Mapping Forest Aboveground Biomass Using Multisource Remotely Sensed Data
Abstract
:1. Introduction
1.1. Importance of Forests in Global Carbon Cycle
1.2. Biomass Estimation on the Ground
1.3. Challenges in Estimating Forest Aboveground Biomass with Remote Sensing
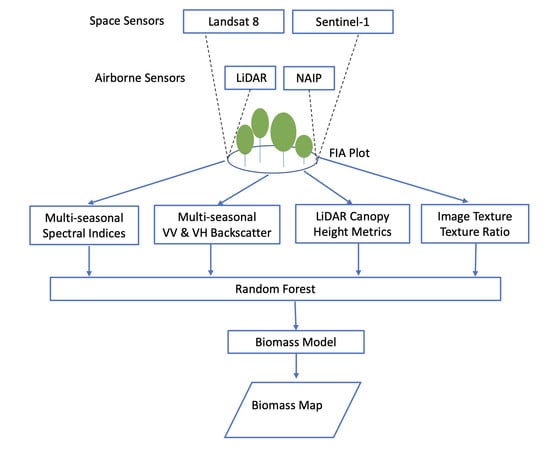
2. Materials and Methods
2.1. Study Area
2.2. Forest Inventory and Analysis Data
2.3. Remote Sensing Data
2.3.1. LiDAR Data
2.3.2. RaDAR Data
2.3.3. Multispectral Data
2.3.4. Very High-Resolution Optical Imagery
2.4. Biomass Model with Random Forest
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC. Summary for Policymakers. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; MassonDelmotte, V., Zhai, P., Pirani, A., Connors, S.L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M.I., et al., Eds.; IPCC: Geneva, Switzerland, 2021. [Google Scholar]
- Luthi, D.; Le Floch, M.; Bereiter, B.; Blunier, T.; Barnola, J.; Siegenthaler, U.; Raynaud, D.; Jouzel, J.; Fischer, H.; Kawamura, K.; et al. High-resolution carbon dioxide concentration record 650,000–800,000 years before present. Nature 2008, 453, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R. Climate Change: Atmospheric Carbon Dioxide. 2020. Available online: http://www.climate.gov/news-features/understanding-climate/climate-change-atmospheric-carbon-dioxide (accessed on 5 February 2022).
- Houghton, R.A. Land-use change and the carbon cycle. Glob. Chang. Biol. 1995, 1, 275–287. [Google Scholar] [CrossRef]
- Tian, H.; Lu, Q.; Ciais, P.; Michalak, A.M.; Canadell, J.G.; Saikawa, E.; Huntzinger, D.N.; Gurney, K.R.; Sitch, S.; Zhang, B.; et al. The terrestrial biosphere as a net source of greenhouse gases to the atmosphere. Nature 2016, 531, 225–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen-Shacham, E.; Walters, G.; Janzen, C.; Maginnis, S. (Eds.) Nature-Based Solutions to Address Global Societal Challenges; IUCN: Gland, Switzerland, 2016; Volume xiii, p. 97. ISBN 978-2-8317-1812-5. [Google Scholar] [CrossRef] [Green Version]
- Le Quere, C.; Andrew, R.M.; Friedlingstein, P.; Sitch, S.; Hauck, J.; Pongratz, J.; Pickers, P.A.; Korsbakken, J.I.; Peters, G.P.; Canadell, J.G.; et al. Global carbon budget 2018. Earth Syst. Sci. Data 2018, 10, 2141–2194. [Google Scholar] [CrossRef] [Green Version]
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.T.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef] [Green Version]
- Choat, B.; Jansen, S.; Brodribb, T.J.; Cochard, H.; Delzon, S.; Bhaskar, R.; Bucci, S.J.; Feild, T.S.; Gleason, S.M.; Hacke, U.G.; et al. Global convergence in the vulnerability of forests to drought. Nature 2012, 491, 751–756. [Google Scholar] [CrossRef] [Green Version]
- Ciais, P.; Reichstein, M.; Viovy, N.; Granier, A.; Ogee, J.; Allard, V.; Aubinet, M.; Buchmann, N.; Bernhofer, C.; Carrara, A.; et al. Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 2005, 437, 529–533. [Google Scholar] [CrossRef]
- Kurz, W.A.; Dymond, C.C.; Stinson, G.; Rampley, G.J.; Neilson, E.T.; Carroll, A.L.; Ebata, T.; Safranyik, L. Mountain pine beetle and forest carbon feedback to climate change. Nature 2008, 452, 987–990. [Google Scholar] [CrossRef]
- Song, C. Optical remote sensing of forest leaf area index and biomass (invited progress report). Prog. Phys. Geogr. 2013, 37, 98–113. [Google Scholar] [CrossRef]
