Integrating Remote Sensing and Landscape Characteristics to Estimate Soil Salinity Using Machine Learning Methods: A Case Study from Southern Xinjiang, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Soil Sampling and Laboratory Analysis
2.3. Satellite Imagery and Preprocessing
2.4. Auxiliary Data
2.5. Modeling Strategies
2.6. Partial Least Squares Path Modeling
2.7. Accuracy Assessment and Uncertainty Assessment
3. Results
3.1. Descriptive Statistics for Estimated EC
3.2. Selection of Independent Variables
3.3. Evaluation of the Accuracy of Estimations
3.4. Mapping of EC and Its Uncertainty
4. Discussion
4.1. Estimation Capabilities of Different Models
4.2. Sensitivity of Water and Vegetation Coverage
4.3. Interaction of the Independent Variables
4.4. Effects of Human Activities on Soil Salinization
5. Conclusions
- Overall, among the 35 factors considered, 9 indices (B6, B7, B10, B11, S6, GARI, GDVI, EVI, and DEM) were significant and contributed to the estimation of soil salinity. The random forest regression model was the best model in this study, with better model performance and accuracy measures (R2 = 0.75, RMSE = 7.33 dS m−1).
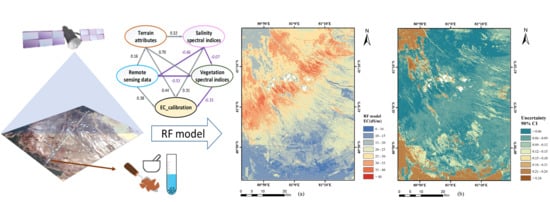
- According to the EC map derived from the random forest model, the northwest and the south of the study area showed relatively low soil salinity. Salt was mainly concentrated in the northwest to the middle of the study area covering the desert.
- Farmland reclamation affected the distribution of soil salinity, resulting in a strong positive correlation between elevation and soil EC. Moreover, the variability of soil salinity was sensitive to moisture and vegetation. Dry season was more conducive for the estimation of soil salinity.
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.; Ding, J.; Yu, D.; Ma, X.; Zhang, Z.; Ge, X.; Teng, D.; Li, X.; Liang, J.; Lizaga, I.; et al. Capability of Sentinel-2 MSI data for monitoring and mapping of soil salinity in dry and wet seasons in the Ebinur Lake region, Xinjiang, China. Geoderma 2019, 353, 172–187. [Google Scholar] [CrossRef]
- Ren, D.; Wei, B.; Xu, X.; Engel, B.; Li, G.; Huang, Q.; Xiong, Y.; Huang, G. Analyzing spatiotemporal characteristics of soil salinity in arid irrigated agro-ecosystems using integrated approaches. Geoderma 2019, 356, 113935. [Google Scholar] [CrossRef]
- Gorji, T.; Sertel, E.; Tanik, A. Monitoring soil salinity via remote sensing technology under data scarce conditions: A case study from Turkey. Ecol. Indic. 2017, 74, 384–391. [Google Scholar] [CrossRef]
- Wicke, B.; Smeets, E.; Dornburg, V.; Vashev, B.; Gaiser, T.; Turkenburg, W.; Faaij, A. The global technical and economic potential of bioenergy from salt-affected soils. Energy Environ. Sci. 2011, 4, 2669–2681. [Google Scholar] [CrossRef] [Green Version]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Gorji, T.; Yildirim, A.; Hamzehpour, N.; Tanik, A.; Sertel, E. Soil salinity analysis of Urmia Lake Basin using Landsat-8 OLI and Sentinel-2A based spectral indices and electrical conductivity measurements. Ecol. Indic. 2020, 112, 106173. [Google Scholar] [CrossRef]
