Transfer Learning Approach for Classification of Histopathology Whole Slide Images
Abstract
:1. Introduction
- i.
- All of the images in the Kimia Path24 database were used for training and testing purposes and were further classified into 24 classes for grayscale histopathology images.
- ii.
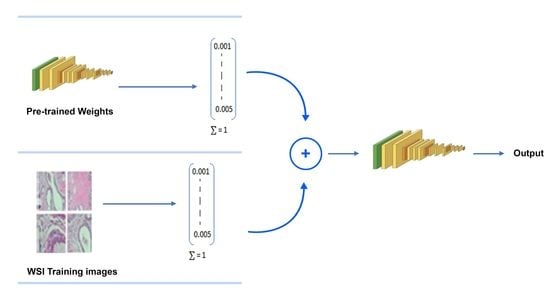
- Training the entire VGG16 and Inception-V3 [8,9] models from scratch after transferring the pre-trained weights of the same model has improved classification accuracy as compared to fine-tuning (by training the last few layers of the base network) or using high level feature extractor techniques for the classification of grayscale images in the Path24 dataset.
- iii.
- The proposed pre-trained CNN models have fully automated the end-to-end structure and do not need any hand-made feature extraction methods.
2. Related Works
3. Material and Methods
3.1. Proposed Model
- Inception-V3 and VGG16 are evaluated for classifying histopathology images automatically.
- The classification effectiveness of purposed pre-trained models is tested by infusing the features vectors from pre-trained network with image pixels normalized. We used grayscale histopathology images.
3.2. Dataset
3.3. Accuracy Calculation
4. Experiments and Results
4.1. Experimental Setup
4.2. Results
4.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep learning for whole slide image analysis: An overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Mehra, R. Effect of layer-wise fine-tuning in magnification-dependent classification of breast cancer histopathological image. Vis. Comput. 2020, 36, 1755–1769. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Huang, Z.; Pan, Z.; Lei, B. Transfer learning with deep convolutional neural network for SAR target classification with limited labeled data. Remote Sens. 2017, 9, 907. [Google Scholar] [CrossRef] [Green Version]
- Babaie, M.; Kalra, S.; Sriram, A.; Mitcheltree, C.; Zhu, S.; Khatami, A.; Rahnamayan, S.; Tizhoosh, H.R. Classification and retrieval of digital pathology scans: A new dataset. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 8–16. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Alhindi, T.J.; Kalra, S.; Ng, K.H.; Afrin, A.; Tizhoosh, H.R. Comparing LBP, HOG and deep features for classification of histopathology images. In Proceedings of the 2018 International Joint Conference On Neural Networks (IJCNN), Rio, Brazil, 8–13 July 2018; pp. 1–7. [Google Scholar]
- Alinsaif, S.; Lang, J. Histological image classification using deep features and transfer learning. In Proceedings of the 2020 17th Conference on Computer and Robot Vision (CRV), Ottawa, ON, Canada, 13–15 May 2020; pp. 101–108. [Google Scholar]
- Caicedo, J.C.; González, F.A.; Romero, E. Content-based histopathology image retrieval using a kernel-based semantic annotation framework. J. Biomed. Inform. 2011, 44, 519–528. [Google Scholar] [CrossRef] [Green Version]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef] [Green Version]
- Pantanowitz, L.; Valenstein, P.N.; Evans, A.J.; Kaplan, K.J.; Pfeifer, J.D.; Wilbur, D.C.; Collins, L.C.; Colgan, T.J. Review of the current state of whole slide imaging in pathology. J. Pathol. Inform. 2011, 2, 36. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Ma, Y.; Shi, H.; Zhao, Y. Size-scalable content-based histopathological image retrieval from database that consists of WSIs. IEEE J. Biomed. Health Inform. 2017, 22, 1278–1287. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Ma, Y.; Shi, H.; Zhao, Y. Histopathological whole slide image analysis using context-based CBIR. IEEE Trans. Med. Imaging 2018, 37, 1641–1652. [Google Scholar] [CrossRef]
- Ghaznavi, F.; Evans, A.; Madabhushi, A.; Feldman, M. Digital imaging in pathology: Whole-slide imaging and beyond. Annu. Rev. Pathol. Mech. Dis. 2013, 8, 331–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madabhushi, A. Digital pathology image analysis: Opportunities and challenges. Imaging Med. 2009, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, R.; Strayer, D.S.; Rubin, E. Rubin’s Pathology: Clinicopathologic Foundations of Medicine; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008. [Google Scholar]
