Spectral-Spatial Offset Graph Convolutional Networks for Hyperspectral Image Classification
Abstract
:1. Introduction
2. Related Works
2.1. Graph Convolutional Network
2.2. Hyperspectral Image Classification
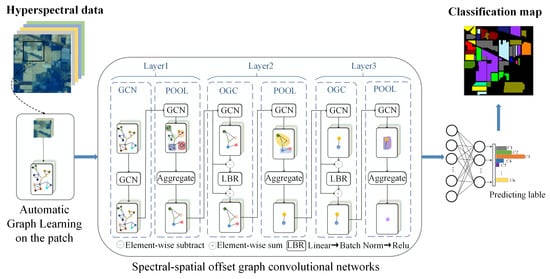
3. Method
3.1. Automatic Graph Learning (AGL)
3.2. Graph Classification Networks
3.2.1. GCN
3.2.2. OGC
3.2.3. Graph Pooling
4. Experiment
4.1. Data Sets
- (1)
- Indian Pines Data Set: The scene over northwestern Indiana, USA was acquired over the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) sensor in 1992. The image consists of 145 × 145 pixels and the spatial resolution is 20 m per pixel.
- (2)
- Pavia University Data Set: The second image comprised by 610 × 340 pixels and each pixel are 1.3 m. It was acquired by the Reflective Optics System Imaging Spectrometer (ROSIS) sensor in 2002. There are 115 bands in the range of 0.43~0.86, and 103 bands without serious noise are selected for experiment. The data set includes nine land cover classes, and a total of 42,776 samples can be referred. As shown in Figure 6 below, the left image is a false color map, the middle column is a ground-truth map, and the right is the corresponding class name. Table 3 lists 9 major land-cover classes in this image, as well as the number of training and testing samples used for our experiments.
- (3)
- Salinas Data Set: The scene over Salinas Valley, California was acquired the AVIRIS sensor. The image consists of 512 × 217 pixels and the spatial resolution is 3.7 m per pixel. There are 204 bands are available after discarding the 20 water absorption bands. The data set contains 16 types of features, and 54,129 samples can be referred. Table 4 lists 16 main land-cover categories involved in this scene, as well as the number of training and testing samples used for our experiments. The false color map and ground-truth map are shown in Figure 7.
4.2. Experimental Settings
4.3. Classification Results
4.3.1. Results on the Indian Pines Data Set
4.3.2. Results on the Pavia University Data Set
4.3.3. Results on the Salinas Data Set
4.4. Impact of Patch Size
4.5. Ablation Study
4.6. Impact of the Number of Labeled Samples
4.7. Running Time
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W.; Feng, F.; Li, H.; Du, Q. Discriminant Analysis-Based Dimension Reduction for Hyperspectral Image Classification: A Survey of the Most Recent Advances and an Experimental Comparison of Different Techniques. IEEE Geosci. Remote Sens. Mag. 2018, 6, 15–34. [Google Scholar] [CrossRef]
- Yin, J.; Qi, C.; Chen, Q.; Qu, J. Spatial-Spectral Network for Hyperspectral Image Classification: A 3-D CNN and Bi-LSTM Framework. Remote Sens. 2021, 13, 2353. [Google Scholar] [CrossRef]
- Yan, H.; Wang, J.; Tang, L.; Zhang, E.; Yan, K.; Yu, K.; Peng, J. A 3D Cascaded Spectral–Spatial Element Attention Network for Hyperspectral Image Classification. Remote Sens. 2021, 13, 2451. [Google Scholar] [CrossRef]
- Pu, S.; Wu, Y.; Sun, X.; Sun, X. Hyperspectral Image Classification with Localized Graph Convolutional Filtering. Remote Sens. 2021, 13, 526. [Google Scholar] [CrossRef]
- Ghamisi, P.; Yokoya, N.; Li, J.; Liao, W.; Liu, S.; Plaza, J.; Rasti, B.; Plaza, A. Advances in Hyperspectral Image and Signal Processing: A Comprehensive Overview of the State of the Art. IEEE Geosci. Remote Sens. Mag. 2017, 5, 37–78. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An Augmented Linear Mixing Model to Address Spectral Variability for Hyperspectral Unmixing. IEEE Trans. Image Process. 2019, 28, 1923–1938. [Google Scholar] [CrossRef] [Green Version]