- Jiang, Q.; Peng, J.; Biswas, A.; Hu, J.; Zhao, R.; He, K.; Shi, Z. Characterising dryland salinity in three dimensions. Sci. Total Environ. 2019, 682, 190–199. [Google Scholar] [CrossRef]
- Ma, Z.; Zhou, L.; Yu, W.; Teng, H.; Shi, Z. Improving TMPA 3B43 V7 datasets using land surface characteristics and ground observations on the Qinghai-Tibet Plateau. Int. J. IEEE Geosci. Remote. Sens. Lett. 2018, 15, 178–182. [Google Scholar] [CrossRef]
- Shi, Z.; Cheng, J.; Huang, M.; Zhou, L. Assessing Reclamation Levels of Coastal Saline Lands with Integrated Stepwise Discriminant Analysis and Laboratory Hyperspectral Data. Pedosphere 2006, 16, 154–160. [Google Scholar] [CrossRef]
- Peng, J.; Ji, W.; Ma, Z.; Li, S.; Chen, S.; Zhou, L.; Shi, Z. Predicting total dissolved salts and soluble ion concentrations in agricultural soils using portable visible near-infrared and mid-infrared spectrometers. Biosyst. Eng. 2016, 152, 94–103. [Google Scholar] [CrossRef]
- Wu, W.; Muhaimeed, A.S.; Al-Shafie, W.M.; Al-Quraishi, A.M.F. Using L-band radar data for soil salinity mapping—a case study in Central Iraq. Environ. Res. Commun. 2019, 1, 081004. [Google Scholar] [CrossRef] [Green Version]
- Davis, E.; Wang, C.; Dow, K. Comparing Sentinel-2 MSI and Landsat 8 OLI in soil salinity detection: A case study of agricultural lands in coastal North Carolina. Int. J. Remote Sens. 2019, 40, 6134–6153. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.; Abulimiti, A.; Cai, L. Quantitative estimation of soil salinity by means of different modeling methods and visible-near infrared (VIS-NIR) spectroscopy, Ebinur Lake Wetland, Northwest China. PeerJ 2018, 6, e4703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeulen, D.; Van Niekerk, A. Machine learning performance for predicting soil salinity using different combinations of geomorphometric covariates. Geoderma 2017, 299, 1–12. [Google Scholar] [CrossRef]
- Dakak, H.; Huang, J.; Zouahri, A.; Douaik, A.; Triantafilis, J. Mapping soil salinity in 3-dimensions using an EM38 and EM4Soil inversion modelling at the reconnaissance scale in central Morocco. Soil Use Manag. 2017, 33, 553–567. [Google Scholar] [CrossRef]
- Hu, J.; Peng, J.; Zhou, Y.; Xu, D.; Zhao, R.; Jiang, Q.; Fu, T.; Shi, Z. Quantitative Estimation of Soil Salinity Using UAV-Borne Hyperspectral and Satellite Multispectral Images. Remote Sens. 2019, 11, 736. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, F.; Ding, J.; Kung, H.; Latif, A.; Johnson, V.C. Estimation of soil salt content (SSC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR), Northwest China, based on a Bootstrap-BP neural network model and optimal spectral indices. Sci. Total Environ. 2018, 615, 918–930. [Google Scholar] [CrossRef]
- Mulder, V.L.; de Bruin, S.; Schaepman, M.E.; Mayr, T.R. The use of remote sensing in soil and terrain mapping—A review. Geoderma 2011, 162, 1–19. [Google Scholar] [CrossRef]
- McBratney, A.B.; Santos, M.L.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Jenny, H. Factors of Soil Formation; McGraw-Hill: New York, NY, USA, 1941. [Google Scholar]