- Zhai, B.; Song, C.; Zhang, H.; Wang, W. Studies on Biomass and productivity of Pinus Tabulaeformis plantation at a Permanent Ecosystem Plot in Taiyue forest region Shanxi Province. J. Beijing For. Univ. 1992, 14, 156–163, (In Chinese with English Abstract). [Google Scholar]
- Gholz, H.L.; Grier, C.C.; Campbell, A.G.; Brown, A.T. Equations for Estimating Biomass and Leaf Area of Plants in the Pacific Northwest; Forest Research Laboratory, School of Forestry, Oregon State University: Corvallis, OR, USA, 1979; Available online: https://ir.library.oregonstate.edu/concern/technical_reports/bn999796n?locale=en (accessed on 5 February 2022).
- Jenkins, J.C.; Chojnacky, D.C.; Heath, L.S.; Birdsey, R.A. Comprehensive Database of Diameter-Based Biomass Regressions for North America Tree Species; General Technical Report NE-319; USDA Forest Service: Washington, DC, USA, 2004.
- Blackard, J.A.; Finco, M.V.; Helmer, E.H.; Holden, G.R.; Hoppus, M.L.; Jacobs, D.M.; Lister, A.J.; Moisen, G.G.; Nelson, M.D.; Riemann, R.; et al. Mapping US forest biomass using nationwide forest inventory data and moderate resolution information. Remote Sens. Environ. 2008, 112, 1658–1677. [Google Scholar] [CrossRef]
- Nelson, R.; Margolis, H.; Montesano, P.; Sun, G.; Cook, B.; Corp, L.; Anderson, H.; deJong, B.; Pellat, F.P.; Fickel, T.; et al. Lidar-based estimates of aboveground biomass in the continental US and Mexico using ground, airborne, and satellite observations. Remote Sens. Environ. 2017, 188, 127–140. [Google Scholar] [CrossRef] [Green Version]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Potter, C.S.; Randerson, J.T.; Field, C.B.; Matson, P.A.; Vitousek, P.M.; Mooney, H.A.; Klooster, S.A. Terrestrial ecosystem production: A process model based on global satellite and surface data. Glob. Biogeochem. Cycles 1993, 7, 811–841. [Google Scholar] [CrossRef]
- Running, S.W.; Nemani, R.R.; Heinsch, F.A.; Zhao, M.; Reeves, M.; Hashimoto, H. A continuous satellite-derived measure of global terrestrial primary production. BioScience 2004, 54, 547–560. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, C.; Band, L.E.; Sun, G. No proportional increase of terrestrial gross carbon sequestration from the greening Earth. J. Geophys. Res.-Biogeosci. 2019, 124, 2540–2553. [Google Scholar] [CrossRef]
- Waring, R.H.; Landsberg, J.J.; Williams, M. Net primary production of forests: A constant fraction of gross primary production? Tree Physiol. 1998, 18, 129–134. [Google Scholar] [CrossRef]
- Song, C.; Woodcock, C.E. Estimating Tree crown size from multiresolution remotely sensed imagery. Photogramm. Eng. Remote Sens. 2003, 69, 1263–1270. [Google Scholar] [CrossRef]
- Kennedy, R.E.; Ohmann, J.; Gregory, M.; Roberts, H.; Yang, Z.; Bell, D.M.; Kane, V.; Hughes, M.J.; Cohen, W.B.; Powell, S.; et al. An empirical, integrated forest biomass monitoring system. Environ. Res. Lett. 2018, 13, 025004. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Pflugmacher, D.; Cohen, W.B.; Kennedy, R.E.; Yang, Z. Using Landsat-derived disturbance and recovery history and lidar to map forest biomass dynamics. Remote Sens. Environ. 2014, 151, 124–137. [Google Scholar] [CrossRef]
- Zhang, X.; Kondragunta, S. Estimating forest biomass in the USA using generalized allometric models and MODIS land products. Geophys. Res. Lett. 2006, 33, L09402. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Woodcock, C.E.; Li, X. The spectral/temporal manifestation of forest succession in optical imagery: The potential of multitemporal imagery. Remote Sens. Environ. 2002, 82, 285–302. [Google Scholar] [CrossRef]
