Mapping Regional Soil Organic Matter Based on Sentinel-2A and MODIS Imagery Using Machine Learning Algorithms and Google Earth Engine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Field Sampling
2.2. Remotely Sensed Imagery
2.2.1. Sentinel-2A and MODIS Satellite Data
2.2.2. Land Cover Dataset
2.3. Variable Datasets Construction
2.3.1. Spectral Index Construction
2.3.2. Terrain Factor
2.4. Methodology
2.4.1. Machine Learning Algorithms
2.4.2. Assessment and Prediction
3. Results and Analysis
3.1. Descriptive Statistics of the SOM Content
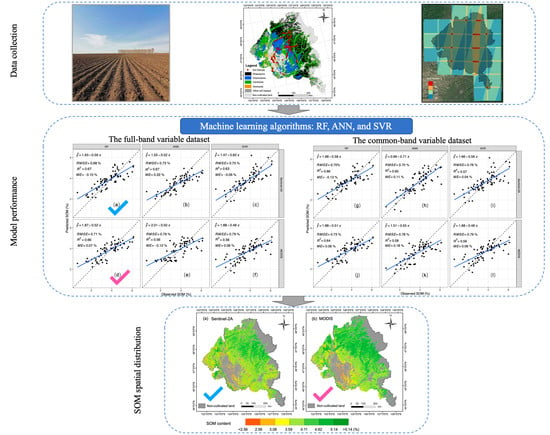
3.2. Model Performance
3.3. Spatial Characteristics of Predicted SOM Map
4. Discussion
4.1. Comparison of Sentinel-2A-Based and MODIS-Based Models
4.2. Applicability of Machine Learning Algorithms
4.3. Advantages of the Google Earth Engine for SOM Prediction
4.4. Limitations and Future Researches
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sparling, G.P.; Wheeler, D.; Vesely, E.-T.; Schipper, L.A. What is Soil Organic Matter Worth? J. Environ. Qual. 2006, 35, 548–557. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.; Li, X.; Huang, L.; Yu, D.; Shi, X.; Chen, H.; Xing, S. Effects of soil map scales on simulating soil organic carbon changes of upland soils in Eastern China. Geoderma 2018, 312, 159–169. [Google Scholar] [CrossRef]
- Manlay, R.J.; Feller, C.; Swift, M. Historical evolution of soil organic matter concepts and their relationships with the fertility and sustainability of cropping systems. Agric. Ecosyst. Environ. 2007, 119, 217–233. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, S.; Yang, Y.; Zhao, R.; Shi, Z.; Rossel, R.A.V. National digital soil map of organic matter in topsoil and its associated uncertainty in 1980’s China. Geoderma 2019, 335, 47–56. [Google Scholar] [CrossRef]
- Guo, L.; Zhao, C.; Zhang, H.; Chen, Y.; Linderman, M.; Zhang, Q.; Liu, Y. Comparisons of spatial and non-spatial models for predicting soil carbon content based on visible and near-infrared spectral technology. Geoderma 2017, 285, 280–292. [Google Scholar] [CrossRef]
- Huang, B.; Sun, W.; Zhao, Y.; Zhu, J.; Yang, R.; Zou, Z.; Ding, F.; Su, J. Temporal and spatial variability of soil organic matter and total nitrogen in an agricultural ecosystem as affected by farming practices. Geoderma 2007, 139, 336–345. [Google Scholar] [CrossRef]
- Nocita, M.; Stevens, A.; Noon, C.; van Wesemael, B. Prediction of soil organic carbon for different levels of soil moisture using Vis-NIR spectroscopy. Geoderma 2013, 199, 37–42. [Google Scholar] [CrossRef]
- Feng, N.; Hongfen, Z.; Rutian, B. Hyperspectral prediction of soil organic matter content in the Reclamation cropland of Coal Mining Areas in the Loess Platesu. Sci. Agric. Sin. 2016, 49, 2126–2135. [Google Scholar]
- Juice, S.M.; Templer, P.H.; Phillips, N.G.; Ellison, A.M.; Pelini, S.L. Ecosystem warming increases sap flow rates of northern red oak trees. Ecosphere 2016, 7, e01221. [Google Scholar] [CrossRef]
- Mishra, U.; Torn, M.S.; Masanet, E.; Ogle, S.M. Improving regional soil carbon inventories: Combining the IPCC carbon inventory method with regression kriging. Geoderma 2012, 189–190, 288–295. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Meersmans, J.; De Ridder, F.; Canters, F.; De Baets, S.; Van Molle, M. A multiple regression approach to assess the spatial distribution of Soil Organic Carbon (SOC) at the regional scale (Flanders, Belgium). Geoderma 2008, 143, 1–13. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Van Raij, B.; Andrade, J.C.; de Cantarella, H.; Quaggio, J.A. Análise Química para Avaliação da Fertilidade de Solos Tropicais; Instituto Agronômico: Campinas, Brazil, 2001.
