A Spatial-Spectral Classification Method Based on Deep Learning for Controlling Pelagic Fish Landings in Chile
Abstract
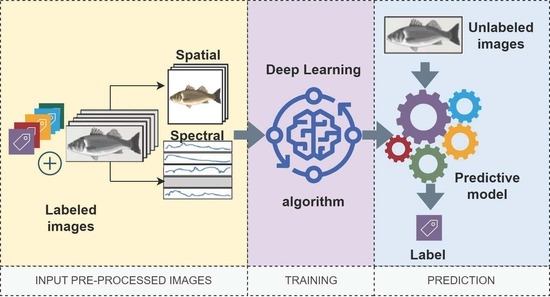
:1. Introduction
- A database of spectral signatures of several fish species;
- A spatial-spectral classification method for the automatic identification of pelagic species
2. Related Work
3. Materials and Methods
3.1. Sample Preparation: Pelagic Fish
3.2. Hyperspectral Imaging Setup and Acquisition
3.3. Data Prepossessing
3.3.1. Extraction of Spectral Signatures
3.3.2. Extraction of RGB Images
3.4. Design of Classifiers for Pelagic Fish Species
3.4.1. Data Augmentation for Improving Classification
3.4.2. Convolutional Neural Network (CNN)
3.4.3. Performance Evaluation
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNN | Convolutional Neural Network |
| DL | Deep Learning |
| FAO | Food and Agriculture Organization |
| HOG | Histogram of Oriented Gradient |
| HS | Hyperspectral |
| HSI | Hyperspectral Imaging |
| NIR | Near-Infrared |
| RGB | Red–Green–Blue |
| SERNAPESCA | Servicio Nacional de Pesca y Acuicultura |
| SVM | Support Vector Machine |
| VIS-NIR | Visible and Near-Infrared |
| HOG | Histogram of Oriented Gradients |
| UdeC | Universidad de Concecpión |
| RoI | Region of Interest |
| SGD | Stochastic Gradient Descent |
References
- Manz, J.; Nsoga, J.; Diazenza, J.B.; Sita, S.; Bakana, G.M.B.; Francois, A.; Ndomou, M.; Gouado, I.; Mamonekene, V. Nutritional composition, heavy metal contents and lipid quality of five marine fish species from Cameroon coast. Heliyon 2023, 9, e14031. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. The Status of World Fisheries and Aquaculture. Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018. [Google Scholar]
- Nahuelhual, L.; Saavedra, G.; Blanco, G.; Wesselink, E.; Campos, G.; Vergara, X. On super fishers and black capture: Images of illegal fishing in artisanal fisheries of southern Chile. Mar. Policy 2018, 95, 36–45. [Google Scholar] [CrossRef]
- Gunnar, S.; Rosenberg, A.A. Combining control measures for more effective management of fisheries under uncertainty: Quotas, effort limitation and protected areas. Philos. Trans. R. Soc. London. Ser. Biol. Sci. 2005, 360, 133–146. [Google Scholar]
- Ye, Y.; Gutierrez, N.L. Ending fishery overexploitation by expanding from local successes to globalized solutions. Nat. Ecol. Evol. 2017, 1, 179. [Google Scholar] [CrossRef]
- Urban, P.; Bekkevold, D.; Degel, H.; Hansen, B.; Jacobsen, M.; Nielsen, A.; Nielsen, E. Scaling from eDNA to biomass: Controlling allometric relationships improves precision in bycatch estimation. ICES J. Mar. Sci. 2023, 80, 1066–1078. [Google Scholar] [CrossRef]
- SERNAPESCA. SERNAPESCA Informes de Gestión. Available online: http://www.sernapesca.cl/informes/resultados-gestion (accessed on 25 November 2019).
- Beaudreau, A.H.; Levin, P.S.; Norman, K.C. Using folk taxonomies to understand stakeholder perceptions for species conservation. Conserv. Lett. 2011, 4, 451–463. [Google Scholar] [CrossRef]
- Rojo, M.M.; Noronha, T.D. Low-technology industries and regional innovation systems: The salmon industry in Chile. J. Spat. Organ. Dyn. 2016, 4, 314–329. [Google Scholar]
- Plotnek, E.; Paredes, F.; Galvez, M.; Pérez-Ramírez, M. From unsustainability to MSC certification: A case study of the artisanal Chilean South Pacific hake fishery. Rev. Fish. Sci. Aquac. 2016, 24, 230–243. [Google Scholar] [CrossRef]
- Schaap, R.J.; Gonzalez-Poblete, E.; Aedo, K.L.S.; Diekert, F. Risk, Restrictive Quotas, and Income Smoothing; Technical Report; CEE-M, University of Montpellier: Montpellier, France, 2022. [Google Scholar]
- Fischer, J. Fish Identification Tools for Biodiversity and Fisheries Assessments: Review and Guidance for Decision-Makers; FAO Fisheries and Aquaculture Technical Paper; FAO: Rome, Italy, 2014; p. 107. [Google Scholar]
- Bendall, C.; Hiebert, S.; Mueller, G. Experiments in Situ Fish Recognition Systems Using Fish Spectral and Spatial Signatures. 1999. Available online: https://pubs.usgs.gov/publication/ofr99104 (accessed on 25 November 2019).