- Luyssaert, S.; Schulze, E.; Borner, A.; Knohl, A.; Hessenmoller, D.; Law, B.E.; Ciais, P.; Grace, J. Old-growth forests as global carbon sinks. Nature 2008, 455, 213–215. [Google Scholar] [CrossRef]
- Sader, S.A.; Waide, R.B.; Lawrence, W.T.; Joyce, A.T. Tropical forest biomass and successional age class relationships to a vegetation index derived from Landsat TM data. Remote Sens. Environ. 1989, 28, 143–156. [Google Scholar] [CrossRef]
- Song, C.; Dannenberg, M.P.; Hwang, T. Optical Remote Sensing of Terrestrial Primary Productivity (invited progress report). Prog. Phys. Geogr. 2013, 37, 834–854. [Google Scholar] [CrossRef]
- Steininger, M.K. Satellite estimation of tropical secondary forest above-ground biomass: Data from Brazil and Bolivia. Int. J. Remote Sens. 2000, 21, 1139–1157. [Google Scholar] [CrossRef]
- Chaparro, D.; Duveiller, G.; Piles, M.; Cescatti, A.; Vall-Llossera, M.; Camps, A.; Entekhabi, D. Sensitivity of L-band vegetation optical depth to carbon stocks in tropical forests: A comparison to higher frequencies and optical indices. Remote Sens. Environ. 2019, 232, 111303. [Google Scholar] [CrossRef]
- Dobson, M.C.; Ulaby, F.T.; LeToan, T.; Beaudoin, A.; Kasischke, E.S.; Christensen, N. Dependence of Radar Backscatteron Coniferous Forest Biomass. IEEE Trans. Geosci. Remote Sens. 1992, 30, 412–415. [Google Scholar] [CrossRef]
- Le Toan, T.; Beaudoin, A.; Riom, J.; Guyon, D. Relating Forest Biomass to SAR Data. IEEE Trans. Geosci. Remote Sens. 1992, 30, 403–411. [Google Scholar] [CrossRef]
- Pulliainen, J.T.; Heiska, K.; Hyyppa, J.; Hallikainen, M.T. Backscattering Properties of Boreal Forests at C- and X-Bands. IEEE Trans. Geosci. Remote Sens. 1994, 32, 1041–1050. [Google Scholar] [CrossRef]
- Cartus, O.; Santoro, M.; Kellndorfer, J. Mapping forest aboveground biomass in the Northeastern United States with ALOS PALSAR dual-polarization L-band. Remote Sens. Environ. 2012, 124, 466–478. [Google Scholar] [CrossRef]
- Hussin, Y.A.; Reich, R.M.; Hoffer, R.M. Estimating Slash Pine Biomass Using Radar Backscatter. IEEE Trans. Geosci. Remote Sens. 1991, 29, 421–425. [Google Scholar] [CrossRef]
- Dubayah, R.; Drake, J.B. Lidar Remote Sensing for Forestry. J. For. 2000, 98, 44–46. [Google Scholar]
- Lefsky, M.A.; Cohen, W.B.; Acker, S.A.; Parker, G.G.; Spies, T.A.; Harding, D. Lidar Remote Sensing of the Canopy Structure and Biophysical Properties of Douglas-Fir Western Hemlock Forests. Remote Sens. Environ. 1999, 70, 330–361. [Google Scholar] [CrossRef]
- Drake, J.B.; Knox, R.G.; Dubayah, R.O.; Clark, D.B.; Condit, R.; Blair, J.B.; Hofton, M. Above-ground biomass estimation in closed canopy Neotropical forests using lidar remote sensing: Factors affecting the generality of relationships. Glob. Ecol. Biogeogr. 2003, 12, 147–159. [Google Scholar] [CrossRef]
- Hakkenberg, C.R.; Song, C.; Peet, R.K.; White, P.S. Forest Structure as a Predictor of Tree Species Diversity in the North Carolina Piedmont. J. Veg. Sci. 2016, 27, 1151–1163. [Google Scholar] [CrossRef]
- Harding, D.J.; Lefsky, M.A.; Parker, G.G.; Blair, J.B. Laser altimeter canopy height profiles: Methods and validation for closed-canopy broadleaf forests. Remote Sens. Environ. 2001, 76, 283–297. [Google Scholar] [CrossRef]