- Lagacherie, P. Digital Soil Mapping: A State of the Art. Digit. Soil Mapp. Ltd. Data 2008, 3–14. [Google Scholar] [CrossRef]
- Bie, S.W.; Beckett, P.H.T. Quality control in soil survey: II. The costs of soil survey. J. Soil Sci. 1971, 22, 453–465. [Google Scholar] [CrossRef]
- Zhao, Z.; Ashraf, M.I.; Meng, F.-R. Model prediction of soil drainage classes over a large area using a limited number of field samples: A case study in the province of Nova Scotia, Canada. Can. J. Soil Sci. 2013, 93, 73–83. [Google Scholar] [CrossRef]
- Matheron, G. Principles of geostatistics. Econ. Geol. 1963, 58, 1246–1266. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, Y.; Shen, C.; Ye, H.; Du, Y. Spatial prediction of soil organic matter using terrain indices and categorical variables as auxiliary information. Geoderma 2012, 171–172, 35–43. [Google Scholar] [CrossRef]
- Tziachris, P.; Aschonitis, V.; Chatzistathis, T.; Papadopoulou, M. Assessment of spatial hybrid methods for predicting soil organic matter using DEM derivatives and soil parameters. Catena 2019, 174, 206–216. [Google Scholar] [CrossRef]
- Schloeder, C.; Zimmerman, N.; Jacobs, M. Comparison of Methods for Interpolating Soil Properties Using Limited Data. Soil Sci. Soc. Am. J. 2001, 65, 470–479. [Google Scholar] [CrossRef]
- Wu, C.; Wu, J.; Luo, Y.; Zhang, L.; Degloria, S.D. Spatial Prediction of Soil Organic Matter Content Using Cokriging with Remotely Sensed Data. Soil Sci. Soc. Am. J. 2009, 73, 1202–1208. [Google Scholar] [CrossRef]
- Dai, F.; Zhou, Q.; Lv, Z.; Wang, X.; Liu, G. Spatial prediction of soil organic matter content integrating artificial neural network and ordinary kriging in Tibetan Plateau. Ecol. Indic. 2014, 45, 184–194. [Google Scholar] [CrossRef]
- Heuvelink, G.; Bierkens, M.F.P. Combining soil maps with interpolations from point observations to predict quantitative soil properties. Geoderma 1992, 55, 1–15. [Google Scholar] [CrossRef]
- McBratney, A.B.; Santos, M.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Q.; Benoy, G.; Chow, T.L.; Xing, Z.; Rees, H.W.; Meng, F.-R. Using artificial neural network models to produce soil organic carbon content distribution maps across landscapes. Can. J. Soil Sci. 2010, 90, 75–87. [Google Scholar] [CrossRef]
- Ward, K.J.; Chabrillat, S.; Neumann, C.; Foerster, S. A remote sensing adapted approach for soil organic carbon prediction based on the spectrally clustered LUCAS soil database. Geoderma 2019, 353, 297–307. [Google Scholar] [CrossRef]
- Webster, R.; Oliver, M.A. Geostatistics for Environmental Scientists; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Guo, P.-T.; Wu, W.; Sheng, Q.-K.; Li, M.-F.; Liu, H.-B.; Wang, Z.-Y. Prediction of soil organic matter using artificial neural network and topographic indicators in hilly areas. Nutr. Cycl. Agroecosyst. 2013, 95, 333–344. [Google Scholar] [CrossRef]
- Takata, Y.; Funakawa, S.; Akshalov, K.; Ishida, N.; Kosaki, T. Spatial prediction of soil organic matter in northern Kazakhstan based on topographic and vegetation information. Soil Sci. Plant Nutr. 2007, 53, 289–299. [Google Scholar] [CrossRef]
- Abrougui, K.; Gabsi, K.; Mercatoris, B.; Khemis, C.; Amami, R.; Chehaibi, S. Prediction of organic potato yield using tillage systems and soil properties by artificial neural network (ANN) and multiple linear regressions (MLR). Soil Tillage Res. 2019, 190, 202–208. [Google Scholar] [CrossRef]
- Adhikari, K.; Hartemink, A.E. Digital Mapping of Topsoil Carbon Content and Changes in the Driftless Area of Wisconsin, USA. Soil Sci. Soc. Am. J. 2015, 79, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.-F.; Shi, X.; Yu, D.; Pan, X.; Wang, H.; Sun, W. Using GIS spatial distribution to predict soil organic carbon in subtropical China. Pedosphere 2004, 14, 425–431. [Google Scholar]
- Bogunovic, I.; Trevisani, S.; Pereira, P.; Vukadinovic, V. Mapping soil organic matter in the Baranja region (Croatia): Geological and anthropic forcing parameters. Sci. Total Environ. 2018, 643, 335–345. [Google Scholar] [CrossRef]
