Biodiversity of Microorganisms Colonizing the Surface of Polystyrene Samples Exposed to Different Aqueous Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Incubation of PS Samples
2.2. Analytical Methods
2.3. Microbial Community Analysis
2.4. Scanning Electron Microscopy (SEM)
2.5. Nucleotide Sequence Accession Numbers
3. Results
3.1. Physicochemical Characteristics of Aqueous Environments
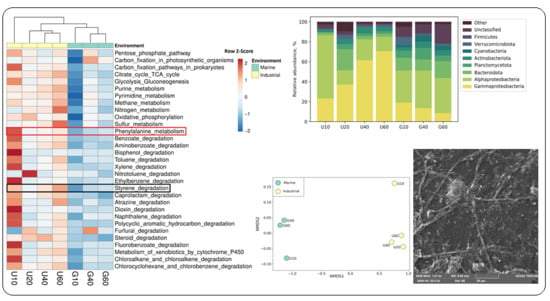
3.2. Analysis of Bacterial Diversity in Biofouling Colonizing PS Specimens in Aqueous Environments
3.3. Prokaryotes Phylogenetic Diversity in Biofilms Formed on PS Samples
3.4. Potential Ability of the Studied Bacterial Communities to Degrade Styrene
3.5. Identification of Bacteria Possessing sty-Operon Genes
3.6. Microscopy of Bacterial Biofilms on PS
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Taxonomic Group, Genus | Industrial | Marine | |||||
|---|---|---|---|---|---|---|---|
| U10 | U20 | U40 | U60 | G10 | G40 | G60 | |
| Actinobacteriota | 2.45 | 4.88 | 6.26 | 4.33 | 3.25 | 6.27 | 5.23 |
| Rhodococcus | 0.01 | 0.01 | 1.19 | 0.04 | 0.04 | 0.00 | 0.01 |
| Microbacterium | 0.05 | 0.00 | 1.82 | 0.03 | 0.11 | 0.01 | 0.00 |
| Mycobacterium | 0.15 | 0.83 | 2.37 | 1.78 | 0.87 | 0.85 | 0.09 |
| Alphaproteobacteria | 63.57 | 14.41 | 20.76 | 14.41 | 32.07 | 37.65 | 35.04 |
| Jannaschia | 0.00 | 0.00 | 0.00 | 0.00 | 0.18 | 0.65 | 1.53 |
| Hyphomonas | 0.01 | 0.03 | 0.04 | 0.02 | 1.56 | 0.19 | 0.03 |
| Limimaricola | 0.00 | 0.00 | 0.00 | 0.00 | 0.24 | 0.81 | 1.15 |
| Ca. Megaira | 0.03 | 1.02 | 0.74 | 0.22 | 0.03 | 0.00 | 0.02 |
| Erythrobacter | 0.03 | 0.03 | 0.03 | 0.00 | 1.26 | 7.20 | 10.82 |
| Methylobacterium | 0.08 | 0.00 | 3.09 | 0.00 | 0.52 | 0.01 | 0.01 |
| Reyranella | 0.17 | 0.97 | 1.96 | 1.47 | 0.00 | 0.00 | 0.00 |
| Brevundimonas | 1.06 | 0.67 | 4.21 | 1.35 | 0.35 | 0.00 | 0.00 |
| Sphingopyxis | 1.51 | 0.17 | 0.13 | 0.17 | 0.01 | 0.00 | 0.01 |
| Novosphingobium | 4.12 | 0.02 | 0.00 | 0.01 | 0.02 | 0.00 | 0.00 |
| Sphingomonas | 4.29 | 0.00 | 0.00 | 0.12 | 0.00 | 0.00 | 0.01 |
| Sphingobium | 6.81 | 0.06 | 0.04 | 0.02 | 1.05 | 0.08 | 0.00 |
| Blastomonas | 40.74 | 0.04 | 0.00 | 0.01 | 0.01 | 0.04 | 0.03 |
| Hyphomicrobium | 0.02 | 0.61 | 1.09 | 1.20 | 0.01 | 0.00 | 0.00 |
| Gammaproteobacteria | 23.09 | 37.08 | 61.53 | 70.54 | 18.95 | 13.49 | 8.51 |
| Pseudomonas | 0.55 | 1.31 | 12.11 | 39.10 | 0.33 | 0.09 | 0.01 |
| Arenimonas | 1.02 | 6.59 | 23.04 | 14.17 | 0.02 | 0.00 | 0.00 |
| Hydrogenophaga | 0.19 | 1.98 | 0.55 | 0.30 | 0.00 | 0.00 | 0.03 |
| Stenotrophomonas | 0.46 | 0.01 | 1.22 | 0.52 | 0.04 | 0.14 | 0.00 |
| Methyloversatilis | 0.48 | 2.66 | 0.06 | 0.19 | 0.00 | 0.00 | 0.00 |
| Acidovorax | 0.47 | 1.82 | 1.73 | 2.75 | 0.00 | 0.00 | 0.00 |
| Dechloromonas | 1.27 | 0.10 | 0.01 | 0.10 | 0.00 | 0.00 | 0.00 |
| Ralstonia | 1.90 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Alcaligenes | 4.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.27 | 0.00 |
| Pseudohongiella | 0.01 | 0.00 | 0.00 | 0.00 | 5.02 | 0.65 | 1.17 |
| Coxiella | 0.00 | 0.00 | 0.00 | 0.00 | 1.16 | 0.99 | 0.08 |
| Planctomycetota | 0.31 | 5.10 | 2.71 | 2.26 | 7.09 | 11.59 | 5.54 |
| Rhodopirellula | 0.00 | 0.00 | 0.00 | 0.00 | 1.40 | 3.17 | 0.35 |