- Baccini, A.; Goetz, S.J.; Walker, W.S.; Laporte, N.T.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.S.A.; Dubayah, R.; Friedl, M.A.; et al. Estimated carbon dioxide emissions from tropical deforestation improved by carbon-density maps. Nat. Clim. Chang. 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Hyde, P.; Nelson, R.; Kimes, D.; Levine, E. Exploring LiDAR–RaDAR synergy—Predicting aboveground biomass in a southwestern ponderosa pine forest using LiDAR, SAR and InSAR. Remote Sens. Environ. 2007, 106, 28–38. [Google Scholar] [CrossRef]
- Qi, W.; Saarela, S.; Armston, J.; Stahl, G.; Dubayah, R. Forest biomass estimation over three distinct forest types using TanDEM-X, InSAR data and simulated GEDI lidar data. Remote Sens. Environ. 2019, 232, 111283. [Google Scholar] [CrossRef]
- Kellndorfer, J.; Walker, W.; Kirsch, K.; Fiske, G.; Bishop, J.; LaPoint, L.; Hoppus, M.; Westfall, J. NACP Aboveground Biomass and CarbonBaseline Data, V. 2 (NBCD 2000), USA 2000; ORNL DAAC: Oak Ridge, TN, USA, 2013. [CrossRef]
- Avitabile, V.; Herold, M.; Heuvelink, G.B.M.; Lewis, S.L.; Phillips, O.L.; Asner, G.P.; Armston, J.; Ashton, P.S.; Banin, L.; Bayol, N.; et al. An integrated pan-tropical biomass map using multiple reference datasets. Glob. Chang. Biol. 2016, 22, 1406–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baccini, A.; Laporte, N.; Goetz, S.J.; Sun, M.; Dong, H. A first map of tropical Africa’s above-ground biomass derived from satellite imagery. Environ. Res. Lett. 2008, 3, 045011. [Google Scholar] [CrossRef] [Green Version]
- Song, C. Estimating Tree Crown Size with Spatial Information of High Resolution Optical Remotely Sensed Imagery. Int. J. Remote Sens. 2007, 28, 3305–3322. [Google Scholar] [CrossRef]
- Bechtold, W.A.; Patterson, P.L. (Eds.) The Enhanced Forest Inventory and Analysis Program—National Sampling Design and Estimation Procedures; U.S. Department of Agriculture, Forest Service, Southern Research Station: Asheville, NC, USA, 2005; 85p.
- McRoberts, R.E.; Bechtold, W.A.; Patterson, P.L.; Scott, C.T.; Reams, G.A. The enhanced forest inventory and analysis program of the USDA Forest Service: Historical perspective and announcement of statistical documentation. J. For. 2005, 103, 304–408. [Google Scholar]
- Yang, L.; Jin, S.; Danielson, P.; Homer, C.; Gass, L.; Bender, S.M.; Case, A.; Costello, C.; Dewitz, J.; Fry, J.; et al. National Land Cover Database: Requirements, research priorities, design, and implementation strategies. ISPRS J. Photogramm. Remote Sens. 2018, 146, 108–123. [Google Scholar] [CrossRef]
- Dannenberg, M.P.; Hakkenberg, C.R.; Song, C. A long-term, consistent land cover history of the southeastern United States. Photogramm. Eng. Remote Sens. 2018, 84, 35–44. [Google Scholar] [CrossRef]
- Sexton, J.O.; Urban, D.L.; Donodue, M.J.; Song, C. Long-term land cover dynamics by multi-temporal classification across the Landsat-5 record. Remote Sens. Environ. 2013, 128, 246–258. [Google Scholar] [CrossRef]
- Fiorella, M.; Ripple, W.J. Analysis of conifer forest regeneration using Landsat Thematic Mapper data. Photogr. Eng. Remote Sens. 1993, 59, 1383–1388. [Google Scholar]
- Song, C.; Dickinson, M.B.; Su, L.; Zhang, S.; Yaussy, D. Estimating Average Tree Crown Size Using Spatial Information from Ikonos and QuickBird Images: Across-Sensor and Across-Site Comparisons. Remote Sens. Environ. 2010, 114, 1099–1107. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, J.G.; Lawler, J.T. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Guan, X.; Liu, L. KnowGRRF. Knowledge-Based Guided Regularized Random Forest, R package version 1.0; 2019. Available online: https://rdrr.io/cran/KnowGRRF/ (accessed on 5 February 2022).