- Rosero-Vlasova, O.A.; Vlassova, L.; Pérez-Cabello, F.; Montorio, R.; Nadal-Romero, E. Modeling soil organic matter and texture from satellite data in areas affected by wildfires and cropland abandonment in Aragón, Northern Spain. J. Appl. Remote Sens. 2018, 12, 042803. [Google Scholar] [CrossRef]
- Bobrovsky, M.; Komarov, A.; Mikhailov, A.; Khanina, L. Modelling dynamics of soil organic matter under different historical land-use management techniques in European Russia. Ecol. Model. 2010, 221, 953–959. [Google Scholar] [CrossRef]
- Alvarez, R.; Steinbach, H.S.; Bono, A. An Artificial Neural Network Approach for Predicting Soil Carbon Budget in Agroecosystems. Soil Sci. Soc. Am. J. 2011, 75, 965–975. [Google Scholar] [CrossRef]
- Gautam, R.; Panigrahi, S.; Franzen, D.; Sims, A. Residual soil nitrate prediction from imagery and non-imagery information using neural network technique. Biosyst. Eng. 2011, 110, 20–28. [Google Scholar] [CrossRef]
- Poggio, L.; Gimona, A.; Spezia, L.; Brewer, M.J. Bayesian spatial modelling of soil properties and their uncertainty: The example of soil organic matter in Scotland using R-INLA. Geoderma 2016, 277, 69–82. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Q.; Sun, D.; Ding, X.; Meng, F.-R. Extended model prediction of high-resolution soil organic matter over a large area using limited number of field samples. Comput. Electron. Agric. 2020, 169, 105172. [Google Scholar] [CrossRef]
- Chen, G.; Li, S.; Knibbs, L.D.; Hamm, N.A.S.; Cao, W.; Li, T.; Guo, J.; Ren, H.; Abramson, M.J.; Guo, Y. A machine learning method to estimate PM2.5 concentrations across China with remote sensing, meteorological and land use information. Sci. Total Environ. 2018, 636, 52–60. [Google Scholar] [CrossRef]
- Subburayalu, S.K.; Slater, B.K. Soil Series Mapping by Knowledge Discovery from an Ohio County Soil Map. Soil Sci. Soc. Am. J. 2013, 77, 1254–1268. [Google Scholar] [CrossRef]
- Wiesmeier, M.; Barthold, F.; Blank, B.; Kögel-Knabner, I. Digital mapping of soil organic matter stocks using Random Forest modeling in a semi-arid steppe ecosystem. Plant Soil 2011, 340, 7–24. [Google Scholar] [CrossRef]
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island—Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, Y.; Chen, Y.; Liu, J.; Zhang, L. Soil organic matter prediction based on remote sensing data and random forest model in Shaanxi Province. J. Nat. Resour. 2017, 32, 1074–1086. [Google Scholar]
- Fernandes, M.M.H.; Coelho, A.P.; Fernandes, C.; Da Silva, M.F.; Marta, C.C.D. Estimation of soil organic matter content by modeling with artificial neural networks. Geoderma 2019, 350, 46–51. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Deiss, L.; Margenot, A.J.; Culman, S.W.; Demyan, M.S. Tuning support vector machines regression models improves prediction accuracy of soil properties in MIR spectroscopy. Geoderma 2020, 365, 114227. [Google Scholar] [CrossRef]
- Ballabio, C. Spatial prediction of soil properties in temperate mountain regions using support vector regression. Geoderma 2009, 151, 338–350. [Google Scholar] [CrossRef]
- Chen, D.; Chang, N.; Xiao, J.; Zhou, Q.; Wu, W. Mapping dynamics of soil organic matter in croplands with MODIS data and machine learning algorithms. Sci. Total Environ. 2019, 669, 844–855. [Google Scholar] [CrossRef]
- Kumar, S.; Lal, R. Mapping the organic carbon stocks of surface soils using local spatial interpolator. J. Environ. Monit. 2011, 13, 3128–3135. [Google Scholar] [CrossRef]
- Qi-Yong, Y.; Zhong-Cheng, J.; Wen-Jun, L.; Hui, L. Prediction of soil organic matter in peak-cluster depression region using kriging and terrain indices. Soil Tillage Res. 2014, 144, 126–132. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, X.; Liu, H.; Zhang, X.; Qu, C.; Hu, W.; Zhang, H. Quantitative analysis of reflectance spectrum of Black soil as affected by soil moisture for prediction of soil moisture in black soil. Acta Pedol. Sin. 2014, 51, 1021–1026. [Google Scholar]
- Dou, X.; Wang, X.; Liu, H.; Zhang, X.; Meng, L.; Pan, Y.; Yu, Z.; Cui, Y. Prediction of soil organic matter using multi-temporal satellite images in the Songnen Plain, China. Geoderma 2019, 356, 113896. [Google Scholar] [CrossRef]