- Babenko, A.; Lempitsky, V. Aggregating local deep features for image retrieval. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1269–1277. [Google Scholar]
- Chatfield, K.; Simonyan, K.; Vedaldi, A.; Zisserman, A. Return of the devil in the details: Delving deep into convolutional nets. arXiv 2014, arXiv:1405.3531. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar]
- Wan, J.; Wang, D.; Hoi, S.C.H.; Wu, P.; Zhu, J.; Zhang, Y.; Li, J. Deep learning for content-based image retrieval: A comprehensive study. In Proceedings of the 22nd ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014; pp. 157–166. [Google Scholar]
- Khatami, A.; Babaie, M.; Khosravi, A.; Tizhoosh, H.R.; Nahavandi, S. Parallel deep solutions for image retrieval from imbalanced medical imaging archives. Appl. Soft Comput. 2018, 63, 197–205. [Google Scholar] [CrossRef]
- Cruz-Roa, A.A.; Ovalle, J.E.A.; Madabhushi, A.; Osorio, F.A.G. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; pp. 403–410. [Google Scholar]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar]
- Zheng, Y.; Jiang, Z.; Xie, F.; Zhang, H.; Ma, Y.; Shi, H.; Zhao, Y. Feature extraction from histopathological images based on nucleus-guided convolutional neural network for breast lesion classification. Pattern Recognit. 2017, 71, 14–25. [Google Scholar] [CrossRef]
- Weston, J.; Bengio, S.; Usunier, N. Large scale image annotation: Learning to rank with joint word-image embeddings. Mach. Learn. 2010, 81, 21–35. [Google Scholar] [CrossRef]
- Seide, F.; Li, G.; Yu, D. Conversational speech transcription using context-dependent deep neural networks. In Proceedings of the Twelfth Annual Conference of the International Speech Communication Association, Florence Italy, 28–31 August 2011. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Domain adaptation for large-scale sentiment classification: A deep learning approach. In Proceedings of the ICML, Bellevue, WA, USA, 28 June–2 July 2011. [Google Scholar]
- Boulanger-Lewandowski, N.; Bengio, Y.; Vincent, P. Modeling temporal dependencies in high-dimensional sequences: Application to polyphonic music generation and transcription. arXiv 2012, arXiv:1206.6392. [Google Scholar]
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 142–158. [Google Scholar] [CrossRef]
- Kumar, M.D.; Babaie, M.; Tizhoosh, H.R. Deep barcodes for fast retrieval of histopathology scans. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar]
- Kieffer, B.; Babaie, M.; Kalra, S.; Tizhoosh, H.R. Convolutional neural networks for histopathology image classification: Training vs. using pre-trained networks. In Proceedings of the 2017 Seventh International Conference on Image Processing Theory, Tools and Applications (IPTA), Montreal, QC, Canada, 28 November–1 December 2017; pp. 1–6. [Google Scholar]
- Saha, M.; Chakraborty, C.; Racoceanu, D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput. Med. Imaging Graph. 2018, 64, 29–40. [Google Scholar] [CrossRef]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Huang, X.; Eric, I.; Chang, C.; Xu, Y. Constrained deep weak supervision for histopathology image segmentation. IEEE Trans. Med. Imaging 2017, 36, 2376–2388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Jia, Z.; Wang, L.-B.; Ai, Y.; Zhang, F.; Lai, M.; Eric, I.; Chang, C. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Sapkota, M.; Xing, F.; Liu, F.; Cui, L.; Yang, L. Pairwise based deep ranking hashing for histopathology image classification and retrieval. Pattern Recognit. 2018, 81, 14–22. [Google Scholar] [CrossRef]
- Le Van, C.; Bao, L.N.; Puri, V.; Thao, N.T.; Le, D.-N. Detecting lumbar implant and diagnosing scoliosis from vietnamese X-ray imaging using the pre-trained api models and transfer learning. CMC Comput. Mater. Contin. 2021, 66, 17–33. [Google Scholar]
- Nguyen, D.T.; Lee, M.B.; Pham, T.D.; Batchuluun, G.; Arsalan, M.; Park, K.R. Enhanced image-based endoscopic pathological site classification using an ensemble of deep learning models. Sensors 2020, 20, 5982. [Google Scholar] [CrossRef] [PubMed]