- Bai, L.; Wang, C.; Zang, S.; Zang, S.; Wu, C.; Luo, J.; Wu, Y. Mapping Soil Alkalinity and Salinity in Northern Songnen Plain, China with the HJ-1 Hyperspectral Imager Data and Partial Least Squares Regression. Sensors 2018, 18, 3855. [Google Scholar] [CrossRef] [Green Version]
- Sultanov, M.; Ibrakhimov, M.; Akramkhanov, A.; Bauer, C.; Conrad, C. Modelling End-of-Season Soil Salinity in Irrigated Agriculture Through Multi-temporal Optical Remote Sensing, Environmental Parameters, and In Situ Information. PFG-J. Photogramm. Remote Sens. Geoinf. Sci. 2019, 86, 221–233. [Google Scholar] [CrossRef]
- Hoa, P.V.; Giang, N.V.; Binh, N.A.; Hai, L.V.H.; Phan, T.; Hasanlou, M.; Bui, D.T. Soil Salinity Mapping Using SAR Sentinel-1 Data and Advanced Machine Learning Algorithms: A Case Study at Ben Tre Province of the Mekong River Delta (Vietnam). Remote Sens. 2019, 11, 128. [Google Scholar] [CrossRef] [Green Version]
- Fathizad, H.; Ali, H.A.M.; Sodaiezadeh, H.; Kerry, R.; Taghizadeh-Mehrjardi, R. Investigation of the spatial and temporal variation of soil salinity using random forests in the central desert of Iran. Geoderma 2020, 365, 114233. [Google Scholar] [CrossRef]
- Masoud, A.A.; Koike, K.; Atwia, M.G.; El-Horiny, M.M.; Gemail, K.S. Mapping soil salinity using spectral mixture analysis of landsat 8 OLI images to identify factors influencing salinization in an arid region. Int. J. Appl. Earth Obs. 2019, 83, 101944. [Google Scholar] [CrossRef]
- Sanches, F.L.F.; Fernandes, A.C.P.; Ferreira, A.R.L.; Cortes, R.M.V.; Pacheco, F.A.L. A partial least squares - Path modeling analysis for the understanding of biodiversity loss in rural and urban watersheds in Portugal. Sci. Total Environ. 2018, 626, 1069–1085. [Google Scholar] [CrossRef]
- Ma, L.; Ma, F.; Li, J.; Ge, J.; Yang, S.; Wu, D.; Feng, J.; Ding, J. Characterizing and modeling regional-scale variations in soil salinity in the arid oasis of Tarim Basin, China. Geoderma 2017, 305, 1–11. [Google Scholar] [CrossRef]
- Peng, J.; Biswas, A.; Jiang, Q.; Zhao, R.; Hu, J.; Hu, B.; Shi, Z. Estimating soil salinity from remote sensing and terrain data in southern Xinjiang Province, China. Geoderma 2019, 337, 1309–1319. [Google Scholar] [CrossRef]
- Yang, R.M.; Guo, W.W. Using Sentinel-1 Imagery for Soil Salinity Prediction Under the Condition of Coastal Restoration. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 1482–1488. [Google Scholar] [CrossRef]
- Chi, Y.; Sun, J.; Liu, W.; Wang, J.; Zhao, M. Mapping coastal wetland soil salinity in different seasons using an improved comprehensive land surface factor system. Ecol. Indic. 2017, 74, 384–391. [Google Scholar] [CrossRef]