- Liu, S.; An, N.; Yang, J.; Dong, S.; Wang, C.; Yin, Y. Prediction of soil organic matter variability associated with different land use types in mountainous landscape in southwestern Yunnan province, China. Catena 2015, 133, 137–144. [Google Scholar] [CrossRef]
- Zhang, X.; Dou, X.; Xie, Y.; Liu, H.; Wang, N.; Wang, X.; Pan, Y. Remote sensing inversion model of soil organic matter in farmland by introducing temporal information. Trans. Chin. Soc. Agric. Eng. 2018, 34, 143–150. [Google Scholar]
- Liu, H.; Pan, Y.; Dou, X.; Zhang, X.; Qiu, Z.; Xu, M.; Xie, Y.; Wang, N. Soil organic matter content inversion model with remote sensing image in field scale of blacksoil area. Trans. Chin. Soc. Agric. Eng. 2018, 34, 127–133. [Google Scholar]
- Meng, X.; Bao, Y.; Liu, J.; Liu, H.; Zhang, X.; Zhang, Y.; Wang, P.; Tang, H.; Kong, F. Regional soil organic carbon prediction model based on a discrete wavelet analysis of hyperspectral satellite data. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102111. [Google Scholar] [CrossRef]
- Bao, Y.; Meng, X.; Ustin, S.; Wang, X.; Zhang, X.; Liu, H.; Tang, H. Vis-SWIR spectral prediction model for soil organic matter with different grouping strategies. Catena 2020, 195, 104703. [Google Scholar] [CrossRef]
- Liu, H.J.; Zhang, B.; Liu, D.W.; Wang, Z.M.; Song, K.S.; Yang, F. Study on Quantitatively Remote Sensing Typical Soils in Songnen Plain, Northeast China. J. Remote Sens. 2008, 12, 647–654. [Google Scholar]
- Rossel, R.V.; Walvoort, D.; McBratney, A.; Janik, L.J.; Skjemstad, J.O. Visible, near infrared, mid infrared or combined diffuse reflectance spectroscopy for simultaneous assessment of various soil properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Chen, F.; Kissel, D.E.; West, L.T.; Adkins, W. Field-Scale Mapping of Surface Soil Organic Carbon Using Remotely Sensed Imagery. Soil Sci. Soc. Am. J. 2000, 64, 746–753. [Google Scholar] [CrossRef] [Green Version]
- Mccarty, G.W.; Reeves, J.B.; Reeves, V.B.; Follett, R.F.; Kimble, J.M. Mid-Infrared and Near-Infrared Diffuse Reflectance Spectroscopy for Soil Carbon Measurement. Soil Sci. Soc. Am. J. 2002, 66, 640–646. [Google Scholar] [CrossRef]
- Summers, D.; Lewis, M.; Ostendorf, B.; Chittleborough, D. Visible near-infrared reflectance spectroscopy as a predictive indicator of soil properties. Ecol. Indic. 2011, 11, 123–131. [Google Scholar] [CrossRef]
- Bilgili, A.V.; van Es, H.M.; Akbas, F.; Durak, A.; Hively, W.D. Visible-near infrared reflectance spectroscopy for assessment of soil properties in a semi-arid area of Turkey. J. Arid. Environ. 2010, 74, 229–238. [Google Scholar] [CrossRef]
- Xiao, W.; Chen, W.; He, T.; Ruan, L.; Guo, J. Multi-Temporal Mapping of Soil Total Nitrogen Using Google Earth Engine across the Shandong Province of China. Sustainability 2020, 12, 10274. [Google Scholar] [CrossRef]
- Liu, F.; Geng, X.; Zhu, A.-X.; Fraser, W.; Waddell, A. Soil texture mapping over low relief areas using land surface feedback dynamic patterns extracted from MODIS. Geoderma 2012, 171–172, 44–52. [Google Scholar] [CrossRef]
- Zhang, B.; Song, X.-F.; Zhang, Y.-H.; Han, D.-M.; Tang, C.-Y.; Yang, L.; Wang, Z.-L. The renewability and quality of shallow groundwater in Sanjiang and Songnen Plain, Northeast China. J. Integr. Agric. 2017, 16, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-C.; Fu, Y.; Liu, L.; Wang, K.; Wang, D.-L. Seasonal Variability in Soil Inorganic Nitrogen Across Borders Between Woodland and Farmland in the Songnen Plain of Northeast China. Pedosphere 2013, 23, 472–481. [Google Scholar] [CrossRef]
- Duan, X.; Xie, Y.; Liu, G.; Gao, X.; Lu, H. Field capacity in black soil region, Northeast China. Chin. Geogr. Sci. 2010, 20, 406–413. [Google Scholar] [CrossRef]
- Song, X.-D.; Yang, F.; Ju, B.; Li, D.-C.; Zhao, Y.-G.; Yang, J.-L.; Zhang, G.-L. The influence of the conversion of grassland to cropland on changes in soil organic carbon and total nitrogen stocks in the Songnen Plain of Northeast China. Catena 2018, 171, 588–601. [Google Scholar] [CrossRef]
- Shi, X.Z.; Yu, D.S.; Xu, S.X.; Warner, E.D.; Wang, H.J.; Sun, W.X.; Zhao, Y.C.; Gong, Z.T. Cross-reference for relating Genetic Soil Classification of China with WRB at different scales. Geoderma 2010, 155, 344–350. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources; World Soil Resources Reports No. 103; FAO: Rome, Italy, 2006. [Google Scholar]