| Planctomicrobium | 0.00 | 0.04 | 0.00 | 0.00 | 3.54 | 2.01 | 0.25 |
| Verrucomicrobiota | 0.39 | 3.82 | 1.00 | 1.66 | 7.18 | 0.80 | 1.70 |
| Pelagicoccus | 0.01 | 0.00 | 0.00 | 0.00 | 4.70 | 0.11 | 0.16 |
| Cerasicoccus | 0.01 | 0.00 | 0.00 | 0.00 | 1.39 | 0.31 | 0.10 |
| Bacteroidota | 2.49 | 20.95 | 3.26 | 3.18 | 12.89 | 11.06 | 17.00 |
| Marinoscillum | 0.02 | 0.00 | 0.00 | 0.00 | 3.22 | 0.37 | 1.73 |
| Maribacter | 0.00 | 0.00 | 0.00 | 0.00 | 0.22 | 1.63 | 1.72 |
| Fulvivirga | 0.00 | 0.00 | 0.00 | 0.00 | 0.61 | 2.13 | 1.99 |
| Tunicatimonas | 0.01 | 0.00 | 0.00 | 0.00 | 0.42 | 1.56 | 3.38 |
| Taeseokella | 0.00 | 0.00 | 0.00 | 0.00 | 1.10 | 0.00 | 0.04 |
| Flavobacterium | 0.24 | 2.36 | 0.91 | 1.02 | 0.00 | 0.00 | 0.00 |
| Chryseolinea | 0.42 | 3.69 | 0.51 | 0.71 | 0.00 | 0.01 | 0.00 |
| OTU | Hit Taxon Name | Hit Strain Name | Accession Number | Similarity, % | % of Total Reads in Each Library | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Marine | Industrial | ||||||||||
| G10 | G40 | G60 | U10 | U20 | U40 | U60 | |||||
| OTU121 | Pseudomonas aeruginosa | JCM 5962T | BAMA01000316 | 100 | 0.05 | 0.01 | 0 | 0 | 0.01 | 0.03 | 1.69 |
| OTU23 | P. putida | NBRC 14164T | AP013070 | 99.8 | 0 | 0 | 0 | 0.05 | 0 | 1.38 | 3.39 |
| OTU4 | P. putida | NBRC 14164T | AP013070 | 100 | 0 | 0 | 0 | 0.07 | 0.01 | 7.99 | 23.29 |
| OTU46 | P. stutzeri | ATCC 17588T | CP002881 | 100 | 0.22 | 0.08 | 0.01 | 0.06 | 0.02 | 0.04 | 5.81 |
| OTU131 | Rhodococcus erythropolis | NBRC 15567T | BCRM01000055 | 100 | 0 | 0 | 0 | 0.004 | 0.01 | 0.83 | 0.02 |
| OTU867 | Sphingopyxis sp. | 113P3 | CP009452 | 100 | 0 | 0 | 0 | 0 | 0.05 | 0.01 | 0.03 |
| OTU167 | S. italica | SC13E-S71T | HE648058 | 99.7 | 0.01 | 0 | 0.01 | 0.59 | 0.01 | 0 | 0.004 |
| OTU550 | S. alaskensis | RB2256T | CP000356 | 99.5 | 0 | 0 | 0 | 0.01 | 0.05 | 0.06 | 0.04 |
| OTU606 | S. fribergensis | Kp5.2T | CP009122 | 100 | 0 | 0 | 0 | 0 | 0.04 | 0.05 | 0.09 |
| OTU362 | Mycolicibacterium hippocampi | BFLP-6T | FN430736 | 100 | 0.43 | 0.49 | 0.04 | 0.01 | 0.004 | 0 | 0.004 |
| OTU71 | Nocardia asteroides | NBRC 15531T | BAFO01000006 | 100 | 0 | 0 | 0 | 0 | 0.64 | 0.15 | 0.11 |
| OTU66 | Altererythrobacter spongiae | HN-Y73T | MG437235 | 99 | 0.36 | 0.37 | 0.62 | 0 | 0 | 0 | 0 |
| OTU878 | Tropicibacter naphthalenivorans | CECT 7648T | CYSE01000022 | 99 | 0.12 | 0.03 | 0.14 | 0 | 0 | 0 | 0 |
| OTU1301 | Marinobacter flavimaris | SW-145T | NR_025799 | 99.6 | 0.018 | 0 | 0 | 0 | 0 | 0 | 0 |







References
- PlasticsEurope. Plastics—The Facts 2019. An Analysis of European Plastics Production, Demand and Waste Data; PlasticsEurope: Brussels, Belgium, 2019; Available online: https://www.plasticseurope.org/application/files/1115/7236/4388/FINAL_web_version_Plastics_the_facts2019_14102019.pdf (accessed on 30 April 2020).
- Law, K.; Moret-Ferguson, S.; Goodwin, D.; Zettler, E.; DeForce, E.; Kukulka, T.; Proskurowski, G. Distribution of surface plastic debris in the eastern Pacific Ocean from an 11-year dataset. Environ. Sci. Technol. 2014, 48, 4732–4738. [Google Scholar] [CrossRef] [PubMed]
- Law, K.L. Plastics in the marine environment. Annu. Rev. Mar. Sci. 2017, 9, 205–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic pollution in the world’s oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef] [Green Version]
