A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
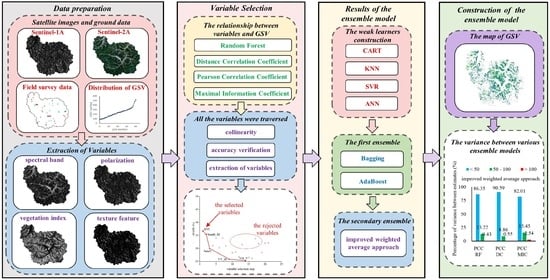
2.2. Framework of This Research
2.3. Ground Data Collection and Processing
2.4. Remote Sensing Images and Pre-Processing
2.5. Extraction of Variables
2.6. Proposed Variable Selection Criterion
2.7. Secondary Ensemble with Improved Weighted Average Approach
2.7.1. Secondary Ensemble Algorithm
2.7.2. Accuracy Evaluation
3. Results
3.1. The Results of Variables Selection
3.2. The Result of the Secondary Ensemble
3.3. Mapping the Forest GSV
4. Discussion
4.1. Variable Selection
4.2. Ensemble Model
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dixon, R.; Solomon, A.; Brown, S.; Houghton, R.; Trexier, M.; Wisniewski, J. Carbon Pools and Flux of Global Forest Ecosystems. Science 1994, 263, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Bonan, G. Forests and Climate Change: Forcings, Feedbacks, and the Climate Benefits of Forests. Science 2008, 320, 1444–1449. [Google Scholar] [CrossRef] [Green Version]
- Solberg, S.; Hansen, E.; Gobakken, T.; Næsset, E.; Zahabu, E. Biomass and InSAR height relationship in a dense tropical forest. Remote Sens. Environ. 2017, 192, 166–175. [Google Scholar] [CrossRef]
- Carnus, J.; Parrotta, J.; Brockerhoff, E.; Arbez, M.; Jactel, H.; Kremer, A.; Lamb, D.; O’Hara, K.; Walters, B. Planted forests and biodiversity. J. For. 2006, 104, 65–77. [Google Scholar]
- Brockerhoff, E.; Jactel, H.; Parrotta, J.; Ferraz, S. Role of eucalypt and other planted forests in biodiversity conservation and the provision of biodiversity-related ecosystem services. For. Ecol. Manag. 2013, 301, 43–50. [Google Scholar] [CrossRef]
- Long, J.; Lin, H.; Wang, G.; Sun, H.; Yan, E. Mapping Growing Stem Volume of Chinese Fir Plantation Using a Saturation-based Multivariate Method and Quad-polarimetric SAR Images. Remote Sens. 2019, 11, 1872. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, Z.; Lin, H.; Wang, G.; Sun, H.; Long, J.; Zhang, M. Estimating the Growing Stem Volume of Chinese Pine and Larch Plantations based on Fused Optical Data Using an Improved Variable Screening Method and Stacking Algorithm. Remote Sens. 2020, 12, 871. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Xu, X.; Wu, F.; Sun, Z.; Xia, H.; Meng, Q.; Huang, W.; Zhou, H.; Gao, J.; Li, W.; et al. Estimating Forest Stock Volume in Hunan Province, China, by Integrating In Situ Plot Data, Sentinel-2 Images, and Linear and Machine Learning Regression Models. Remote Sens. 2020, 12, 186. [Google Scholar] [CrossRef] [Green Version]
- Hollaus, M.; Wagner, W.; Schadauer, K.; Maier, B.; Gabler, K. Growing stock estimation for alpine forests in Austria: A robust lidar-based approach. Can. J. For. Res. 2009, 39, 1387–1400. [Google Scholar] [CrossRef]
- Ploton, P.; Barbier, N.; Couteron, P.; Antin, C.M.; Ayyappan, N.; Balachandran, N.; Barathan, N.; Bastin, J.-F.; Chuyong, G.; Dauby, G.; et al. Toward a general tropical forest biomass prediction model from very high resolution optical satellite images. Remote Sens. Environ. 2017, 200, 140–153. [Google Scholar] [CrossRef]
- Wang, H.; Wang, C.; Wu, H. Using GF-2 Imagery and the Conditional Random Field Model for Urban Forest Cover Mapping. Remote Sens. Lett. 2016, 7, 378–387. [Google Scholar] [CrossRef]
- Lu, D.; Batistella, M. Exploring TM Image Texture and Its Relationships with Biomass Estimation in Rondônia, Brazilian Amazon. Acta Amaz. 2005, 35, 249–257. [Google Scholar] [CrossRef]
- Stanczyk, U. Feature Evaluation by Filter, Wrapper, and Embedded Approaches. Stud. Comput. Intell. 2015, 584, 29–44. [Google Scholar] [CrossRef]
- Sandri, M.; Zuccolotto, P. Variable Selection Using Random Forests. In Data Analysis, Classification and the Forward Search; Studies in Classification, Data Analysis, and Knowledge Organization; Springer: Berlin/Heidelberg, Germany, 2006; pp. 263–270. ISBN 978-3-540-35977-7. [Google Scholar]
- Wolf, L.; Bileschi, S. Combining Variable Selection with Dimensionality Reduction. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–26 June 2005. [Google Scholar] [CrossRef] [Green Version]