- O’Kelly, B.C. Accurate determination of moisture content of organic soils using the oven drying method. Dry. Technol. 2004, 22, 1767–1776. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Methods of Soil Analysis: Part 3 Chemical Methods, 5.3; Soil Science Society of America: Madison, WI, USA, 1996; pp. 961–1010. [Google Scholar]
- Friedl, M.A.; Sulla-Menashe, D.; Tan, B.; Schneider, A.; Ramankutty, N.; Sibley, A.; Huang, X. MODIS Collection 5 global land cover: Algorithm refinements and characterization of new datasets. Remote Sens. Environ. 2010, 114, 168–182. [Google Scholar] [CrossRef]
- Huang, N.; He, J.-S.; Niu, Z. Estimating the spatial pattern of soil respiration in Tibetan alpine grasslands using Landsat TM images and MODIS data. Ecol. Indic. 2013, 26, 117–125. [Google Scholar] [CrossRef]
- Bao, N.; Wu, L.; Ye, B.; Yang, K.; Zhou, W. Assessing soil organic matter of reclaimed soil from a large surface coal mine using a field spectroradiometer in laboratory. Geoderma 2017, 288, 47–55. [Google Scholar] [CrossRef]
- Frazier, B.E.; Cheng, Y. Remote sensing of soils in the Eastern Palouse region with landsat thematic mapper. Remote Sens. Environ. 1989, 28, 317–325. [Google Scholar] [CrossRef]
- Liu, H.-J.; Ning, D.-H.; Kang, R.; Jin, H.-N.; Zhang, X.-L.; Sheng, L. A Study on Predicting Model of Organic Matter Contend Incorporating Soil Moisture Variation. Spectrosc. Spectr. Anal. 2017, 37, 566–570. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Zhang, C.-T.; Yang, Y. Can the spatial prediction of soil organic matter be improved by incorporating multiple regression confidence intervals as soft data into BME method? Catena 2019, 178, 322–334. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Gao, B.-C. NDWI—A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Kheir, R.B.; Greve, M.H.; Bøcher, P.K.; Greve, M.B.; Larsen, R.; McCloy, K. Predictive mapping of soil organic carbon in wet cultivated lands using classification-tree based models: The case study of Denmark. J. Environ. Manag. 2010, 91, 1150–1160. [Google Scholar] [CrossRef]
- Li, Y. Can the spatial prediction of soil organic matter contents at various sampling scales be improved by using regression kriging with auxiliary information? Geoderma 2010, 159, 63–75. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, S.; Yang, Y.; Zhao, R.; Shi, Z.; Rossel, R.V. Baseline map of soil organic matter in China and its associated uncertainty. Geoderma 2019, 335, 47–56. [Google Scholar] [CrossRef]
- Yang, L.; He, X.; Shen, F.; Zhou, C.; Zhu, A.-X.; Gao, B.; Chen, Z.; Li, M. Improving prediction of soil organic carbon content in croplands using phenological parameters extracted from NDVI time series data. Soil Tillage Res. 2020, 196, 104465. [Google Scholar] [CrossRef]
- Jin, X.; Du, J.; Liu, H.; Wang, Z.; Song, K. Remote estimation of soil organic matter content in the Sanjiang Plain, Northest China: The optimal band algorithm versus the GRA-ANN model. Agric. For. Meteorol. 2016, 218–219, 250–260. [Google Scholar] [CrossRef]
- Bowers, S.; Hanks, A. Reflection of Radiant Energy from Soil. Soil Sci. 1965, 100, 130–138. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yao, F.; Li, L.; Zhang, W. Relationships between water indexes and soil moisture/crop physiological indexes using ground-based remote sensing and field experiments. Trans. Chin. Soc. Agric. Eng. 2010, 26, 151–155. [Google Scholar]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, N.N.; Angiuli, E.; Gamba, P.; Gaughan, A.; Lisini, G.; Stevens, F.R.; Tatem, A.J.; Trianni, G. Multitemporal settlement and population mapping from Landsat using Google Earth Engine. Int. J. Appl. Earth Obs. Geoinf. 2015, 35, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Gong, P.; Qi, S.; Liu, C.; Xiong, T. Mapping bamboo with regional phenological characteristics derived from dense Landsat time series using Google Earth Engine. Int. J. Remote Sens. 2019, 40, 9541–9555. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, H.; Li, Z.; Hackman, K.O.; Liu, C.; Andriamiarisoa, R.L.; Raherivelo, T.N.A.N.; Li, Y.; Gong, P. Automatic High-Resolution Land Cover Production in Madagascar Using Sentinel-2 Time Series, Tile-Based Image Classification and Google Earth Engine. Remote Sens. 2020, 12, 3663. [Google Scholar] [CrossRef]