- Hossain, E.; Alam, S.M.S.; Ali, A.A.; Amin, M.A. Fish activity tracking and species identification in underwater video. In Proceedings of the 2016 5th International Conference on Informatics, Electronics and Vision (ICIEV), Dhaka, Bangladesh, 13–14 May 2016; pp. 62–66. [Google Scholar] [CrossRef]
- Spampinato, C.; Giordano, D.; Di Salvo, R.; Chen-Burger, Y.H.J.; Fisher, R.B.; Nadarajan, G. Automatic fish classification for underwater species behavior understanding. In Proceedings of the 1st ACM International Workshop on Analysis and Retrieval of Tracked Events and Motion in Imagery Streams, ARTEMIS ’10, New York, NY, USA, 30 September–1 October 2010; pp. 45–50. [Google Scholar] [CrossRef]
- Hu, J.; Li, D.; Duan, Q.; Han, Y.; Chen, G.; Si, X. Fish Species Classification by Color, Texture and Multi-class Support Vector Machine Using Computer Vision. Comput. Electron. Agric. 2012, 88, 133–140. [Google Scholar] [CrossRef]
- White, D.; Svellingen, C.; Strachan, N. Automated measurement of species and length of fish by computer vision. Fish. Res. 2006, 80, 203–210. [Google Scholar] [CrossRef]
- Storbeck, F.; Daan, B. Fish species recognition using computer vision and a neural network. Fish. Res. 2001, 51, 11–15. [Google Scholar] [CrossRef]
- Cheng, J.H.; Sun, D.W. Hyperspectral imaging as an effective tool for quality analysis and control of fish and other seafoods: Current research and potential applications. Trends Food Sci. Technol. 2014, 37, 78–91. [Google Scholar] [CrossRef]
- Cheng, J.H.; Qu, J.H.; Sun, D.W.; Zeng, X.A. Visible/near-infrared hyperspectral imaging prediction of textural firmness of grass carp (ctenopharyngodon idella) as affected by frozen storage. Food Res. Int. 2014, 56, 190–198. [Google Scholar] [CrossRef]
- Sivertsen, A.H.; Heia, K.; Hindberg, K.; Godtliebsen, F. Automatic nematode detection in cod fillets (Gadus morhua L.) by hyperspectral imaging. J. Food Eng. 2012, 111, 675–681. [Google Scholar] [CrossRef]
- Costa, C.; Antonucci, F.; Menesatti, P.; Pallottino, F.; Boglione, C.; Cataudella, S. An advanced colour calibration method for fish freshness assessment: A comparison between standard and passive refrigeration modalities. Food Bioprocess Technol. 2013, 6, 2190–2195. [Google Scholar] [CrossRef]
- Ramírez, D.; Pezoa, J.E. Spectral vision system for discriminating small pelagic species caught by small-scale fishing. In Proceedings of the Infrared Sensors, Devices, and Applications VIII; LeVan, P.D., Wijewarnasuriya, P., D’Souza, A.I., Eds.; International Society for Optics and Photonics, SPIE: San Diego, CA, USA, 2018; Volume 10766, pp. 169–179. [Google Scholar] [CrossRef]
- Rathi, D.; Jain, S.; Indu, S. Underwater fish species classification using convolutional neural network and deep learning. In Proceedings of the 2017 9th International Conference on Advances in Pattern Recognition (ICAPR), Bangalore, India, 27–30 December 2017; pp. 1–6. [Google Scholar]
- Deep, B.V.; Dash, R. Underwater fish species recognition using deep learning techniques. In Proceedings of the 2019 6th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 7–8 March 2019; pp. 665–669. [Google Scholar]
- Ulucan, O.; Karakaya, D.; Turkan, M. A large-scale dataset for fish segmentation and classification. In Proceedings of the 2020 Innovations in Intelligent Systems and Applications Conference (ASYU), Istanbul, Turkey, 15–17 October 2020; pp. 1–5. [Google Scholar]
- Alsmadi, M.K.; Almarashdeh, I. A survey on fish classification techniques. J. King Saud-Univ.-Comput. Inf. Sci. 2022, 34, 1625–1638. [Google Scholar] [CrossRef]
- Deka, J.; Laskar, S.; Baklial, B. Automated Freshwater Fish Species Classification using Deep CNN. J. Inst. Eng. India Ser. B 2023, 104, 1–19. [Google Scholar] [CrossRef]
- Song, H.; Yang, W.W.; Dai, S.; Du, L.; Sun, Y.C. Using dual-channel CNN to classify hyperspectral image based on spatial-spectral information. Math. Biosci. Eng. 2020, 17, 3450–3477. [Google Scholar] [CrossRef]