- Campbell, M.J.; Dennison, P.E.; Kerr, K.L.; Brewer, S.C.; Anderegg, W.R.L. Scaled biomass estimation in woodland ecosystems: Testing the invidual and combined capacities of satellite multispectral and lidar data. Remote Sens. Environ. 2021, 262, 112511. [Google Scholar] [CrossRef]
- Chen, Q. Modeling aboveground tree woody biomass using national-scale allometric methods and airborne lidar. ISPRS J. Photogramm. Remote Sens. 2015, 106, 95–106. [Google Scholar] [CrossRef]
- Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; Queinnec, M.; Luther, J.E.; Bolton, D.K.; White, J.C.; Wulder, M.A.; van Lier, O.R.; Hermosilla, T. Modelling lidar-derived estimates of forest attributes over space and time: A review of approaches and future trends. Remote Sens. Environ. 2021, 260, 112477. [Google Scholar] [CrossRef]
- Huang, H.; Li, C.; Wang, X.; Zhou, X.; Gong, P. Integration of multi-resource remotely sensed data and allometric models for forest aboveground biomass estimation in China. Remote Sens. Environ. 2019, 221, 225–234. [Google Scholar] [CrossRef]
- Babcock, C.; Finley, A.O.; Andersen, H.; Pattison, R.; Cook, B.D.; Morton, D.C.; Alonzo, M.; Nelson, R.; Gregoire, T.; Ene, L.; et al. Geostatistical estimation of forest biomass in interior Alaska combining Landsat-derived tree cover, sampled airborne lidar and field observations. Remote Sens. Environ. 2018, 212, 212–230. [Google Scholar] [CrossRef] [Green Version]
- Grier, C.C.; Logan, R.S. Old-growth Psudotsuga menziesii of a western Oregon watershed: Biomass distribution and production budgets. Ecol. Monogr. 1977, 47, 373–400. [Google Scholar] [CrossRef]
- Dong, J.; Kaufmann, R.K.; Myneni, R.B.; Tucker, C.J.; Kauppi, P.; Liski, J.; Buermann, W.; Alexeyev, V.; Hughes, M.K. Remote sensing of boreal and temperate forest woody biomass: Carbon pools, sources, and sinks. Remote Sens. Environ. 2003, 84, 393–410. [Google Scholar] [CrossRef] [Green Version]
- Piao, S.L.; Fang, J.Y.; Zhu, B.; Tan, K. Forest biomass carbon stocks in China over the past 2 decades: Estimation based on integrated inventory and satellite data. J. Geophys. Res. 2005, 110, G01006. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.L.; Cohen, W.B.; Healey, S.P.; Kennedy, R.E.; Moisen, G.G.; Pierce, K.B.; Ohmann, J.L. Quantification of live aboveground forest biomass dynamics with Landsat time-series and field inventory data: A comparison of empirical modeling approaches. Remote Sens. Environ. 2010, 114, 1053–1068. [Google Scholar] [CrossRef]
- Rignot, E.; Way, J.; Williams, C.; Viereck, L. Radar estimates of aboveground biomass in Boreal forest of Interior Alaska. IEEE Trans. Geosci. Remote Sens. 1994, 32, 1117–1124. [Google Scholar] [CrossRef] [Green Version]
- Luckman, A.; Baker, J.; Kuplich, T.M.; da Costa Freitas Yanasse, C.; Frery, A.C. A Study of the relationship between Radar backscatter and regenerating tropicl forest biomass for spaceborne SAR instruments. Remote Sens. Environ. 1997, 60, 1–13. [Google Scholar] [CrossRef]