- Benediktsson, J.A.; Palmason, J.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Mura, M.D.; Benediktsson, J.A.; Waske, B.; Bruzzone, L. Morphological Attribute Profiles for the Analysis of Very High Resolution Images. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3747–3762. [Google Scholar] [CrossRef]
- Pan, C.; Gao, X.; Wang, Y.; Li, J. Markov Random Fields Integrating Adaptive Interclass-Pair Penalty and Spectral Similarity for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2520–2534. [Google Scholar] [CrossRef]
- Rodriguez, P.; Wiles, J.; Elman, J. A Recurrent Neural Network that Learns to Count. Connect. Sci. 1999, 11, 5–40. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Generative Adversarial Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5046–5063. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Zhang, Y.; Shen, Q. Spectral-spatial classification of hyperspectral imagery using a dual-channel convolutional neural network. Remote Sens. Lett. 2017, 8, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral–Spatial Residual Network for Hyperspectral Image Classification: A 3-D Deep Learning Framework. IEEE Trans. Geosci. Remote Sens. 2018, 56, 847–858. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017, arXiv:1609.02907. Available online: http://arxiv.org/abs/1609.02907 (accessed on 12 August 2021).
- Qin, A.; Shang, Z.; Tian, J.; Wang, Y.; Zhang, T.; Tang, Y.Y. Spectral–Spatial Graph Convolutional Networks for Semisupervised Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2019, 16, 241–245. [Google Scholar] [CrossRef]
- Mou, L.; Lu, X.; Li, X.; Zhu, X.X. Nonlocal Graph Convolutional Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 8246–8257. [Google Scholar] [CrossRef]
- Sha, A.; Wang, B.; Wu, X.; Zhang, L. Semisupervised Classification for Hyperspectral Images Using Graph Attention Networks. IEEE Geosci. Remote Sens. Lett. 2021, 18, 157–161. [Google Scholar] [CrossRef]
- Wan, S.; Gong, C.; Zhong, P.; Du, B.; Zhang, L.; Yang, J. Multiscale Dynamic Graph Convolutional Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3162–3177. [Google Scholar] [CrossRef] [Green Version]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph Convolutional Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 5966–5978. [Google Scholar] [CrossRef]
- Ying, R.; He, R.; Chen, K.; Eksombatchai, P.; Hamilton, W.L.; Leskovec, J. Graph Convolutional Neural Networks for Web-Scale Recommender Systems. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, New York, NY, USA, 19–23 August 2018; pp. 974–983. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Young, T.; Huang, M.; Zhao, H.; Xu, J.; Zhu, X. Commonsense Knowledge Aware Conversation Generation with Graph Attention. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, Stockholm, Sweden, 13–19 July 2018; pp. 4623–4629. [Google Scholar] [CrossRef] [Green Version]
- Gori, M.; Monfardini, G.; Scarselli, F. A new model for learning in graph domains. In Proceedings of the 2005 IEEE International Joint Conference on Neural Networks, Montreal, QC, Canada, 31 July–4 August 2005; Volume 2, pp. 729–734. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, P.; Zhu, W. Deep Learning on Graphs: A Survey. IEEE Trans. Knowl. Data Eng. 2020, 14, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Bruna, J.; Zaremba, W.; Szlam, A.; LeCun, Y. Spectral Networks and Locally Connected Networks on Graphs. arXiv 2014, arXiv:1312.6203. Available online: http://arxiv.org/abs/1312.6203 (accessed on 12 August 2021).