- Yu, H.; Liu, M.; Du, B.; Wang, Z.; Hu, L.; Zhang, B. Mapping Soil Salinity/Sodicity by using Landsat OLI Imagery and PLSR Algorithm over Semiarid West Jilin Province, China. Sensors 2019, 107, 105517. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zucca, C.; Muhaimeed, A.S.; Ziadat, F.; Nangia, V.; Payne, W.B. Soil salinity prediction and mapping by machine learning regression in Central Mesopotamia, Iraq. Land Degrad. Dev. 2018, 29, 4005–4014. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, F.; Wang, C.; Wu, S.; Liu, J.; Xu, A.; Pan, K.; Pan, X. Estimating the soil salinity over partially vegetated surfaces from multispectral remote sensing image using non-negative matrix factorization. Geoderma 2019, 354, 113887. [Google Scholar] [CrossRef]
- Wang, F.; Shi, Z.; Biswas, A.; Yang, S.; Ding, J. Multi-algorithm comparison for predicting soil salinity. Geoderma 2020, 365, 114211. [Google Scholar] [CrossRef]
- Nouri, H.; Borujeni, S.C.; Alaghmand, S.; Anderson, S.J.; Sutton, P.C.; Parvazian, S.; Beecham, S. Soil Salinity Mapping of Urban Greenery Using Remote Sensing and Proximal Sensing Techniques; The Case of Veale Gardens within the Adelaide Parklands. Sustainability 2018, 10, 2826. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ding, J.; Yu, D.; Teng, D.; He, B.; Chen, X.; Ge, X.; Zhang, Z.; Wang, Y.; Yang, X.; et al. Machine learning-based detection of soil salinity in an arid desert region, Northwest China: A comparison between Landsat-8 OLI and Sentinel-2 MSI. Sci. Total Environ. 2020, 707, 136092. [Google Scholar] [CrossRef]
- Cheng, J.L.; Shi, Z.; Zhu, Y.W. Assessment and mapping of environmental quality in agricultural soils of Zhejiang Province, China. J. Environ. Sci. 2007, 19, 50–54. [Google Scholar] [CrossRef]
- Sabri, E.M.; Boukdir, A.; Karaoui, I.; Arioua, A.; Messlouhi, R.; Idrissi, A.E.A. Modelling soil salinity in Oued El Abid watershed, Morocco. E3S Web Conf. 2018, 37, 04002. [Google Scholar] [CrossRef] [Green Version]
- Taghadosi, M.M.; Hasanlou, M.; Eftekhari, K. Soil salinity mapping using dual-polarized SAR Sentinel-1 imagery. Int. J. Remote Sens. 2018, 40, 237–252. [Google Scholar] [CrossRef]
- Zhu, L.; Walker, J.P.; Tsang, L.; Huang, H.; Ye, N.; Rudiger, C. Soil moisture retrieval from time series multi-angular radar data using a dry down constraint. Remote Sens. Environ. 2019, 231, 111237. [Google Scholar] [CrossRef]
- Alifu, H.; Vuillaume, J.F.; Johnson, B.A.; Hirabayashi, Y. Machine-learning classification of debris-covered glaciers using a combination of Sentinel-1/-2 (SAR/optical), Landsat 8 (thermal) and digital elevation data. Geomorphology 2020, 369, 1–18. [Google Scholar] [CrossRef]
- Qiu, S.; Zhu, Z.; He, B. Fmask 4.0: Improved cloud and cloud shadow detection in Landsats 4–8 and Sentinel-2 imagery. Remote Sens. Environ. 2019, 231, 111205. [Google Scholar] [CrossRef]
- Muller, E.; DeÂcamps, H. Modeling soil moisture-reflectance. Remote Sens. Environ. 2000, 76, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Taghizadeh-Mehrjardi, R.; Minasny, B.; Sarmadian, F.; Malone, B.P. Digital mapping of soil salinity in Ardakan region, central Iran. Geoderma 2014, 213, 15–28. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Kumar, K.; Hari, P.K.S.; Arora, M.K. Estimation of water cloud model vegetation parameters using a genetic algorithm. Hydrol. Sci. J. 2012, 57, 776–789. [Google Scholar] [CrossRef] [Green Version]