- Jambeck, J.R.; Geyer, R.; Wilcox, C.; Siegler, T.R.; Perryman, M.; Andrady, A.; Narayan, R.; Law, K.L. Plastic waste inputs from land into the ocean. Science 2015, 347, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Distribution and importance of microplastics in the marine environment: A review of the sources, fate, effects, and potential solutions. Environ. Int. 2017, 102, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Amaral-Zettler, L.A.; Zettler, E.R.; Slikas, B.; Boyd, G.D.; Melvin, D.W.; Morrall, C.E.; Proskurowski, G.; Mincer, T.J. The biogeography of the Plastisphere: Implications for policy. Front. Ecol. Environ. 2015, 13, 541–546. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Osborn, A.M.; Duhaime, M.B. Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE 2016, 11, e0159289. [Google Scholar] [CrossRef]
- Ivar do Sul, J.A.; Tagg, A.S.; Labrenz, M. Exploring the common denominator between microplastics and microbiology: A scientometric approach. Scientometrics 2018, 117, 2145–2157. [Google Scholar] [CrossRef]
- Costerton, J.W.; Geesey, G.G.; Cheng, K.J. How bacteria stick. Sci. Am. 1978, 238, 86–95. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Marine biofilms: A successful microbial strategy with economic implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef] [Green Version]
- Briand, J.-F.; Barani, A.; Garnier, C.; Réhel, K.; Urvois, F.; LePoupon, C.; Bouchez, A.; Debroas, D.; Bressy, C. Spatio-temporal variations of marine biofilm communities colonizing artificial substrata including antifouling coatings in contrasted French coastal environments. Microb. Ecol. 2017, 74, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Kesy, K.; Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in Baltic Sea mesocosms. Front. Microbiol. 2019, 10, 1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018, 8, 2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogonowski, M.; Motiei, A.; Ininbergs, K.; Hell, E.; Gerdes, Z.; Udekwu, K.I.; Bacsik, Z.; Gorokhova, E. Evidence for selective bacterial community structuring on microplastics. Environ. Microbiol. 2018, 20, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Dussud, C.; Meistertzheim, A.L.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M.L.; et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef]
- Hoellein, T.; Rojas, M.; Pink, A.; Gasior, J.; Kelly, J. Anthropogenic litter in urban freshwater ecosystems: Distribution and microbial interactions. PLoS ONE 2014, 9, e98485. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Zheng, Y.; Yanful, E.K.; Bassi, A.S. A review of plastic waste biodegradation. Crit. Rev. Biotechnol. 2005, 25, 243–250. [Google Scholar] [CrossRef]
- Krueger, M.C.; Seiwert, B.; Prager, A.; Zhang, S.W.; Abel, B.; Harms, H.; Schlosser, D. Degradation of polystyrene and selected analogues by biological Fenton chemistry approaches: Opportunities and limitations. Chemosphere 2017, 173, 520–528. [Google Scholar] [CrossRef]
- Webb, H.; Arnott, J.; Crawford, R.; Ivanova, E. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 2013, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamiya, K.; Sakasita, G.; Ooi, T.; Kinoshita, S. Enzymatic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121. J. Ferment. Bioeng. 1997, 84, 480–482. [Google Scholar] [CrossRef]