- Strobl, C.; Malley, J.; Tutz, G. An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol. Methods 2009, 14, 323–348. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cutler, A. Remembering Leo Breiman. Ann. Appl. Stat. 2010, 4, 1621–1633. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.J.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009; pp. 267–268. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Xu, S.; Zhao, Y.; Wang, M.; Shi, X. Comparison of multivariate methods for estimating selected soil properties from intact soil cores of paddy fields by Vis–NIR spectroscopy. Geoderma 2018, 310, 29–43. [Google Scholar] [CrossRef]
- Chen, G.; Ge, Z. SVM-tree and SVM-forest algorithms for imbalanced fault classification in industrial processes. IFAC J. Syst. Control. 2019, 8, 100052. [Google Scholar] [CrossRef]
- Yong, L. Supervised classification of multispectral remote sensing image using BP neural network. J. Infrared Millim. Waves 1998, 2, 153–156. [Google Scholar]
- Rudiyanto; Minasny, B.; Setiawan, B.I.; Saptomo, S.K.; McBratney, A.B. Open digital mapping as a cost-effective method for mapping peat thickness and assessing the carbon stock of tropical peatlands. Geoderma 2018, 313, 25–40. [Google Scholar] [CrossRef]
- Gomez, C.; Rossel, R.A.V.; McBratney, A.B. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, J.; Wang, J.; Ge, X. Prediction of soil organic matter in northwestern China using fractional-order derivative spectroscopy and modified normalized difference indices. Catena 2019, 185, 104257. [Google Scholar] [CrossRef]
- Wang, B.; Waters, C.; Orgill, S.; Gray, J.; Cowie, A.; Clark, A.; Liu, D.L. High resolution mapping of soil organic carbon stocks using remote sensing variables in the semi-arid rangelands of eastern Australia. Sci. Total Environ. 2018, 630, 367–378. [Google Scholar] [CrossRef]
- Mishra, U.; Lal, R.; Liu, D.; Van Meirvenne, M. Predicting the Spatial Variation of the Soil Organic Carbon Pool at a Regional Scale. Soil Sci. Soc. Am. J. 2010, 74, 906–914. [Google Scholar] [CrossRef]
- Zhao, M.-S.; Rossiter, D.G.; Li, D.-C.; Zhao, Y.-G.; Liu, F.; Zhang, G.-L. Mapping soil organic matter in low-relief areas based on land surface diurnal temperature difference and a vegetation index. Ecol. Indic. 2014, 39, 120–133. [Google Scholar] [CrossRef]
- Jain, M.; Mondal, P.; DeFries, R.S.; Small, C.; Galford, G.L. Mapping cropping intensity of smallholder farms: A comparison of methods using multiple sensors. Remote Sens. Environ. 2013, 134, 210–223. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xiao, X.; Qin, Y.; Wang, J.; Xu, X.; Hu, Y.; Qiao, Z. Mapping cropping intensity in China using time series Landsat and Sentinel-2 images and Google Earth Engine. Remote Sens. Environ. 2020, 239, 111624. [Google Scholar] [CrossRef]
- Li, L.; Li, N.; Lu, D.; Chen, Y. Mapping Moso bamboo forest and its on-year and off-year distribution in a subtropical region using time-series Sentinel-2 and Landsat 8 data. Remote Sens. Environ. 2019, 231, 111265. [Google Scholar] [CrossRef]
- Mahmoudzadeh, H.; Matinfar, H.R.; Taghizadeh-Mehrjardi, R.; Kerry, R. Spatial prediction of soil organic carbon using machine learning techniques in western Iran. Geoderma Reg. 2020, 21, e00260. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, Q.; Huang, J.Z.; Ng, M.K.; Li, X. Stratified sampling for feature subspace selection in random forests for high dimensional data. Pattern Recognit. 2013, 46, 769–787. [Google Scholar] [CrossRef]
- Menze, B.H.; Kelm, B.M.; Masuch, R.; Himmelreich, U.; Bachert, P.; Petrich, W.; Hamprecht, F.A. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009, 10, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heung, B.; Bulmer, C.E.; Schmidt, M.G. Predictive soil parent material mapping at a regional-scale: A Random Forest approach. Geoderma 2014, 214–215, 141–154. [Google Scholar] [CrossRef]