- Defferrard, M.; Bresson, X.; Vandergheynst, P. Convolutional Neural Networks on Graphs with Fast Localized Spectral Filtering. Adv. Neural Inform. Process. Syst. 2016, 29, 3837–3845. Available online: https://proceedings.neurips.cc/paper/2016/hash/04df4d434d481c5bb723be1b6df1ee65-Abstract.html (accessed on 1 October 2021).
- Xu, B.; Shen, H.; Cao, Q.; Cen, K.; Cheng, X. Graph Convolutional Networks using Heat Kernel for Semi-supervised Learning. arXiv 2020, arXiv:2007.16002. Available online: http://arxiv.org/abs/2007.16002 (accessed on 1 October 2021).
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs. Adv. Neural Inform. Process. Syst. 2017, 30, 1024–1034. Available online: https://proceedings.neurips.cc/paper/2017/hash/5dd9db5e033da9c6fb5ba83c7a7ebea9-Abstract.html (accessed on 1 October 2021).
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. Available online: http://arxiv.org/abs/1710.10903 (accessed on 1 October 2021).
- Monti, F.; Boscaini, D.; Masci, J.; Rodola, E.; Svoboda, J.; Bronstein, M.M. Geometric Deep Learning on Graphs and Manifolds Using Mixture Model CNNs. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5115–5124. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Du, Q. Robust Joint Sparse Representation Based on Maximum Correntropy Criterion for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 7152–7164. [Google Scholar] [CrossRef]
- Peng, J.; Sun, W.; Du, Q. Self-Paced Joint Sparse Representation for the Classification of Hyperspectral Images. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1183–1194. [Google Scholar] [CrossRef]
- Liu, S.; Du, Q.; Tong, X.; Samat, A.; Bruzzone, L. Unsupervised Change Detection in Multispectral Remote Sensing Images via Spectral-Spatial Band Expansion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 3578–3587. [Google Scholar] [CrossRef]
- Licciardi, G.; Marpu, P.R.; Chanussot, J.; Benediktsson, J.A. Linear Versus Nonlinear PCA for the Classification of Hyperspectral Data Based on the Extended Morphological Profiles. IEEE Geosci. Remote Sens. Lett. 2012, 9, 447–451. [Google Scholar] [CrossRef] [Green Version]
- Izenman, A.J. Linear Discriminant Analysis. In Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning; Izenman, A.J., Ed.; Springer: New York, NY, USA, 2013; pp. 237–280. [Google Scholar] [CrossRef]
- He, X.; Niyogi, P. Locality Preserving Projections. In Advances in Neural Information Processing Systems 16; Thrun, S., Saul, L.K., Schölkopf, B., Eds.; MIT Press: Cambridge, MA, USA, 2004; pp. 153–160. Available online: http://papers.nips.cc/paper/2359-locality-preserving-projections.pdf (accessed on 25 September 2020).
- He, X.; Cai, D.; Yan, S.; Zhang, H.-J. Neighborhood preserving embedding. In Proceedings of the Tenth IEEE International Conference on Computer Vision (ICCV’05), Washington, DC, USA, 17–21 October 2005; Volume 1, pp. 1208–1213. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, K.; Dong, Y.; Wu, K.; Hu, X. Semisupervised Classification Based on SLIC Segmentation for Hyperspectral Image. IEEE Geosci. Remote Sens. Lett. 2020, 17, 1440–1444. [Google Scholar] [CrossRef]
- Jia, S.; Deng, B.; Zhu, J.; Jia, X.; Li, Q. Local Binary Pattern-Based Hyperspectral Image Classification with Superpixel Guidance. IEEE Trans. Geosci. Remote Sens. 2018, 56, 749–759. [Google Scholar] [CrossRef]
- Ding, Y.; Pan, S.; Chong, Y. Robust Spatial–Spectral Block-Diagonal Structure Representation with Fuzzy Class Probability for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 1747–1762. [Google Scholar] [CrossRef]
- Zhao, Z.-Q.; Zheng, P.; Xu, S.-T.; Wu, X. Object Detection with Deep Learning: A Review. IEEE Trans. Neural Networks Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.K.; Zhang, X.; Huang, X. Hyperspectral Image Classification with Deep Learning Models. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Audebert, N.; Le Saux, B.; Lefevre, S. Deep Learning for Classification of Hyperspectral Data: A Comparative Review. IEEE Geosci. Remote Sens. Mag. 2019, 7, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep Learning-Based Classification of Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Jia, X. Spectral–Spatial Classification of Hyperspectral Data Based on Deep Belief Network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Hong, D.; Han, Z.; Yao, J.; Gao, L.; Zhang, B.; Plaza, A.; Chanussot, J. Spectral Former: Rethinking Hyperspectral Image Classification with Transformers. arXiv 2021, arXiv:2107.02988. Available online: http://arxiv.org/abs/2107.02988 (accessed on 1 October 2021).