- Lyklema, J.; Norde, W.; van Loosdrecht, M.C.M.; Zehnder, A.J.B. Adhesion of bacteria to polystyrene surfaces. Colloids Surf. 1989, 39, 175–187. [Google Scholar] [CrossRef]
- Mor, R.; Sivan, A. Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber. Biodegradation 2008, 19, 851–858. [Google Scholar] [CrossRef]
- Tischler, D. Microbial Styrene Degradation. SpringerBriefs in Microbiology; Springer: Berlin/Heidelberg, Germany, 2015; p. 108. [Google Scholar] [CrossRef]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2017, 38, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, A.; Pourbabaee, A.A.; Alikhani, H.A.; Shabani, F.; Esmaeili, E. Biodegradation of low-density polyethylene (LDPE) by mixed culture of Lysinibacillus xylanilyticus and Aspergillus niger in soil. PLoS ONE 2013, 8, e71720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Chowdhuri, U.R.; Kundu, P.P. Bio-degradation of polyethylene waste by simultaneous use of two bacteria: Bacillus licheniformis for production of bio-surfactant and Lysinibacillus fusiformis for bio-degradation. RSC Adv. 2016, 6, 2982–2992. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Wu, W.-M.; Zhao, J.; Song, Y.; Gao, L.; Yang, R.; Jiang, L. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 2. Role of gut microorganisms. Environ. Sci. Technol. 2015, 49, 12087–12093. [Google Scholar] [CrossRef]
- Oelschlägel, M.; Zimmerling, J.; Tischler, D. A Review: The styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds. Front. Microbiol. 2018, 9, 490. [Google Scholar] [CrossRef]
- Mooney, A.; Ward, P.G.; O’Connor, K.E. Microbial degradation of styrene: Biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl. Microbiol. Biotechnol. 2006, 72, 1–10. [Google Scholar] [CrossRef]
- Warhurst, A.M.; Fewson, C.A. Microbial metabolism and biotransformation of styrene. J. Appl. Bacteriol. 1994, 77, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Grbić-Galić, D.; Churchman-Eisel, N.; Mraković, I. Microbial transformation of styrene by anaerobic consortia. J. Appl. Bacteriol. 1990, 69, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Tourova, T.P.; Sokolova, D.S.; Nazina, T.N.; Laptev, A.B. Phylogenetic diversity of microbial communities from the surface of polyethylene terephthalate materials exposed to different water environments. Microbiology 2020, 89, 96–106. [Google Scholar] [CrossRef]
- Laptev, A.B.; Nikolaev, E.V.; Kurchev, E.V.; Goryashnik, Y.S. Features of biodegradation of thermoplastics based on polyesters in different climatic zones. Trudy VIAM 2019, 7, 84–91. (In Russian) [Google Scholar] [CrossRef]
- Bonch-Osmolovskaya, E.A.; Miroshnichenko, M.L.; Lebedinsky, A.V.; Chernyh, N.A.; Nazina, T.N.; Ivoilov, V.S.; Belyaev, S.S.; Boulygina, E.S.; Lysov, Y.P.; Perov, A.N.; et al. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl. Environ. Microbiol. 2003, 69, 6143–6151. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [Green Version]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Community Ecology Package. R Package. Version 2.5–6. 2019. Available online: https://cran.ism.ac.jp/web/packages/vegan/vegan.pdf (accessed on 1 September 2019).