- Kuter, S. Completing the machine learning saga in fractional snow cover estimation from MODIS Terra reflectance data: Random forests versus support vector regression. Remote Sens. Environ. 2021, 255, 112294. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, Y. (Eds.) Ensemble Machine Learning: Methods and Applications; Springer: Berlin, Germany, 2012. [Google Scholar]
- Boulesteix, A.-L.; Janitza, S.; Kruppa, J.; König, I.R. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 493–507. [Google Scholar] [CrossRef] [Green Version]
- Benke, K.; Norng, S.; Robinson, N.; Chia, K.; Rees, D.; Hopley, J. Development of pedotransfer functions by machine learning for prediction of soil electrical conductivity and organic carbon content. Geoderma 2020, 366, 114210. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can we open the black box of AI? Nature 2016, 538, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Burges, C.J. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Hou, J.; Huang, C.; Zhang, Y.; Guo, J. On the Value of Available MODIS and Landsat8 OLI Image Pairs for MODIS Fractional Snow Cover Mapping Based on an Artificial Neural Network. IEEE Trans. Geosci. Remote Sens. 2020, 58, 4319–4334. [Google Scholar] [CrossRef]
- Suykens, J.A. Advances in Learning Theory: Methods, Models, and Applications; IOS Press: Amsterdam, The Netherlands, 2003; Volume 190. [Google Scholar]
- Huang, H.; Chen, Y.; Clinton, N.; Wang, J.; Wang, X.; Liu, C.; Gong, P.; Yang, J.; Bai, Y.; Zheng, Y.; et al. Mapping major land cover dynamics in Beijing using all Landsat images in Google Earth Engine. Remote Sens. Environ. 2017, 202, 166–176. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Kumar, L.; Mutanga, O. Google Earth Engine Applications Since Inception: Usage, Trends, and Potential. Remote Sens. 2018, 10, 1509. [Google Scholar] [CrossRef] [Green Version]
- Žížala, D.; Minařík, R.; Zádorová, T. Soil Organic Carbon Mapping Using Multispectral Remote Sensing Data: Prediction Ability of Data with Different Spatial and Spectral Resolutions. Remote Sens. 2019, 11, 2947. [Google Scholar] [CrossRef]
- Gallo, B.C.; Demattê, J.A.M.; Rizzo, R.; Safanelli, J.L.; Mendes, W.D.S.; Lepsch, I.F.; Sato, M.V.; Romero, D.J.; Lacerda, M.P.C. Multi-Temporal Satellite Images on Topsoil Attribute Quantification and the Relationship with Soil Classes and Geology. Remote Sens. 2018, 10, 1571. [Google Scholar] [CrossRef]
- Diek, S.; Fornallaz, F.; Schaepman, M.E.; De Jong, R. Barest Pixel Composite for Agricultural Areas Using Landsat Time Series. Remote Sens. 2017, 9, 1245. [Google Scholar] [CrossRef] [Green Version]
- Blasch, G.; Spengler, D.; Itzerott, S.; Wessolek, G. Organic Matter Modeling at the Landscape Scale Based on Multitemporal Soil Pattern Analysis Using RapidEye Data. Remote Sens. 2015, 7, 11125–11150. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, S.; Holden, N.M. Determination of Soil Organic Matter and Carbon Fractions in Forest Top Soils using Spectral Data Acquired from Visible-Near Infrared Hyperspectral Images. Soil Sci. Soc. Am. J. 2012, 76, 586–596. [Google Scholar] [CrossRef]
- Ataieyan, P.; Moghaddam, P.A.; Sepehr, E. Estimation of Soil Organic Carbon using Artificial Neural Network and Multiple Linear Regression Models based on Color Image Processing. J. Agric. Mach. 2018, 8, 137–148. [Google Scholar]
- Selige, T.; Böhner, J.; Schmidhalter, U. High resolution topsoil mapping using hyperspectral image and field data in multivariate regression modeling procedures. Geoderma 2006, 136, 235–244. [Google Scholar] [CrossRef]
- Silvero, N.E.Q.; Demattê, J.A.M.; Amorim, M.T.A.; dos Santos, N.V.; Rizzo, R.; Safanelli, J.L.; Poppiel, R.R.; Mendes, W.D.S.; Bonfatti, B.R. Soil variability and quantification based on Sentinel-2 and Landsat-8 bare soil images: A comparison. Remote Sens. Environ. 2021, 252, 112117. [Google Scholar] [CrossRef]
- Lin, C.; Zhu, A.-X.; Wang, Z.; Wang, X.; Ma, R. The refined spatiotemporal representation of soil organic matter based on remote images fusion of Sentinel-2 and Sentinel-3. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102094. [Google Scholar] [CrossRef]