- Chen, L.; Wei, Z.; Xu, Y. A lightweight spectral–spatial feature extraction and fusion network for hyperspectral image classification. Remote. Sens. 2020, 12, 1395. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, D.; Gu, J.; Süsstrunk, S. What is the space of spectral sensitivity functions for digital color cameras? In Proceedings of the 2013 IEEE Workshop on Applications of Computer Vision (WACV), Clearwater Beach, FL, USA, 15–17 January 2013; pp. 168–179. [Google Scholar] [CrossRef]
- Liu, Y.; Pu, H.; Sun, D.W. Efficient extraction of deep image features using convolutional neural network (CNN) for applications in detecting and analysing complex food matrices. Trends Food Sci. Technol. 2021, 113, 193–204. [Google Scholar] [CrossRef]
- Naseri, M.N.; Agrawal, A.P. Impact of transfer learning on siamese networks for face recognition with few images per class. In Proceedings of the 2021 Asian Conference on Innovation in Technology (ASIANCON), Pune, India, 27–29 August 2021; pp. 1–5. [Google Scholar]
- Siri, C.S. Enhancing cartoon recognition in real time: Comparative analysis of CNN, ResNet50, and VGG16 deep learning models. In Proceedings of the 2023 2nd International Conference on Augmented Intelligence and Sustainable Systems (ICAISS), Trichy, India, 23–25 August 2023; pp. 114–121. [Google Scholar]
- Soares, L.; Botelho, S.; Nagel, R.; Drews, P.L. A visual inspection proposal to identify corrosion levels in marine vessels using a deep neural network. In Proceedings of the 2021 Latin American Robotics Symposium (LARS), 2021 Brazilian Symposium on Robotics (SBR), and 2021 Workshop on Robotics in Education (WRE), Natal, Brazil, 11–15 October 2021; pp. 222–227. [Google Scholar]
- Mujtaba, D.F.; Mahapatra, N.R. A study of feature importance in fish species prediction neural networks. In Proceedings of the 2022 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 14–16 December 2022; pp. 1547–1550. [Google Scholar]
- Mujtaba, D.F.; Mahapatra, N.R. Fish species classification with data augmentation. In Proceedings of the 2021 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 15–17 December 2021; pp. 1588–1593. [Google Scholar]
- Yin, C.; Zhu, Y.; Liu, S.; Fei, J.; Zhang, H. Enhancing network intrusion detection classifiers using supervised adversarial training. J. Supercomput. 2020, 76, 6690–6719. [Google Scholar] [CrossRef]
- Ben Tamou, A.; Benzinou, A.; Nasreddine, K. Targeted data augmentation and hierarchical classification with deep learning for fish species identification in underwater images. J. Imaging 2022, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, S.; Singh, R. Convolutional neural network: An overview and application in image classification. In Proceedings of the 3rd International Conference on Sustainable Computing: SUSCOM 2021; Springer: Singapore, 2022; pp. 145–153. [Google Scholar]
- Ahmed, F.; Basak, B.; Chakraborty, S.; Karmokar, T.; Reza, A.W.; Imam, O.T.; Arefin, M.S. Developing a classification CNN model to classify different types of fish. In Proceedings of the Intelligent Computing & Optimization: Proceedings of the 5th International Conference on Intelligent Computing and Optimization 2022 (ICO2022); Springer: Cham, Germany, 2022; pp. 529–539. [Google Scholar]
- Zhang, P.; He, J.; Huang, W.; Zhang, J.; Yuan, Y.; Chen, B.; Yang, Z.; Xiao, Y.; Yuan, Y.; Wu, C.; et al. Water Pipeline Leak Detection Based on a Pseudo-Siamese Convolutional Neural Network: Integrating Handcrafted Features and Deep Representations. Water 2023, 15, 1088. [Google Scholar] [CrossRef]
- Guo, X.; Zhao, X.; Liu, Y.; Li, D. Underwater sea cucumber identification via deep residual networks. Inf. Process. Agric. 2019, 6, 307–315. [Google Scholar] [CrossRef]
- Garbin, C.; Zhu, X.; Marques, O. Dropout vs. batch normalization: An empirical study of their impact to deep learning. Multimed. Tools Appl. 2020, 79, 12777–12815. [Google Scholar] [CrossRef]