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER. Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.J.; Crécy-Lagard, V.D.; Zallot, R. Gene Graphics: A genomic neighborhood data visualization web application. Bioinformatics 2018, 34, 1406–1408. [Google Scholar] [CrossRef] [PubMed]
- Syranidou, E.; Karkanorachaki, K.; Amorotti, F.; Repouskou, E.; Kroll, K.; Kolvenbach, B.; Corvini, P.F.-X.; Fava, F.; Kalogerakis, N. Development of tailored indigenous marine consortia for the degradation of naturally weathered polyethylene films. PLoS ONE 2017, 12, e0183984. [Google Scholar] [CrossRef] [Green Version]
- Otto, K.; Hofstetter, K.; Röthlisberger, M.; Witholt, B.; Schmid, A. Biochemical characterization of StyAB from Pseudomonas sp. strain VLB120 as a two-component flavin-diffusible monooxygenase. J. Bacteriol. 2004, 186, 5292–5302. [Google Scholar] [CrossRef] [Green Version]
- Kohler, K.A.; Ruckert, C.; Schatschneider, S.; Vorholter, F.J.; Szczepanowski, R.; Blank, L.M.; Niehaus, K.; Goesmann, A.; Puhler, A.; Kalinowski, J.; et al. Complete genome sequence of Pseudomonas sp. strain VLB120 a solvent tolerant, styrene degrading bacterium, isolated from forest soil. J. Biotechnol. 2013, 168, 729–730. [Google Scholar] [CrossRef]
- Lee, O.O.; Chung, H.C.; Yang, J.; Wang, Y.; Dash, S.; Wang, H.; Qian, P.-Y. Molecular techniques revealed highly diverse microbial communities in natural marine biofilms on polystyrene dishes for invertebrate larval settlement. Microb. Ecol. 2014, 68, 81–93. [Google Scholar] [CrossRef]
- Pu, W.; Cui, C.; Guo, C.; Wu, Z.L. Characterization of two styrene monooxygenases from marine microbes. Enzym. Microb. Technol. 2018, 112, 29–34. [Google Scholar] [CrossRef]
- Elifantz, H.; Horn, G.; Ayon, M.; Cohen, Y.; Minz, D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013, 85, 348–357. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slightom, R.N.; Buchan, A. Surface colonization by marine roseobacters: Integrating genotype and phenotype. Appl. Environ. Microbiol. 2009, 75, 6027–6037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner-Döbler, I.; Biebl, H. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 2006, 60, 255–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catão, E.C.P.; Pollet, T.; Misson, B.; Garnier, C.; Ghiglione, J.-F.; Barry-Martinet, R.; Maintenay, M.; Bressy, C.; Briand, J.-F. Shear stress as a major driver of marine biofilm communities in the NW Mediterranean Sea. Front. Microbiol. 2019, 10, 1768. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Blache, Y.; Stokes, K.R.; Briand, J.F. Marine biofilms on artificial surfaces: Structure and dynamics. Environ. Microbiol. 2013, 15, 2879–2893. [Google Scholar] [CrossRef]
- Izumi, H.; Sagulenko, E.; Webb, R.I.; Fuerst, J.A. Isolation and diversity of planctomycetes from the sponge Niphates sp., seawater, and sediment of Moreton Bay, Australia. Antonie Leeuwenhoek 2013, 104, 533–546. [Google Scholar] [CrossRef]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.A.; Fong, A.A.; Thomas, K.A.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef] [Green Version]
- Rickard, A.H.; Gilbert, P.; Handley, P.S. Influence of growth environment on coaggregation between freshwater biofilm bacteria. J. Appl. Microbiol. 2004, 96, 1367–1373. [Google Scholar] [CrossRef]