- Hu, W.; Huang, Y.; Wei, L.; Zhang, F.; Li, H.-C. Deep Convolutional Neural Networks for Hyperspectral Image Classification. J. Sens. 2015, 2015, e258619. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wu, G.; Zhang, F.; Du, Q. Hyperspectral Image Classification Using Deep Pixel-Pair Features. IEEE Trans. Geosci. Remote Sens. 2017, 55, 844–853. [Google Scholar] [CrossRef]
- Makantasis, K.; Karantzalos, K.; Doulamis, A.; Doulamis, N. Deep supervised learning for hyperspectral data classification through convolutional neural networks. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4959–4962. [Google Scholar] [CrossRef]
- Slavkovikj, V.; Verstockt, S.; De Neve, W.; Van Hoecke, S.; Van de Walle, R. Hyperspectral Image Classification with Convolutional Neural Networks. In Proceedings of the 23rd ACM international conference on Multimedia, New York, NY, USA, 26–30 October 2015; pp. 1159–1162. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Li, S.; Fang, L.; Lu, T. Hyperspectral Image Classification with Deep Feature Fusion Network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3173–3184. [Google Scholar] [CrossRef]
- Gong, Z.; Zhong, P.; Yu, Y.; Hu, W.; Li, S. A CNN With Multiscale Convolution and Diversified Metric for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3599–3618. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Unsupervised Spectral–Spatial Feature Learning via Deep Residual Conv–Deconv Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kwon, H. Going Deeper with Contextual CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2017, 26, 4843–4855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Zou, J.; Yao, C.; Zhao, X.; Li, T.; Bai, G. HSI-CNN: A Novel Convolution Neural Network for Hyperspectral Image. In Proceedings of the 2018 International Conference on Audio, Language and Image Processing (ICALIP), Shanghai, China, 16–17 July 2018; pp. 464–469. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–Spatial Classification of Hyperspectral Imagery with 3D Convolutional Neural Network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Huang, R.; Guo, S.; Li, L.; Zhu, M.; Yang, S.; Jiao, L. NAS-Guided Lightweight Multiscale Attention Fusion Network for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 8754–8767. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Pereira, N.S.; Plaza, J.; Plaza, A. Ghostnet for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 1–16. [Google Scholar] [CrossRef]
- Bai, J.; Ding, B.; Xiao, Z.; Jiao, L.; Chen, H.; Regan, A.C. Hyperspectral Image Classification Based on Deep Attention Graph Convolutional Network. IEEE Trans. Geosci. Remote Sens. 2021, 1–16. [Google Scholar] [CrossRef]
- Ding, Y.; Guo, Y.; Chong, Y.; Pan, S.; Feng, J. Global Consistent Graph Convolutional Network for Hyperspectral Image Classification. IEEE Trans. Instrum. Meas. 2021, 70, 5501516. [Google Scholar] [CrossRef]
- Saha, S.; Mou, L.; Zhu, X.X.; Bovolo, F.; Bruzzone, L. Semisupervised Change Detection Using Graph Convolutional Network. IEEE Geosci. Remote Sens. Lett. 2021, 18, 607–611. [Google Scholar] [CrossRef]
- Ouyang, S.; Li, Y. Combining Deep Semantic Segmentation Network and Graph Convolutional Neural Network for Semantic Segmentation of Remote Sensing Imagery. Remote Sens. 2021, 13, 119. [Google Scholar] [CrossRef]
- Wan, S.; Gong, C.; Pan, S.; Yang, J. Multi-Level Graph Convolutional Network with Automatic Graph Learning for Hyperspectral Image Classification. arXiv 2020, arXiv:2009.09196. Available online: http://arxiv.org/abs/2009.09196 (accessed on 1 October 2021).