- Ji, Y.; Huang, J.; Ju, Y.; Guo, S.; Cairong, Y. Forest structure dependency analysis of L-band SAR backscatter. PeerJ 2020, 8, e10055. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Motoda, H. Feature Extraction Construction and Selection: A Data Mining Perspective. J. Am. Stat. Assoc. 1999, 94, 1390. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction of Variable and Feature Selection. J. Mach. Learn. Res. Spec. Issue Var. Feature Sel. 2003, 3, 1157–1182. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Yu, S.; Zhao, F.; Tian, L.; Zhao, Z. Comparison of machine learning algorithms for forest parameter estimations and application for forest quality assessments. For. Ecol. Manag. 2019, 434, 224–234. [Google Scholar] [CrossRef]
- Hilario, M.; Kalousis, A. Approaches to dimensionality reduction in proteomic biomarker studies. Brief. Bioinform. 2008, 9, 102–118. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Galiano, V.; Luque-Espinar, J.A.; Chica-Olmo, M.; Mendes, M. Feature selection approaches for predictive modelling of groundwater nitrate pollution: An evaluation of filters, embedded and wrapper methods. Sci. Total Environ. 2017, 624, 661–672. [Google Scholar] [CrossRef]
- Zhang, C.; Denka, S.; Cooper, H.; Mishra, D.R. Quantification of sawgrass marsh aboveground biomass in the coastal Everglades using object-based ensemble analysis and Landsat data. Remote Sens. Environ. 2018, 204, 366–379. [Google Scholar] [CrossRef]
- Crowther, T.; Glick, H.; Covey, K.; Bettigole, C.; Maynard, D.; Thomas, S.; Smith, J.; Hintler, G.; Duguid, M.; Amatulli, G.; et al. Mapping tree density at a global scale. Nature 2015, 525, 201–205. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Investigating the robustness of the new Landsat-8 Operational Land Imager derived texture metrics in estimating plantation forest aboveground biomass in resource constrained areas. ISPRS J. Photogramm. Remote Sens. 2015, 108, 12–32. [Google Scholar] [CrossRef]
- Pu, R.; Cheng, J. Mapping forest leaf area index using reflectance and textural information derived from WorldView-2 imagery in a mixed natural forest area in Florida, USA. Int. J. Appl. Earth Obs. Geoinf. 2015, 42, 11–23. [Google Scholar] [CrossRef]
- Cooner, A.; Shao, Y.; Campbell, J. Detection of Urban Damage Using Remote Sensing and Machine Learning Algorithms: Revisiting the 2010 Haiti Earthquake. Remote Sens. 2016, 8, 868. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gong, W.; Xing, Y.; Hu, X.; Gong, J. Estimation of the forest stand mean height and aboveground biomass in Northeast China using SAR Sentinel-1B, multispectral Sentinel-2A, and DEM imagery. ISPRS J. Photogramm. Remote Sens. 2019, 151, 277–289. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman and Hall/CRC: London, UK, 1984; ISBN 978-0-412-04841-8. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Basak, D.; Srimanta, P.; Patranbis, D.C. Support vector regression. Neural Inform. Process. Lett. Rev. 2007, 11, 67–80. [Google Scholar]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design, and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef]
- Breiman, L. Bagging Predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Grossmann, E. AdaTree: Boosting a Weak Classifier into a Decision Tree. In Proceedings of the 2004 Conference on Computer Vision and Pattern Recognition Workshop, Washington, DC, USA, 27 June–2 July 2004. [Google Scholar]
- Wang, J.; Xu, J.; Peng, Y.; Wang, H.; Shen, J. Prediction of forest unit volume based on hybrid feature selection and ensemble learning. Evol. Intell. 2020, 13, 21–32. [Google Scholar] [CrossRef]
- Tao, Y.; Peng, Y.; Jiang, Q.; Yucui, L.I.; Fang, S.; Gong, Y. Remote Detection of Critical Growth Stages in Rapeseed Using Vegetation Spectral and Stacking Combination Method. J. Geomat. 2019, 44, 20–23. [Google Scholar]
- Huete, A.; Didan, K.; van Leeuwen, W.; Vermote, E. Global-scale analysis of vegetation indices for moderate resolution monitoring of terrestrial vegetation. Remote Sens. 1999, 141–151. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D.; Harlan, J. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA/GSFC Type III, Final Report; NASA/GSFC: Greenbelt, MD, USA, 1974. Available online: https://ntrs.nasa.gov/citations/19740022555 (accessed on 11 November 2021).