- Sari, C.T.; Gunduz-Demir, C. Unsupervised feature extraction via deep learning for histopathological classification of colon tissue images. IEEE Trans. Med. Imaging 2018, 38, 1139–1149. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, H.; Gan, C.; Lin, H.; Dou, Q.; Tsougenis, E.; Huang, Q.; Cai, M.; Heng, P.-A. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans. Cybern. 2019, 50, 3950–3962. [Google Scholar] [CrossRef]
- Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U. Brain tumor classification in MRI image using convolutional neural network. Math. Biosci. Eng 2020, 17, 6203. [Google Scholar] [CrossRef]
- Sun, X.; Wu, P.; Hoi, S.C. Face detection using deep learning: An improved faster RCNN approach. Neurocomputing 2018, 299, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Hameed, Z.; Zahia, S.; Garcia-Zapirain, B.; Javier Aguirre, J.; María Vanegas, A. Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors 2020, 20, 4373. [Google Scholar] [CrossRef] [PubMed]




| Classes | Amount of Data | Precision | Recall | F1-Score | |||
|---|---|---|---|---|---|---|---|
| Inception-V3 | VGG16 | Inception-V3 | VGG16 | Inception-V3 | VGG16 | ||
| c0 | 64 | 0.83 | 0.70 | 0.84 | 0.81 | 0.84 | 0.75 |
| c1 | 65 | 0.92 | 0.94 | 1.00 | 0.95 | 0.96 | 0.95 |
| c2 | 65 | 0.80 | 0.80 | 0.91 | 0.85 | 0.85 | 0.82 |
| c3 | 75 | 0.64 | 0.66 | 0.91 | 0.83 | 0.75 | 0.73 |
| c4 | 15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| c5 | 40 | 0.80 | 0.52 | 0.20 | 0.42 | 0.32 | 0.47 |
| c6 | 70 | 0.90 | 0.83 | 0.89 | 0.86 | 0.89 | 0.85 |
| c7 | 50 | 0.63 | 0.70 | 0.74 | 0.56 | 0.68 | 0.62 |
| c8 | 60 | 0.79 | 0.79 | 0.77 | 0.77 | 0.78 | 0.78 |
| c9 | 60 | 0.93 | 0.76 | 0.87 | 0.87 | 0.90 | 0.81 |
| c10 | 70 | 0.90 | 0.83 | 0.90 | 0.83 | 0.90 | 0.83 |
| c11 | 70 | 0.87 | 0.82 | 0.87 | 0.90 | 0.87 | 0.86 |
| c12 | 70 | 0.76 | 0.70 | 0.93 | 0.87 | 0.84 | 0.78 |
| c13 | 60 | 0.85 | 0.84 | 0.87 | 0.77 | 0.86 | 0.80 |
| c14 | 60 | 0.97 | 0.84 | 0.97 | 0.93 | 0.97 | 0.88 |
| c15 | 30 | 0.00 | 0.93 | 0.00 | 0.43 | 0.00 | 0.59 |
| c16 | 45 | 0.81 | 0.76 | 0.64 | 0.56 | 0.72 | 0.64 |
| c17 | 45 | 0.65 | 0.68 | 0.93 | 0.80 | 0.76 | 0.73 |
| c18 | 25 | 0.00 | 0.89 | 0.00 | 0.32 | 0.00 | 0.47 |
| c19 | 25 | 0.91 | 1.00 | 0.40 | 0.52 | 0.56 | 0.68 |
| c20 | 65 | 0.68 | 0.74 | 1.00 | 0.98 | 0.81 | 0.85 |
| c21 | 65 | 0.80 | 0.77 | 0.91 | 0.83 | 0.85 | 0.80 |
| c22 | 65 | 0.84 | 0.80 | 0.65 | 0.74 | 0.73 | 0.77 |
| c23 | 65 | 0.78 | 0.82 | 0.94 | 0.71 | 0.85 | 0.76 |
| Paper | Model | Method | |||
|---|---|---|---|---|---|
| Babaie et al. [7] | CNN | Train from scratch | 64.98 | 64.75 | 42.07 |
| Kieffer et al. [37] | VGG-16 | Feature Extractor | 65.21 | 64.96 | 42.36 |
| Kieffer et al. [37] | VGG-16 | Fine-tuning | 63.85 | 66.23 | 42.29 |
| Kieffer et al. [37] | Inception-V3 | Feature Extractor | 70.94 | 72.24 | 50.54 |
| Kieffer et al. [37] | Inception-v3 | Fine-tuning | 74.87 | 76.10 | 56.98 |
| Simonyan et al. [8] | VGG-16 base model | Train from scratch | 69.89 | 71.09 | 49.68 |
| Szegedy et al. [9] | Inception-V3 base model | Train from scratch | 72.65 | 73.00 | 53.03 |
| Proposed model | VGG-16 | Feature Extractor | 77.41 | 71.27 | 55.17 |
| Proposed model | Inception-V3 | Feature Extractor | 79.90 | 71.33 | 57.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.; Shaikh, A.; Alshahrani, H.; Alghamdi, A.; Alrizq, M.; Baber, J.; Bakhtyar, M. Transfer Learning Approach for Classification of Histopathology Whole Slide Images. Sensors 2021, 21, 5361. https://doi.org/10.3390/s21165361
Ahmed S, Shaikh A, Alshahrani H, Alghamdi A, Alrizq M, Baber J, Bakhtyar M. Transfer Learning Approach for Classification of Histopathology Whole Slide Images. Sensors. 2021; 21(16):5361. https://doi.org/10.3390/s21165361
Chicago/Turabian StyleAhmed, Shakil, Asadullah Shaikh, Hani Alshahrani, Abdullah Alghamdi, Mesfer Alrizq, Junaid Baber, and Maheen Bakhtyar. 2021. "Transfer Learning Approach for Classification of Histopathology Whole Slide Images" Sensors 21, no. 16: 5361. https://doi.org/10.3390/s21165361