, W.; Dunman, P.M.; Kuechenmeister, L.J.; Weaver, K.E. Characterization of the Effects of an RpoC Mutation That Confers Resistance to the Fst Peptide Toxin-Antitoxin System Toxin. J. Bacteriol. 2013, 195, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Preis, H.; Wiedemann, C.; Brantl, S. BsrG/SR4 from Bacillus Subtilis- the First Temperature-Dependent Type I Toxin-Antitoxin System: BsrG/SR4 - Toxin-Antitoxin System in B. Subtilis. Mol. Microbiol. 2012, 83, 579–598. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Brantl, S. One Antitoxin—Two Functions: SR4 Controls Toxin MRNA Decay and Translation. Nucleic Acids Res. 2013, 41, 9870–9880. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Brantl, S.; Strahl, H. Against the Mainstream: The Membrane-Associated Type I Toxin BsrG from B Acillus Subtilis Interferes with Cell Envelope Biosynthesis without Increasing Membrane Permeability: Cellular Toxicity of B. Subtilis Type I Toxin BsrG. Mol. Microbiol. 2015, 98, 651–666. [Google Scholar] [CrossRef]
- Brantl, S.; Müller, P. Toxin–Antitoxin Systems in Bacillus Subtilis. Toxins 2019, 11, 262. [Google Scholar] [CrossRef]
- Sharma, C.M.; Hoffmann, S.; Darfeuille, F.; Reignier, J.; Findeiß, S.; Sittka, A.; Chabas, S.; Reiche, K.; Hackermüller, J.; Reinhardt, R.; et al. The Primary Transcriptome of the Major Human Pathogen Helicobacter Pylori. Nature 2010, 464, 250–255. [Google Scholar] [CrossRef]
- Arnion, H.; Korkut, D.N.; Masachis Gelo, S.; Chabas, S.; Reignier, J.; Iost, I.; Darfeuille, F. Mechanistic Insights into Type I Toxin Antitoxin Systems in Helicobacter Pylori: The Importance of MRNA Folding in Controlling Toxin Expression. Nucleic Acids Res. 2017, 45, 4782–4795. [Google Scholar] [CrossRef]
- El Mortaji, L.; Tejada-Arranz, A.; Rifflet, A.; Boneca, I.G.; Pehau-Arnaudet, G.; Radicella, J.P.; Marsin, S.; De Reuse, H. A Peptide of a Type I Toxin−antitoxin System Induces Helicobacter Pylori Morphological Transformation from Spiral Shape to Coccoids. Proc. Natl. Acad. Sci. USA 2020, 117, 31398–31409. [Google Scholar] [CrossRef]
- Kawano, M.; Oshima, T.; Kasai, H.; Mori, H. Molecular Characterization of Long Direct Repeat (LDR) Sequences Expressing a Stable MRNA Encoding for a 35-Amino-Acid Cell-Killing Peptide and a Cis-Encoded Small Antisense RNA in Escherichia Coli. Mol. Microbiol. 2002, 45, 333–349. [Google Scholar] [CrossRef]
- Pinel-Marie, M.-L.; Brielle, R.; Felden, B. Dual Toxic-Peptide-Coding Staphylococcus Aureus RNA under Antisense Regulation Targets Host Cells and Bacterial Rivals Unequally. Cell Rep. 2014, 7, 424–435. [Google Scholar] [CrossRef]
- Pinel-Marie, M.-L.; Brielle, R.; Riffaud, C.; Germain-Amiot, N.; Polacek, N.; Felden, B. RNA Antitoxin SprF1 Binds Ribosomes to Attenuate Translation and Promote Persister Cell Formation in Staphylococcus Aureus. Nat. Microbiol. 2021, 6, 209–220. [Google Scholar] [CrossRef]
- Krantz, B.A. A Phenylalanine Clamp Catalyzes Protein Translocation through the Anthrax Toxin Pore. Science 2005, 309, 777–781. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The Structure-Mechanism Relationship and Mode of Actions of Antimicrobial Peptides: A Review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]
- Alix, E.; Blanc-Potard, A.-B. Hydrophobic Peptides: Novel Regulators within Bacterial Membrane: Regulatory Membrane Peptides in Bacteria. Mol. Microbiol. 2009, 72, 5–11. [Google Scholar] [CrossRef]
- Cheng, H.-Y.; Soo, V.W.C.; Islam, S.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K. Toxin GhoT of the GhoT/GhoS Toxin/Antitoxin System Damages the Cell Membrane to Reduce Adenosine Triphosphate and to Reduce Growth under Stress: GhoT Damages the Membrane to Increase Resistance. Environ. Microbiol. 2014, 16, 1741–1754. [Google Scholar] [CrossRef]
- Nicolas, I.; Bordeau, V.; Bondon, A.; Baudy-Floc’h, M.; Felden, B. Novel Antibiotics Effective against Gram-Positive and -Negative Multi-Resistant Bacteria with Limited Resistance. PLoS Biol. 2019, 17, e3000337. [Google Scholar] [CrossRef]
- Booth, J.A.; Suganthan, R.; Gaustad, P.; Bjørås, M. Development of DinQ from Escherichia Coli as an Anti-Cell-Envelope Antibiotic. Int. J. Antimicrob. Agents 2015, 45, 196–197. [Google Scholar] [CrossRef]




 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nonin-Lecomte, S.; Fermon, L.; Felden, B.; Pinel-Marie, M.-L. Bacterial Type I Toxins: Folding and Membrane Interactions. Toxins 2021, 13, 490. https://doi.org/10.3390/toxins13070490
Nonin-Lecomte S, Fermon L, Felden B, Pinel-Marie M-L. Bacterial Type I Toxins: Folding and Membrane Interactions. Toxins. 2021; 13(7):490. https://doi.org/10.3390/toxins13070490
Chicago/Turabian StyleNonin-Lecomte, Sylvie, Laurence Fermon, Brice Felden, and Marie-Laure Pinel-Marie. 2021. "Bacterial Type I Toxins: Folding and Membrane Interactions" Toxins 13, no. 7: 490. https://doi.org/10.3390/toxins13070490