- Douaoui, A.E.K.; Nicolas, H.; Walter, C. Detecting salinity hazards within a semiarid context by means of combining soil and remote-sensing data. Geoderma 2006, 134, 217–230. [Google Scholar] [CrossRef]
- Abbas, A.; Khan, S. Using Remote Sensing Techniques for Appraisal of Irrigated Soil Salinity. In Proceedings of International Congress on Modelling and Simulation; Oxley, L., Kulasiri, D., Eds.; Modelling & Simulation Soc Australia & New Zealand Inc.: Christchurch, New Zealand, 2007; pp. 2632–2638. [Google Scholar]
- Bannari, A.; Guedon, A.M.; El-Harti, A.; Cherkaoui, F.Z.; El-Chmari, A. Characterization of Slightly and Moderately Saline and Sodic Soils in Irrigated Agricultural Land using Simulated Data of Advanced Land Imaging (EO-1) Sensor. Commun. Soil Sci. Plant Anal. 2008, 39, 2795–2811. [Google Scholar] [CrossRef]
- Alexakis, D.D.; Daliakopoulos, I.N.; Panagea, I.S.; Tsanis, I.K. Assessing soil salinity using WorldView-2 multispectral images in Timpaki, Crete, Greece. Geocarto Int. 2016, 33, 321–338. [Google Scholar] [CrossRef]
- Gitelson, A.; Kaufman, Y.; Merzlyak, M. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Wu, W.; Al-Shafie, W.M.; Mhaimeed, A.S.; Ziadat, F.; Nangia, V.; Payne, W.B. Soil Salinity Mapping by Multiscale Remote Sensing in Mesopotamia, Iraq. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 4442–4452. [Google Scholar] [CrossRef]
- Goel, N.S.; Qin, W. Influences of canopy architecture on relationships between various vegetation indices and LAI and Fpar: A computer simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Scudiero, E.; Skaggs, T.H.; Corwin, D.L. Regional scale soil salinity evaluation using Landsat 7, western San Joaquin Valley, California, USA. Geoderma Reg. 2014, 2–3, 82–90. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, B. Prediction of soil salinity with soil-reflected spectra: A comparison of two regression methods. Sci. Rep. 2019, 9, 5067–5075. [Google Scholar] [CrossRef] [PubMed]
- Nurmemet, I.; Sagan, V.; Ding, J.; Halik, U.; Abliz, A.; Yakup, Z. A WFS-SVM Model for Soil Salinity Mapping in Keriya Oasis, Northwestern China Using Polarimetric Decomposition and Fully PolSAR Data. Remote Sens. 2018, 10, 598. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Dai, Z.; He, Z.; Ma, J.; Wang, Y.; Wang, Y. Learning Traffic as Images: A Deep Convolutional Neural Network for Large-Scale Transportation Network Speed Prediction. Sensors 2017, 17, 818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso-Monsalve, S.; Suárez-Cetrulo, A.L.; Cervantes, A.; Quintana, D. Convolution on neural networks for high-frequency trend prediction of cryptocurrency exchange rates using technical indicators. Expert Syst. Appl. 2020, 149, 113250–113264. [Google Scholar] [CrossRef]
- Henseler, J.; Hubona, G.; Ray, P.A. Using PLS path modeling in new technology research: Updated guidelines. Ind. Manag. Data Syst. 2016, 116, 2–20. [Google Scholar] [CrossRef]