- Wan, S.; Gong, C.; Zhong, P.; Pan, S.; Li, G.; Yang, J. Hyperspectral Image Classification with Context-Aware Dynamic Graph Convolutional Network. IEEE Trans. Geosci. Remote Sens. 2021, 59, 597–612. [Google Scholar] [CrossRef]
- Chung, F.R.K.; Graham, F.C. Spectral Graph Theory; American Mathematical Society: Providence, RI, USA, 1997. [Google Scholar]
- Hammond, D.K.; VanderGheynst, P.; Gribonval, R. Wavelets on graphs via spectral graph theory. Appl. Comput. Harmon. Anal. 2011, 30, 129–150. [Google Scholar] [CrossRef] [Green Version]
- Ying, R.; You, J.; Morris, C.; Ren, X.; Hamilton, W.L.; Leskovec, J. Hierarchical Graph Representation Learning with Differentiable Pooling. arXiv 2019, arXiv:1806.08804. Available online: http://arxiv.org/abs/1806.08804 (accessed on 17 August 2021).
- Blanzieri, E.; Melgani, F. Nearest Neighbor Classification of Remote Sensing Images with the Maximal Margin Principle. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1804–1811. [Google Scholar] [CrossRef]
- Gao, L.; Li, J.; Khodadadzadeh, M.; Plaza, A.; Zhang, B.; He, Z.; Yan, H. Subspace-Based Support Vector Machines for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2015, 12, 349–353. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]













| Layer | Module | Input Tensor Size | Output Tensor Size |
|---|---|---|---|
| 1 | GCN | 32 × D × 49 | 32 × 32 × 49 |
| POOL | 32 × 32 × 49 | 32 × 32 × 16 | |
| 2 | OGC | 32 × 32 × 16 | 32 × 32 × 16 |
| POOL | 32 × 32 × 16 | 32 × 32 × 4 | |
| 3 | OGC | 32 × 32 × 4 | 32 × 32 × 4 |
| POOL | 32 × 32 × 4 | 32 × 32 × 1 | |
| 4 | FC | 32 × 32 | 32 × C |
| Class | Class Name | Train | Test |
|---|---|---|---|
| 1 | Alfalfa | 15 | 31 |
| 2 | Corn-notill | 50 | 1378 |
| 3 | Corn-mintill | 50 | 780 |
| 4 | Corn | 50 | 187 |
| 5 | Grass-pasture | 50 | 433 |
| 6 | Grass-trees | 50 | 680 |
| 7 | Grass-pasture-mowed | 15 | 13 |
| 8 | Hay-windrowed | 50 | 428 |
| 9 | Oats | 15 | 5 |
| 10 | Soybean-notill | 50 | 922 |
| 11 | Soybean-mintill | 50 | 2405 |
| 12 | Soybean-clean | 50 | 543 |
| 13 | Wheat | 50 | 155 |
| 14 | Woods | 50 | 1215 |
| 15 | Buildings-Grass-Trees-Drives | 50 | 336 |
| 16 | Stone-Steel-Towers | 50 | 43 |
| Total | 695 | 9554 |
| Class | Class Name | Train | Test |
|---|---|---|---|