- Zeng, C.; Yang, L.; Zhu, A.-X.; Rossiter, D.G.; Liu, J.; Liu, J.; Qin, C.; Wang, D. Mapping soil organic matter concentration at different scales using a mixed geographically weighted regression method. Geoderma 2016, 281, 69–82. [Google Scholar] [CrossRef]
- Kumar, S.; Lal, R.; Liu, D.; Rafiq, R. Estimating the spatial distribution of organic carbon density for the soils of Ohio, USA. J. Geogr. Sci. 2013, 23, 280–296. [Google Scholar] [CrossRef]
- Guo, P.-T.; Li, M.-F.; Luo, W.; Tang, Q.-F.; Liu, Z.-W.; Lin, Z.-M. Digital mapping of soil organic matter for rubber plantation at regional scale: An application of random forest plus residuals kriging approach. Geoderma 2015, 237–238, 49–59. [Google Scholar] [CrossRef]
- Zhang, S.-W.; Shen, C.-Y.; Chen, X.-Y.; Ye, H.-C.; Huang, Y.-F.; Lai, S. Spatial Interpolation of Soil Texture Using Compositional Kriging and Regression Kriging with Consideration of the Characteristics of Compositional Data and Environment Variables. J. Integr. Agric. 2013, 12, 1673–1683. [Google Scholar] [CrossRef] [Green Version]
- Jeong, G.; Oeverdieck, H.; Park, S.J.; Huwe, B.; Ließ, M. Spatial soil nutrients prediction using three supervised learning methods for assessment of land potentials in complex terrain. Catena 2017, 154, 73–84. [Google Scholar] [CrossRef]
- Piccini, C.; Marchetti, A.; Francaviglia, R. Estimation of soil organic matter by geostatistical methods: Use of auxiliary information in agricultural and environmental assessment. Ecol. Indic. 2014, 36, 301–314. [Google Scholar] [CrossRef]







| Satellite | Sentinel-2A | MOD09A1 | ||||
|---|---|---|---|---|---|---|
| Band Name | Band | Central Wavelength/(um) | Spatial Resolution/(m) | Band | Central Wavelength/(um) | Spatial Resolution/(m) |
| Coastal | 1 | 433 | 60 | X | X | X |
| Blue | 2 | 490 | 10 | 3 | 469 | 500 |
| Green | 3 | 560 | 10 | 4 | 555 | 500 |
| Red | 4 | 665 | 10 | 1 | 645 | 500 |
| Red edge-1 | 5 | 705 | 20 | X | X | X |
| Red edge-2 | 6 | 740 | 20 | X | X | X |
| Red edge-3 | 7 | 783 | 20 | X | X | X |
| NIR-1 | 8 | 842 | 10 | 2 | 858.5 | 500 |
| NIR-2 | 8A | 865 | 20 | X | X | X |
| Water vapour | 9 | 945 | 60 | X | X | X |
| SWIR-1 | 11 | 1610 | 20 | 6 | 1640 | 500 |
| SWIR-2 | 12 | 2190 | 20 | 7 | 2130 | 500 |
| Mid-infrared | X | X | X | 5 | 1692.5 | 500 |
| Variable Dataset | Full-Band | Common-Band | ||
|---|---|---|---|---|
| Variable Name | Sentinel-2A | MODIS | Sentinel-2A | MODIS |
| Coastal | √ b1 | X | X | X |
| Blue | √ b2 | √ b3 | √ b2 | √ b3 |
| Green | √ b3 | √ b4 | √ b3 | √ b4 |
| Red | √ b4 | √ b1 | √ b4 | √ b1 |
| Red edge-1 | √ b5 | X | X | X |
| Red edge-2 | √ b6 | X | X | X |
| Red edge-3 | √ b7 | X | X | X |
| NIR-1 | √ b8 | √ b2 | √ b8 | √ b2 |
| NIR-2 | √ b8A | X | X | X |
| Water vapour | √ b9 | X | X | X |
| SWIR-1 | √ b11 | √ b6 | √ b11 | √ b6 |
| SWIR-2 | √ b12 | √ b7 | √ b12 | √ b7 |
| Mid-infrared | X | √ b5 | X | X |
| √ | √ | √ | √ | |
| NDVI | √ | √ | √ | √ |
| EVI | √ | √ | √ | √ |
| RVI | √ | √ | √ | √ |
| DVI | √ | √ | √ | √ |
| NDWI | √ | √ | √ | √ |
| Elevation | √ | √ | √ | √ |
| Soil Dataset | N | SOM | |||
|---|---|---|---|---|---|
| Minimum % | Maximum % | SD % | Mean % | ||
| Whole dataset | 281 | 0.43 | 7.90 | 1.21 | 3.85 |
| Training dataset | 207 | 0.43 | 7.90 | 1.22 | 3.91 |
| Validation dataset | 74 | 0.64 | 6.12 | 1.17 | 3.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Zhang, M.; Yang, H.; Jin, Y.; Zhang, X.; Liu, H. Mapping Regional Soil Organic Matter Based on Sentinel-2A and MODIS Imagery Using Machine Learning Algorithms and Google Earth Engine. Remote Sens. 2021, 13, 2934. https://doi.org/10.3390/rs13152934
Zhang M, Zhang M, Yang H, Jin Y, Zhang X, Liu H. Mapping Regional Soil Organic Matter Based on Sentinel-2A and MODIS Imagery Using Machine Learning Algorithms and Google Earth Engine. Remote Sensing. 2021; 13(15):2934. https://doi.org/10.3390/rs13152934
Chicago/Turabian StyleZhang, Meiwei, Meinan Zhang, Haoxuan Yang, Yuanliang Jin, Xinle Zhang, and Huanjun Liu. 2021. "Mapping Regional Soil Organic Matter Based on Sentinel-2A and MODIS Imagery Using Machine Learning Algorithms and Google Earth Engine" Remote Sensing 13, no. 15: 2934. https://doi.org/10.3390/rs13152934