- Danks, N.; Sharma, P.; Sarstedt, M. Model selection uncertainty and multimodel inference in partial least squares structural equation modeling (PLS-SEM). J. Bus. Res. 2020, 113, 13–24. [Google Scholar] [CrossRef]
- Oliveira, C.F.; do Valle Junior, R.F.; Valera, C.A.; Rodrigues, V.S.; Fernandes, L.F.S.; Pacheco, F.A.L. The modeling of pasture conservation and of its impact on stream water quality using Partial Least Squares-Path Modeling. Sci. Total Environ. 2019, 697, 134081. [Google Scholar] [CrossRef]
- Huang, J.; Wu, J.; Fang, Y.; Wu, J.; Huang, J. Comparison of partial least square regression, support vector machine, and deep-learning techniques for estimating soil salinity from hyperspectral data. J. Appl. Remote Sens. 2018, 12, 022204. [Google Scholar]
- Pan, L.; Politis, D.N. Bootstrap prediction intervals for linear, nonlinear and nonparametric autoregressions. J. Stat. Plan. Inference 2016, 177, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Xue, J.; Chen, S.; Zhou, Y.; Liang, Z.; Wang, N.; Shi, Z. Fine-Resolution Mapping of Soil Total Nitrogen across China Based on Weighted Model Averaging. Remote Sens. 2020, 12, 85. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Buces, N.; Siebe, C.; Cram, S.; Palacio, J.L. Mapping soil salinity using a combined spectral response index for bare soil and vegetation: A case study in the former lake Texcoco, Mexico. J. Arid Environ. 2006, 65, 644–667. [Google Scholar] [CrossRef]
- Zhang, T.; Qi, J.; Gao, Y.; Ouyang, Z.; Zeng, S.; Zhao, B. Detecting soil salinity with MODIS time series VI data. Ecol. Indic. 2015, 52, 480–489. [Google Scholar] [CrossRef]
- Shahabi, M.; Jafarzadeh, A.A.; Neyshabouri, M.R.; Ghorbani, M.A.; Kamran, K.V. Spatial modeling of soil salinity using multiple linear regression, ordinary kriging and artificial neural network methods. Arch. Agron. Soil Sci. 2016, 63, 151–160. [Google Scholar] [CrossRef]
- Ma, L.; Yang, S.; Simayi, Z.; Gu, Q.; Li, J.; Yang, X.; Ding, J. Modeling variations in soil salinity in the oasis of Junggar Basin, China. Land Degrad. Dev. 2018, 29, 551–562. [Google Scholar] [CrossRef]
- Patel, N.R.; Mukund, A.; Parida, B.R. Satellite-derived vegetation temperature condition index to infer root zone soil moisture in semi-arid province of Rajasthan, India. Geocarto Int. 2019, 1–17. [Google Scholar] [CrossRef]
- Hajj, M.E.; Baghdadi, N.; Zribi, M.; Belaud, G.; Cheviron, B.; Courault, D.; Charron, F. Soil moisture retrieval over irrigated grassland using X-band SAR data. Remote Sens. Environ. 2016, 176, 202–218. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Meng, Q.; Yao, S.; Wang, Q.; Zeng, J.; Zhao, S.; Ma, J. Soil Moisture Retrieval from the Chinese GF-3 Satellite and Optical Data over Agricultural Fields. Sensors 2018, 18, 2675. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Koganti, T.; Santos, F.A.M.; Triantafilis, J. Mapping soil salinity and a fresh-water intrusion in three-dimensions using a quasi-3d joint-inversion of DUALEM-421S and EM34 data. Sci. Total Environ. 2017, 577, 395–404. [Google Scholar] [CrossRef]