| 1 | Asphalt | 50 | 6581 |
| 2 | Meadows | 50 | 18,599 |
| 3 | Gravel | 50 | 2049 |
| 4 | Trees | 50 | 3014 |
| 5 | Painted metal sheets | 50 | 1295 |
| 6 | Bare Soil | 50 | 4979 |
| 7 | Bitumen | 50 | 1280 |
| 8 | Self-Blocking Bricks | 50 | 3632 |
| 9 | Shadows | 50 | 897 |
| Total | 450 | 42,326 |
| Class | Class Name | Train | Test |
|---|---|---|---|
| 1 | Brocoli_green_weeds_1 | 50 | 1959 |
| 2 | Brocoli_green_weeds_2 | 50 | 3676 |
| 3 | Fallow | 50 | 1926 |
| 4 | Fallow_rough_plow | 50 | 1344 |
| 5 | Fallow_smooth | 50 | 2628 |
| 6 | Stubble | 50 | 3909 |
| 7 | Celery | 50 | 3529 |
| 8 | Grapes_untrained | 50 | 11,221 |
| 9 | Soil_vinyard_develop | 50 | 6153 |
| 10 | Corn_senesced_green_weeds | 50 | 3228 |
| 11 | Lettuce_romaine_4wk | 50 | 1018 |
| 12 | Lettuce_romaine_5wk | 50 | 1877 |
| 13 | Lettuce_romaine_6wk | 50 | 866 |
| 14 | Lettuce_romaine_7wk | 50 | 1020 |
| 15 | Vinyard_untrained | 50 | 7218 |
| 16 | Vinyard_vertical_trellis | 50 | 1757 |
| Total | 800 | 53,329 |
| Class | KNN | SVM | 2D-CNN | CNN-PPF | GCN | miniGCN | MDGCN | FuNet-C | SSOGCN |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 15.54 | 16.89 | 12.03 | 16.22 | 18.92 | 17.57 | 93.54 | 29.59 | 20.95 |
| 2 | 45.79 | 68.94 | 70.6 | 67.49 | 75.47 | 68.07 | 65.82 | 78.81 | 87.52 |
| 3 | 54.87 | 57.56 | 68.25 | 55.38 | 62.05 | 53.97 | 83.33 | 84.49 | 93.46 |
| 4 | 63.64 | 79.68 | 99.52 | 78.07 | 86.63 | 66.84 | 96.25 | 96.26 | 100.0 |
| 5 | 84.30 | 89.15 | 94.48 | 89.61 | 88.68 | 77.37 | 79.44 | 97.92 | 98.38 |
| 6 | 87.65 | 91.32 | 100.0 | 92.50 | 94.85 | 93.38 | 92.05 | 99.12 | 98.68 |
| 7 | 92.31 | 92.31 | 100.0 | 100.0 | 100.0 | 100.0 | 23.07 | 100.0 | 100.0 |
| 8 | 89.72 | 95.09 | 97.1 | 96.73 | 97.20 | 98.36 | 100.0 | 100.0 | 100.0 |
| 9 | 80.00 | 100.0 | 100.0 | 100.0 | 100.0 | 80.00 | 0.00 | 100.0 | 100.0 |
| 10 | 67.68 | 77.66 | 75.58 | 74.51 | 80.48 | 69.52 | 73.53 | 85.25 | 92.41 |
| 11 | 49.94 | 59.09 | 70.8 | 63.58 | 59.58 | 63.04 | 88.77 | 78.50 | 90.19 |
| 12 | 44.94 | 62.80 | 65.72 | 78.08 | 79.56 | 64.64 | 67.77 | 79.74 | 95.76 |
| 13 | 96.13 | 98.06 | 100.0 | 100.0 | 98.71 | 98.06 | 100.0 | 100.0 | 99.35 |
| 14 | 74.65 | 80.00 | 89.15 | 84.44 | 80.41 | 86.17 | 92.02 | 96.30 | 97.12 |
| 15 | 52.98 | 71.43 | 84.27 | 76.19 | 80.06 | 69.64 | 96.43 | 89.29 | 99.11 |