- Tucker, C. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Cohen, W.; Maiersperger, T.; Gower, S.; Turner, D. An improved strategy for regression of biophysical variables and Landsat ETM+ data. Remote Sens. Environ. 2003, 84, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Huete, A. A soil adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 17, 37–53. [Google Scholar] [CrossRef]
- Vafaei, S.; Soosani, J.; Adeli, K.; Fadaei, H.; Naghavi, H.; Pham, T.; Tien Bui, D. Improving Accuracy Estimation of Forest Aboveground Biomass Based on Incorporation of ALOS-2 PALSAR-2 and Sentinel-2A Imagery and Machine Learning: A Case Study of the Hyrcanian Forest Area (Iran). Remote Sens. 2018, 10, 172. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, Y.; Ren, C.; Zhang, B.; Wang, Z. Optimal Combination of Predictors and Algorithms for Forest Above-Ground Biomass Mapping from Sentinel and SRTM Data. Remote Sens. 2019, 11, 414. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Mausel, P.; Brondízio, E.; Moran, E. Relationships between forest stand parameters and Landsat TM spectral responses in the Brazilian Amazon Basin. For. Ecol. Manag. 2004, 198, 149–167. [Google Scholar] [CrossRef]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Hudak, A.T.; Crookston, N.; Evans, J.; Hall, D.; Falkowski, M. Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Kutia, M.; Sarkissian, A.; Lin, H.; Jiangping, L.; Sun, H.; Wang, G. Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method. Sensors 2020, 20, 7248. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Smith, A.R.; Kutia, M.; Wang, G.; Liu, H.; Sun, H. A Modified kNN Method for Mapping the Leaf Area Index in Arid and Semi-Arid Areas of China. Remote Sens. 2020, 12, 1884. [Google Scholar] [CrossRef]
- Chirici, G.; Barbati, A.; Corona, P.; Marchetti, M.; Travaglini, D.; Maselli, F.; Bertini, R. Non-parametric and parametric methods using satellite images for estimating growing stock volume in alpine and Mediterranean forest ecosystems. Remote Sens. Environ. 2008, 112, 2686–2700. [Google Scholar] [CrossRef] [Green Version]
- Maltamo, M.; Malinen, J.; Packalen, P.; Suvanto, A.; Kangas, J. Nonparametric estimation of stem volume using airborne laser scanning, aerial photography, and stand-register data. Can. J. For. Res. 2006, 36, 426–436. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R. Comparison of support vector machine, neural network, and CART algorithms for the land-cover classification using limited training data points. ISPRS J. Photogramm. Remote Sens. 2012, 70, 78–87. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Pu, R.; Zhang, Z. Mapping Robinia Pseudoacacia Forest Health Conditions by Using Combined Spectral, Spatial, and Textural Information Extracted from IKONOS Imagery and Random Forest Classifier. Remote Sens. 2015, 7, 9020–9044. [Google Scholar] [CrossRef] [Green Version]
- Ali, I.; Greifeneder, F.; Stamenkovic, J.; Neumann, M.; Notarnicola, C. Review of Machine Learning Approaches for Biomass and Soil Moisture Retrievals from Remote Sensing Data. Remote Sens. 2015, 7, 16398–16421. [Google Scholar] [CrossRef] [Green Version]
- Esteban, J.; Mcroberts, R.; Fernández-Landa, A.; Tomé, J.; Nӕsset, E. Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data. Remote Sens. 2019, 11, 1944. [Google Scholar] [CrossRef] [Green Version]
- Fanos, A.M.; Pradhan, B.; Alamri, A.; Lee, C.-W. Machine Learning-Based and 3D Kinematic Models for Rockfall Hazard Assessment Using LiDAR Data and GIS. Remote Sens. 2020, 12, 1755. [Google Scholar] [CrossRef]