- Schuberth, F.; Rademaker, M.E.; Henseler, J. Estimating and assessing second-order constructs using PLS-PM: The case of composites of composites. Ind. Manag. Data Syst. 2020, 120, 2211–2241. [Google Scholar] [CrossRef]
- Racetin, I.; Krtalic, A.; Srzic, V.; Zovko, M. Characterization of short-term salinity fluctuations in the Neretva River Delta situated in the southern Adriatic Croatia using Landsat-5 TM. Ecol. Indic. 2020, 110, 105924. [Google Scholar] [CrossRef]
- Thiam, S.; Villamor, G.B.; Kyei-Baffour, N.; Matty, F. Soil salinity assessment and coping strategies in the coastal agricultural landscape in Djilor district, Senegal. Land Use Policy 2019, 88, 104191. [Google Scholar] [CrossRef]
- Elia, S.; Todd, H.S.; Dennis, L.C. Regional-scale soil salinity assessment using Landsat ETM+ canopy reflectance. Remote Sens. Environ. 2015, 169, 335–343. [Google Scholar]
- Xu, H.; Chen, C.; Zheng, H.; Luo, G.; Yang, L.; Wang, W.; Wu, S.; Ding, J. AGA-SVR-based selection of feature subsets and optimization of parameter in regional soil salinization monitoring. Int. J. Remote Sens. 2020, 41, 4470–4495. [Google Scholar] [CrossRef]
- Ding, J.; Yu, D. Monitoring and evaluating spatial variability of soil salinity in dry and wet seasons in the Werigan–Kuqa Oasis, China, using remote sensing and electromagnetic induction instruments. Geoderma 2014, 235–236, 316–322. [Google Scholar] [CrossRef]







| Sensor | Spectral Band | Wavelength (μm) | Resolution (m) |
|---|---|---|---|
| OLI | Band 1—Coastal/Aerosol | 0.433–0.453 | 30 |
| Band 2—Blue | 0.450–0.515 | 30 | |
| Band 3—Green | 0.525–0.600 | 30 | |
| Band 4—Red | 0.630–0.680 | 30 | |
| Band 5—Near Infrared (NIR) | 0.845–0.885 | 30 | |
| Band 6—Short Wavelength Infrared (SWIR) | 1.560–1.600 | 30 | |
| Band 7—Short Wavelength Infrared (SWIR) | 2.100–2.300 | 30 | |
| Band 8—Panchromatic | 0.500–0.680 | 15 | |
| Band 9—Cirrus | 1.360–1.390 | 30 | |
| TRIS | Band 10—Long Wavelength Infrared | 10.30–11.30 | 100 |
| Band 11—Long Wavelength Infrared | 11.50–12.50 | 100 |
| Parameter | Value |
|---|---|
| Pass direction | Ascending |
| Mode | Interferometric wide (IW) |
| Polarization | VH, VV |
| Temporal resolution | 12 days |
| Spatial resolution | 20 m |
| Wavelength | 5.6 cm |
| Radiometric stability | 0.5 dB (3ơ) |
| Radiometric accuracy | 1 dB (3ơ) |
| Phase error | 5° |
| Orbit number | 17,635 |
| Swath | 250 km |
| Auxiliary Data | Land Surface Parameters | Abbreviation | Formulations | References |
|---|---|---|---|---|
| Remote sensing data | Bands 1–7 Band 10 Band 11 | B1–B7 B10 B11 | Landsat-8 | |
| Brightness index | BI | [(B4)2 + (B5)2]0.5 | [46] | |
| Salinity spectral indices | Salinity index | SI | (B4 × B2)0.5 | [46] |
| Salinity index 1 | SI1 | (B4 × B3)0.5 | [33] | |
| Salinity index 3 | SI3 | [(B4)2 + (B3)2]0.5 | [33] | |
| Salinity index I | S1 | B2/B4 | [47] | |
| Salinity index II | S2 | (B2 − B4)/(B2 + B4) | [47] | |
| Salinity index III | S3 | B3 × B4/B2 | [48] | |
| Salinity index IV | S4 | B2 × B4/B3 | [48] | |
| Salinity index V | S5 | B4 × B5/B3 | [48] | |
| Salinity index VI | S6 | B6/B7 | [49] | |