- Liao, Z.; He, B.; Quan, X.; van Jijk, A.I.J.M.; Qiu, S.; Yin, C. Biomass estimation in dense tropical forest using multiple information from single-baseline P-band PolInSAR data. Remote Sens. Environ. 2019, 221, 489–507. [Google Scholar] [CrossRef]
- Quegan, S.; Le Toan, T.; Cave, J.; Dall, J.; Exbrayat, J.; Minh, D.H.T.; Lomas, M.; D’alessandro, M.M.; Paillou, P.; Papathanassiou, K.; et al. The European Space Agency BIOMASS mission: Measuring forest aboveground. Remote Sens. Environ. 2019, 227, 44–60. [Google Scholar] [CrossRef] [Green Version]
- Amini, J.; Sumantyo, J.T.S. Employing a Method on SAR and Optical Images for Forest Biomass Estimation. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3026–4020. [Google Scholar] [CrossRef]
- Cutler, M.E.J.; Boyd, D.S.; Foody, G.M.; Vetrivel, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. Remote Sens. 2012, 70, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Duncanson, L.I.; Niemann, K.O.; Wulder, M.A. Integration of GLAS and Landsat TM data for aboveground biomass estimation. Can. J. Remote Sens. 2010, 36, 129–141. [Google Scholar] [CrossRef]
- Nelson, R.; Ranson, K.J.; Sun, G.; Kimes, D.S.; Kharuk, V.; Montesano, P. Estimating Siberian timber volume using MODIS and ICESat/GLAS. Remote Sens. Environ. 2009, 113, 691–701. [Google Scholar] [CrossRef]
- Phua, M.; Johari, S.A.; Wong, O.C.; Ioki, K.; Mahali, M.; Nilus, R.; Coomes, D.A.; Maycock, C.R.; Hashim, M. Synergistic use of Landsat 8 OLI image and airborne LiDAR data for aboveground biomass estimation in tropical lowland rainforests. For. Ecol. Manag. 2017, 406, 163–171. [Google Scholar] [CrossRef]
- Brovkina, O.; Novotny, J.; Cienciala, E.; Zemek, F.; Russ, R. Mapping forest aboveground biomass using airborne hyperspectraland LiDAR data in the mountainous conditions of Central Europe. Ecol. Eng. 2017, 100, 2019–2230. [Google Scholar] [CrossRef]
- Andersen, H.; Strunk, J.; Temesgen, H.; Atwood, D.; Winterberger, K. Using multilevel remote sensing and ground data to estimate forest biomass resources in remote regions: A case study in the boreal forests of interior Alaska. Can. J. Remote Sens. 2011, 37, 596–611. [Google Scholar] [CrossRef]





| Predictor | Description |
|---|---|
| LiDAR | |
| ZQ25 | 25th Percentile Height of the LiDAR point cloud |
| ZQ50 | 50th Percentile Height of the LiDAR point cloud |
| ZQ75 | 75th Percentile Height of the LiDAR point cloud |
| ZQ85 | 85th Percentile Height of the LiDAR point cloud |
| ZQ95 | 95th Percentile Height of the LiDAR point cloud |
| SD_ZQ25 | Standard Deviation of ZQ25 within 3 × 3 window |
| SD_ZQ50 | Standard Deviation of ZQ50 within 3 × 3 window |
| SD_ZQ75 | Standard Deviation of ZQ75 within 3 × 3 window |
| SD_ZQ85 | Standard Deviation of ZQ85 within 3 × 3 window |
| SD_ZQ95 | Standard Deviation of ZQ95 within 3 × 3 window |
| RaDAR-Sentinel-1C | |
| VV_winter | VV Polarization, Leaf-off Conditions |
| VH_winter | VH Polarization, Leaf-off Conditions |
| VV_summer | VV Polarization, Leaf-on Conditions |
| VH_summer | VH Polarization, Leaf-on Conditions |