| 16 | 93.02 | 93.02 | 100.00 | 97.67 | 95.35 | 90.70 | 83.72 | 100.0 | 100.0 |
| OA(%) | 61.06 | 71.20 | 78.93 | 73.42 | 74.7 | 71.33 | 80.53 | 85.54 | 92.51 |
| AA(%) | 68.32 | 77.06 | 83.32 | 79.41 | 81.12 | 74.83 | 77.23 | 87.83 | 92.06 |
| KA(%) | 56.29 | 67.60 | 76.10 | 69.91 | 71.47 | 67.42 | 81.11 | 83.52 | 91.45 |
| Class | KNN | SVM | 2D-CNN | CNN-PPF | GCN | miniGCN | MDGCN | FuNet-C | SSOGCN |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 67.45 | 63.07 | 76.10 | 57.73 | 64.03 | 78.62 | 62.80 | 85.34 | 95.00 |
| 2 | 70.88 | 82.59 | 83.52 | 83.56 | 83.42 | 86.77 | 88.67 | 95.33 | 99.09 |
| 3 | 67.01 | 81.80 | 75.40 | 82.97 | 78.09 | 70.82 | 93.76 | 91.65 | 84.58 |
| 4 | 88.12 | 90.44 | 97.51 | 89.28 | 89.25 | 88.49 | 81.89 | 96.95 | 95.92 |
| 5 | 98.92 | 99.61 | 99.46 | 99.00 | 98.61 | 98.53 | 97.17 | 99.92 | 99.77 |
| 6 | 69.05 | 82.43 | 78.53 | 54.35 | 86.10 | 81.74 | 99.22 | 90.76 | 97.23 |
| 7 | 87.97 | 92.11 | 94.77 | 93.52 | 90.47 | 90.16 | 85.19 | 97.89 | 98.28 |
| 8 | 71.97 | 78.85 | 84.55 | 62.89 | 79.87 | 83.07 | 79.79 | 88.79 | 96.26 |
| 9 | 100.0 | 99.89 | 100.0 | 100.0 | 100.0 | 100.0 | 53.33 | 100.0 | 99.89 |
| OA(%) | 73.26 | 80.44 | 83.65 | 75.35 | 81.14 | 84.69 | 84.24 | 92.93 | 97.08 |
| AA(%) | 80.15 | 85.64 | 87.76 | 80.36 | 85.54 | 86.47 | 82.37 | 94.07 | 96.22 |
| KA(%) | 65.96 | 74.96 | 78.79 | 67.64 | 75.84 | 79.99 | 79.57 | 90.66 | 96.11 |
| Class | KNN | SVM | 2D-CNN | CNN-PPF | miniGCN | MDGCN | FuNet-C | SSOGCN |
|---|---|---|---|---|---|---|---|---|
| 1 | 88.42 | 97.13 | 95.61 | 85.15 | 97.60 | 100.0 | 100.0 | 100.0 |
| 2 | 88.25 | 97.08 | 97.03 | 99.97 | 99.89 | 100.0 | 99.97 | 100.0 |
| 3 | 93.82 | 91.49 | 96.0 | 94.65 | 93.87 | 63.11 | 99.22 | 99.88 |
| 4 | 90.48 | 99.56 | 99.55 | 99.63 | 99.03 | 97.06 | 99.55 | 99.75 |
| 5 | 83.87 | 92.84 | 96.88 | 97.41 | 97.45 | 99.74 | 97.34 | 97.26 |
| 6 | 82.02 | 99.72 | 99.39 | 99.82 | 99.97 | 100.0 | 100.0 | 100.0 |
| 7 | 87.79 | 99.3 | 99.12 | 99.60 | 99.63 | 100.0 | 99.43 | 100.0 |
| 8 | 50.84 | 57.58 | 51.38 | 92.06 | 67.13 | 85.47 | 64.46 | 83.12 |
| 9 | 81.75 | 97.69 | 97.40 | 99.58 | 99.38 | 100.0 | 99.93 | 99.80 |
| 10 | 97.40 | 85.0 | 90.68 | 89.71 | 92.13 | 98.66 | 97.15 | 97.84 |