- Limoli, D.; Jones, C.; Wozniak, D. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Oelschlägel, M.; Zimmerling, J.; Schlömann, M.; Tischler, D. Styrene oxide isomerase of Sphingopyxis sp. Kp5.2. Microbiology (UK) 2014, 160, 2481–2491. [Google Scholar] [CrossRef]
- Tischler, D.; Kermer, R.; Gröning, J.A.D.; Kaschabek, S.R.; van Berkel, W.J.H.; Schlömann, M. StyA1 and StyA2B from Rhodococcus opacus 1CP: A multifunctional styrene monooxygenase system. J. Bacteriol. 2010, 192, 5220–5227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, K.; Buckley, C.M.; Hartmans, S.; Dobson, A.D. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 1995, 61, 544–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Qiao, J.; Liu, Y.; Wu, Z.-L. Styrene monooxygenase from Pseudomonas sp. LQ26 catalyzes the asymmetric epoxidation of both conjugated and unconjugated alkenes. J. Mol. Catal. B Enzym. 2010, 67, 236–241. [Google Scholar] [CrossRef]
- Gassner, G.T. The styrene monooxygenase system. Methods Enzymol. 2019, 620, 423–453. [Google Scholar] [CrossRef]





| Habitat, Specimen | Total Salinity, mg/L | pH | Water Temperature, °C | Total Hardness, mg-eq/L | Total Iron, mg/L | Alkalinity, mg-eq/kg | COD 1, mg O₂/dm3 |
|---|---|---|---|---|---|---|---|
| Industrial Water | |||||||
| U10 | 1098 | 8.5 ± 0.1 | 24.0–27.0 | 13.8 | 0.45 | 6.4 | 101 |
| U20 | 1100 | 8.5 ± 0.1 | 21.5–25.5 | 12.6 | 0.37 | 6.5 | 103 |
| U40 | 918 | 8.4 ± 0.1 | 19.0–23.0 | 9.8 | 0.21 | 4.3 | 72 |
| U60 | 676 | 8.1 ± 0.1 | 14.0–18.0 | 6.7 | 0.61 | 2.4 | 90 |
| Black Sea 2, G10, G40, G60 | 18,600 | 7.8 ± 0.1 | 26.8–25.1 | 66.9 | Nd 3 | Nd | Nd |
| Parameter | Libraries of 16S rRNA Gene Fragments | ||||||
|---|---|---|---|---|---|---|---|
| Industrial | Marine | ||||||
| U10 | U20 | U40 | U60 | G10 | G40 | G60 | |
| Duration of exposure, days | 10 | 20 | 40 | 60 | 10 | 40 | 60 |
| Number of sequences (reads) | 29,466 | 56,405 | 37,552 | 29,546 | 27,243 | 24,261 | 44,303 |
| Number of OTUs | 474 | 703 | 525 | 483 | 558 | 443 | 569 |
| Shannon‒Weaver diversity index (H) | 3.5 | 5.7 | 4.6 | 4.1 | 5.2 | 4.3 | 5.2 |
| Inverse Simpson diversity index (1/S) | 8.1 | 138.9 | 32.6 | 17.1 | 68.5 | 14.0 | 54.6 |
| Evenness (H/Hmax) | 0.56 | 0.87 | 0.72 | 0.67 | 0.82 | 0.71 | 0.81 |
| Berger‒Parker dominance index (D) | 0.32 | 0.04 | 0.10 | 0.19 | 0.06 | 0.21 | 0.09 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tourova, T.; Sokolova, D.; Nazina, T.; Grouzdev, D.; Kurshev, E.; Laptev, A. Biodiversity of Microorganisms Colonizing the Surface of Polystyrene Samples Exposed to Different Aqueous Environments. Sustainability 2020, 12, 3624. https://doi.org/10.3390/su12093624
Tourova T, Sokolova D, Nazina T, Grouzdev D, Kurshev E, Laptev A. Biodiversity of Microorganisms Colonizing the Surface of Polystyrene Samples Exposed to Different Aqueous Environments. Sustainability. 2020; 12(9):3624. https://doi.org/10.3390/su12093624
Chicago/Turabian StyleTourova, Tatyana, Diyana Sokolova, Tamara Nazina, Denis Grouzdev, Eugeni Kurshev, and Anatoly Laptev. 2020. "Biodiversity of Microorganisms Colonizing the Surface of Polystyrene Samples Exposed to Different Aqueous Environments" Sustainability 12, no. 9: 3624. https://doi.org/10.3390/su12093624