- Jose, J.A.; Kumar, C.S.; Sureshkumar, S. Tuna classification using super learner ensemble of region-based CNN-grouped 2D-LBP models. Inf. Process. Agric. 2022, 9, 68–79. [Google Scholar] [CrossRef]
- Salazar, J.J.; Garland, L.; Ochoa, J.; Pyrcz, M.J. Fair train-test split in machine learning: Mitigating spatial autocorrelation for improved prediction accuracy. J. Pet. Sci. Eng. 2022, 209, 109885. [Google Scholar] [CrossRef]
- Ahmed, M.A.; Hossain, M.S.; Rahman, W.; Uddin, A.H.; Islam, M.T. An advanced Bangladeshi local fish classification system based on the combination of deep learning and the internet of things (IoT). J. Agric. Food Res. 2023, 14, 100663. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Chen, S.; Webb, G.I.; Liu, L.; Ma, X. A novel selective naïve Bayes algorithm. Knowl.-Based Syst. 2020, 192, 105361. [Google Scholar] [CrossRef]
- Chauhan, V.K.; Dahiya, K.; Sharma, A. Problem formulations and solvers in linear SVM: A review. Artif. Intell. Rev. 2019, 52, 803–855. [Google Scholar] [CrossRef]
- Pant, N.; Bal, B.K. Improving Nepali ocr performance by using hybrid recognition approaches. In Proceedings of the 2016 7th International Conference on Information, Intelligence, Systems & Applications (IISA), Chalkidiki, Greece, 13–15 July 2016; pp. 1–6. [Google Scholar]
- Nugroho, K.A. A comparison of handcrafted and deep neural network feature extraction for classifying optical coherence tomography (OCT) images. In Proceedings of the 2018 2nd International Conference on Informatics and Computational Sciences (ICICoS), Semarang, Indonesia, 30–31 October 2018; pp. 1–6. [Google Scholar]
- Kernbach, J.M.; Staartjes, V.E. Foundations of machine learning-based clinical prediction modeling: Part II—Generalization and overfitting. Mach. Learn. Clin. Neurosci. Found. Appl. 2022, 134, 15–21. [Google Scholar]










| Species | Image | Provided Samples | Augmented Samples | Number of Samples |
|---|---|---|---|---|
| Engraulis ringens (Anchoveta) |  | 58 | 942 | 1000 |
| Merluccius gayi (Merluza) |  | 75 | 925 | 1000 |
| Normanichthys crockeri (Mote) |  | 341 | 659 | 1000 |
| Stromateus stellatus (Pampanito) |  | 42 | 958 | 1000 |
| Strangomera bentincki (Sardina) |  | 128 | 872 | 1000 |
| Total number of samples | 5000 |
| Classifier | Accuracy (%) | Precision (%) | Recall (%) | F1-Value (%) |
|---|---|---|---|---|
| CNN (Spectral) | 92.90 | 95.11 | 92.90 | 93.99 |
| CNN (RGB) | 57.10 * | 55.53 * | 57.10 * | 56.30 * |
| CNN (RGB + Spectral) | 94.26 | 95.58 | 94.26 | 94.92 |
| SVM (Spectral) | 87.10 | 87.13 * | 87.10 | 87.11 |
| SVM (RGB) | 57.66 * | 59.02 * | 57.66 * | 58.33 * |
| SVM (RGB + Spectral) | 74.94 * | 76.90 * | 74.94 * | 75.91 * |
| SVM (HOG) | 62.58 * | 64.52 * | 62.58 * | 63.54 * |
| SVM (HOG + Spectral) | 87.68 | 87.71 * | 87.68 | 87.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pezoa, J.E.; Ramírez, D.A.; Godoy, C.A.; Saavedra, M.F.; Restrepo, S.E.; Coelho-Caro, P.A.; Flores, C.A.; Pérez, F.G.; Torres, S.N.; Urbina, M.A. A Spatial-Spectral Classification Method Based on Deep Learning for Controlling Pelagic Fish Landings in Chile. Sensors 2023, 23, 8909. https://doi.org/10.3390/s23218909
Pezoa JE, Ramírez DA, Godoy CA, Saavedra MF, Restrepo SE, Coelho-Caro PA, Flores CA, Pérez FG, Torres SN, Urbina MA. A Spatial-Spectral Classification Method Based on Deep Learning for Controlling Pelagic Fish Landings in Chile. Sensors. 2023; 23(21):8909. https://doi.org/10.3390/s23218909
Chicago/Turabian StylePezoa, Jorge E., Diego A. Ramírez, Cristofher A. Godoy, María F. Saavedra, Silvia E. Restrepo, Pablo A. Coelho-Caro, Christopher A. Flores, Francisco G. Pérez, Sergio N. Torres, and Mauricio A. Urbina. 2023. "A Spatial-Spectral Classification Method Based on Deep Learning for Controlling Pelagic Fish Landings in Chile" Sensors 23, no. 21: 8909. https://doi.org/10.3390/s23218909