| 11 | 78.09 | 92.81 | 98.13 | 93.03 | 97.35 | 48.78 | 99.71 | 99.45 |
| 12 | 87.16 | 98.74 | 99.89 | 99.73 | 99.89 | 100.0 | 100.0 | 99.94 |
| 13 | 88.34 | 99.10 | 99.65 | 99.77 | 99.54 | 100.0 | 100.0 | 100.0 |
| 14 | 80.29 | 90.81 | 98.63 | 93.24 | 98.14 | 99.63 | 98.92 | 100.0 |
| 15 | 63.52 | 52.01 | 66.42 | 23.04 | 70.16 | 81.84 | 76.20 | 91.99 |
| 16 | 89.53 | 93.66 | 97.10 | 98.01 | 98.41 | 93.36 | 98.69 | 99.94 |
| OA(%) | 76.05 | 81.84 | 83.41 | 85.98 | 87.85 | 91.72 | 88.84 | 95.05 |
| AA(%) | 83.22 | 90.28 | 92.68 | 91.52 | 94.35 | 91.70 | 95.66 | 98.06 |
| KA(%) | 73.52 | 79.85 | 81.62 | 84.27 | 86.5 | 90.77 | 87.61 | 94.49 |
| Data Set | Without OGC | Without AGL | Without Graph Pooling | SSOGCN |
|---|---|---|---|---|
| Indian Pines | 90.12 | 91.38 | 86.65 | 92.51 |
| Pavia University | 94.91 | 94.04 | 93.47 | 97.08 |
| Salinas | 93.74 | 93.85 | 88.60 | 95.05 |
| Methods | Time (s) | Params (K) |
|---|---|---|
| 2D-CNN | 178.06 | 30.0 |
| CNN-PPF | 61.86 | 44.82 |
| GCN | 198.12 | 4.36 |
| miniGCN | 776.86 | 28.70 |
| MDGCN | 322.01 | 13.07 |
| FuNet-C | 678.13 | 148.59 |
| SSOGCN | 597.05 | 23.41 |
| Methods | Time (s) | Params (K) |
|---|---|---|
| 2D-CNN | 167.65 | 29.10 |
| CNN-PPF | 149.18 | 31.50 |
| GCN | 410.38 | 2.27 |
| miniGCN | 2925.70 | 15.19 |
| MDGCN | 447.87 | 6.81 |
| FuNet-C | 2753.58 | 77.44 |
| SSOGCN | 1471.03 | 12.22 |
| Methods | Time (s) | Params (K) |
|---|---|---|
| 2D-CNN | 283.94 | 30.00 |
| CNN-PPF | 243.28 | 45.32 |
| miniGCN | 1273.54 | 29.22 |
| MDGCN | 458.48 | 13.31 |
| FuNet-C | 5002.85 | 150.264 |
| SSOGCN | 1183.93 | 23.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Luo, H.; Song, W.; Mei, H.; Su, C. Spectral-Spatial Offset Graph Convolutional Networks for Hyperspectral Image Classification. Remote Sens. 2021, 13, 4342. https://doi.org/10.3390/rs13214342
Zhang M, Luo H, Song W, Mei H, Su C. Spectral-Spatial Offset Graph Convolutional Networks for Hyperspectral Image Classification. Remote Sensing. 2021; 13(21):4342. https://doi.org/10.3390/rs13214342
Chicago/Turabian StyleZhang, Minghua, Hongling Luo, Wei Song, Haibin Mei, and Cheng Su. 2021. "Spectral-Spatial Offset Graph Convolutional Networks for Hyperspectral Image Classification" Remote Sensing 13, no. 21: 4342. https://doi.org/10.3390/rs13214342