| SD_VV_winter | Standard Deviation of the VV Polarization, Leaf-off Conditions |
| SD_VH_winter | Standard Deviation of the VH Polarization, Leaf-off Conditions |
| SD_VV_summer | Standard Deviation of the VV Polarization, Leaf-on Conditions |
| SD_VH_summer | Standard Deviation of the VH Polarization, Leaf-on Conditions |
| Multispectral-Landsat 8 | |
| B_winter | Brightness TCT Component, Leaf-off Conditions |
| G_winter | Greenness TCT Component, Leaf-off Conditions |
| W_winter | Wetness TCT Component, Leaf-off Conditions |
| EVI_winter | Enhanced Vegetation Index, Leaf-off Conditions |
| NDVI_winter | Normalized Difference Vegetation Index, Leaf-off Conditions |
| SI_winter | Structural Index, Leaf-off Conditions |
| B_summer | Brightness TCT Component, Leaf-on Conditions |
| G_summer | Greenness TCT Component, Leaf-on Conditions |
| W_summer | Wetness TCT Component, Leaf-on Conditions |
| EVI_summer | Enhanced Vegetation Index, Leaf-on Conditions |
| NDVI_summer | Normalized Difference Vegetation Index, Leaf-on Conditions |
| SI_summer | Structural Index, Leaf-on Conditions |
| Very High Resolution-NAIP | |
| T1×1 | Local texture at 1 m spatial resolution |
| R2/3 | Ratio of the local texture at 2 m to that at 3 m resolution |
| SD_T1×1 | Standard deviation of T1×1 |
| SD_R2/3 | Standard deviation of R2/3 |
| Dataset | R2 | RMSE (Mg/ha) |
|---|---|---|
| All Data | 0.598 | 19.5 |
| LiDAR | 0.595 | 19.6 |
| RaDAR | 0.065 | 29.7 |
| Multispectral | 0.065 | 29.7 |
| Very High Resolution | −0.027 | - |
| Tasseled Cap Components | 0.123 | 28.8 |
| Spectral Indices | 0.043 | 30.1 |
| Sample Size | Mean R2 | Min R2 | Max R2 | Std. Dev. R2 |
|---|---|---|---|---|
| One-Third (75) | 0.567 | 0.396 | 0.725 | 0.08894 |
| Half (113) | 0.586 | 0.413 | 0.669 | 0.05041 |
| Two-Thirds (150) | 0.598 | 0.513 | 0.679 | 0.03585 |
| All Data Points (227) | 0.611 | 0.598 | 0.626 | 0.00625 |
| Sample Size | Mean RMSE | Min RMSE | Max RMSE | Std. Dev. RMSE |
| One-Third (75) | 20.2 | 14.5 | 24.5 | 2.60506 |
| Half (113) | 19.8 | 15.6 | 23.0 | 1.89259 |
| Two-Thirds (150) | 19.4 | 16.7 | 21.8 | 1.22976 |
| All Data Points (227) | 19.2 | 18.8 | 19.5 | 0.15180 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehlers, D.; Wang, C.; Coulston, J.; Zhang, Y.; Pavelsky, T.; Frankenberg, E.; Woodcock, C.; Song, C. Mapping Forest Aboveground Biomass Using Multisource Remotely Sensed Data. Remote Sens. 2022, 14, 1115. https://doi.org/10.3390/rs14051115
Ehlers D, Wang C, Coulston J, Zhang Y, Pavelsky T, Frankenberg E, Woodcock C, Song C. Mapping Forest Aboveground Biomass Using Multisource Remotely Sensed Data. Remote Sensing. 2022; 14(5):1115. https://doi.org/10.3390/rs14051115
Chicago/Turabian StyleEhlers, Dekker, Chao Wang, John Coulston, Yulong Zhang, Tamlin Pavelsky, Elizabeth Frankenberg, Curtis Woodcock, and Conghe Song. 2022. "Mapping Forest Aboveground Biomass Using Multisource Remotely Sensed Data" Remote Sensing 14, no. 5: 1115. https://doi.org/10.3390/rs14051115