- Li, X.; Lin, H.; Long, J.; Xu, X. Mapping the Growing Stem Volume of the Coniferous Plantations in North China Using Multispectral Data from Integrated GF-2 and Sentinel-2 Images and an Optimized Feature Variable Selection Method. Remote Sens. 2021, 13, 2740. [Google Scholar] [CrossRef]











| Tree Species | Number of Plots | The Range of GSV | The Average GSV | STD |
|---|---|---|---|---|
| Larch | 38 | 86.17~405.56 | 208.38 | 81.84 |
| Chinese pine | 43 | 91.97~514.96 | 253.32 | 112.75 |
| Sensors | Acquisition Date | Spectral Bands/Polarizations |
|---|---|---|
| Sentinel-1A (level-1GRD) | 19 September 2017 | VH, VV |
| Sentinel-2A (level-1C) | 22 September 2017 | Band2, Band3, Band4, Band5, Band6, Band7, Band8, Band8A |
| Variable Type | Variable Name | Description of Variables | Sensors |
|---|---|---|---|
| Vegetation Index | Enhanced Vegetation Index (EVI) | 2.5 × (Band8 − Band4)/(Band8 + 6 Band4 − 7.5 × Band2 + 1) | Sentinel-2A |
| Enhanced Vegetation Index-2 (EVI-2) | 2.5 × (Band8 − Band4)/(Band8 + 2.4 × Band4 + 1) | Sentinel-2A | |
| Normalized Difference Vegetation Index (NDVI) | (Band8 − Band4)/(Band8 + Band4) | Sentinel-2A | |
| Ratio Vegetation Index (RVI) | Band8/Band4 | Sentinel-2A | |
| Spectral Vegetation Index (SVI) | Band4/Band8 | Sentinel-2A | |
| Soil Adjusted Vegetation Index (SAVI) | (1 + L) × (Band8 − Band4)/(Band8 + Band4 + L) L = 0.5 in most conditions | Sentinel-2A | |
| Spectral reflection | Spectral bands | Band2, Band3, Band4, Band5, Band6, Band7, Band8, Band8A | Sentinel-2A |
| Features of SAR | Backscattering coefficient | VH, VV, VH/VV | Sentinel-1A |
| Texture features | Mean, Variance, Contrast, Entropy, Homogeneity, Dissimilarity, Entropy, Second moment, Correlation | Gray Level Co-occurrence Matrix (GLCM) with size of 3 × 3 | Sentinel-1A Sentinel-2A |
| Variable Selection Criterion | Method of Ranking | Models | The First Selected Variable | Number of Variables | Number of Operations |
|---|---|---|---|---|---|
| Forward | FORW | CART | band4 | 2 | 295 |
| FORW | KNN | band4 | 10 | 1034 | |
| FORW | ANN | EVI_2 | 4 | 486 | |
| FORW | SVR | EVI | 6 | 672 | |
| The proposed criterion for variable selection | RF | CART | EVI_2 | 6 | 41 |
| RF | KNN | EVI_2 | 6 | 27 | |
| RF | ANN | EVI_2 | 4 | 51 | |
| RF | SVR | EVI_2 | 6 | 43 | |
| DC | CART | band4_M | 4 | 67 | |
| DC | KNN | band4_M | 5 | 26 | |
| DC | ANN | band4_M | 4 | 58 | |
| DC | SVR | band4_M | 7 | 64 | |
| MIC | CART | band4_M | 4 | 95 | |
| MIC | KNN | band4_M | 9 | 37 | |
| MIC | ANN | band4_M | 5 | 46 | |
| MIC | SVR | band4_M | 5 | 44 | |
| PCC | CART | RVI | 7 | 20 | |
| PCC | KNN | RVI | 5 | 21 | |
| PCC | ANN | RVI | 4 | 65 | |
| PCC | SVR | RVI | 7 | 59 |
| Ranking Methods | First Ensemble (Bagging and AdaBoost) | Secondary Ensemble (IWA) | |||||
|---|---|---|---|---|---|---|---|
| Number of Variables | rRMSE (%) | R2 | Number of Models | Number of Related Variables | rRMSE (%) | R2 | |
| RF | 4~6 | 21.91~30.28 | 0.47~0.72 | 3 | 7 | 20.14 | 0.77 |
| DC | 4~7 | 23.41~28.89 | 0.52~0.68 | 8 | 14 | 21.34 | 0.74 |
| MIC | 4~9 | 23.60~31.49 | 0.43~0.68 | 5 | 13 | 19.89 | 0.77 |
| PCC | 4~7 | 21.93~28.83 | 0.52~0.72 | 8 | 15 | 18.89 | 0.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Lin, H.; Liu, Z.; Ye, Z.; Li, X.; Long, J. A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest. Remote Sens. 2021, 13, 4631. https://doi.org/10.3390/rs13224631
Xu X, Lin H, Liu Z, Ye Z, Li X, Long J. A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest. Remote Sensing. 2021; 13(22):4631. https://doi.org/10.3390/rs13224631
Chicago/Turabian StyleXu, Xiaodong, Hui Lin, Zhaohua Liu, Zilin Ye, Xinyu Li, and Jiangping Long. 2021. "A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest" Remote Sensing 13, no. 22: 4631. https://doi.org/10.3390/rs13224631