| Salinity index VII | S7 | (B6 − B7)/(B6 + B7) | [49] | |
| Salinity index VII | S8 | (B3 + B4)/2 | [47] | |
| Salinity index IX | S9 | (B3 + B4 + B5)/2 | [47] | |
| Soil salinity index | SI_T | B4/B5 × 100 | [50] | |
| Vegetation spectral indices | Normalized difference vegetation index | NDVI | (B5 − B4)/(B5 + B4) | [51] |
| Green atmospherically resistant vegetation index | GARI | {B5 − [B3 + γ × (B2 − B4)]}/{B5 + [B3 + γ × (B2 − B4)]} | [51] | |
| Extended NDVI | ENDVI | (B5 + B7 − B4)/(B5 + B7 + B4) | [36] | |
| Generalized difference vegetation index | GDVI | (B52 − B42)/(B52 + B42) | [52] | |
| Non-linear vegetation index | NLI | (B52 − B4)/(B52 + B4) | [53] | |
| Canopy response salinity index | CRSI | (B5 × B4 − B3 × B2)/(B5 × B4 + B3 × B2)0.5 | [52] | |
| Enhanced vegetation index | EVI | g × (B5 − B4)/(B5 + C1 × B4 − C2 × B2 + L) | [54] | |
| Terrain attributes | Elevation | DEM | [44] | |
| Slope | S | SAGA GIS | ||
| Aspect | A | SAGA GIS | ||
| Radar data | Backscattering coefficients of VH band | VH | (ơ0 − ơveg_VH0)/L | [46] |
| Backscattering coefficients of VV band | VV | (ơ0 − ơveg_VV0)/L | [46] |
| Datasets | Amount | Min | Max | Mean | Median | SD | Kutosis | Skewness | CV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Bare soil | 68 | 4.10 | 44.80 | 20.47 | 17.40 | 9.76 | 0.56 | 1.09 | 48% |
| Vegetation covered | 35 | 2.09 | 46.70 | 18.77 | 14.58 | 11.48 | 0.04 | 0.85 | 61% | |
| Entire datasets | 103 | 2.09 | 46.70 | 19.89 | 16.73 | 10.35 | 0.27 | 0.93 | 52% | |
| Testing | Bare soil | 33 | 2.09 | 42.60 | 18.45 | 16.43 | 9.57 | 0.27 | 0.79 | 52% |
| Vegetation covered | 15 | 5.90 | 41.50 | 18.61 | 16.37 | 8.51 | 3.28 | 1.58 | 46% | |
| Entire datasets | 48 | 2.09 | 42.60 | 18.50 | 16.40 | 9.16 | 0.68 | 0.94 | 50% | |
| Total | Bare soil | 101 | 2.09 | 44.80 | 19.81 | 16.73 | 9.70 | 0.46 | 0.98 | 49% |
| Vegetation covered | 50 | 2.09 | 46.70 | 18.72 | 15.71 | 10.59 | 0.45 | 0.96 | 57% | |
| Entire study area | 151 | 2.09 | 46.70 | 19.45 | 16.67 | 9.98 | 0.39 | 0.94 | 51% | |
| Category | Factor | R | Category | Factor | R |
|---|---|---|---|---|---|
| Remote sensing data | B1 | 0.15 | Salinity spectral indices | S6 | −0.44 ** |
| B2 | 0.19 * | S7 | 0.03 | ||
| B3 | 0.21 ** | S8 | 0.19 * | ||
| B4 | 0.14 | S9 | 0.19 * | ||
| B5 | 0.18 * | SI_T | −0.01 | ||
| B6 | 0.26 ** | Vegetation spectral indices | NDVI | −0.01 | |
| B7 | 0.23 ** | GARI | −0.23 * | ||
| B10 | 0.40 ** | ENDVI | 0.09 | ||
| B11 | 0.39 ** | GDVI | −0.76 ** | ||
| BI | 0.14 | NLI | 0.22 ** | ||
| Salinity spectral indices | SI | 0.18 * | CRSI | −0.21 * | |
| SI1 | 0.19 * | EVI | 0.23 ** | ||
| SI3 | 0.18 * | Terrain attributes | DEM | 0.41 ** | |
| S1 | 0.22 ** | S | 0.17 * | ||
| S2 | 0.22 ** | A | −0.06 | ||
| S3 | 0.14 | Radar data | VH | −0.03 | |
| S4 | 0.14 | VV | 0.01 | ||
| S5 | −0.04 |
| Models | Total | Bootstrap | Vegetation Covered | Bare Soil | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | R2 | RMSE | R2 | RMSE | R2 | RMSE | |
| PLSR | 0.52 | 7.32 | 0.42 | 8.28 | 0.58 | 6.77 | 0.51 | 7.60 |
| CNN | 0.53 | 6.44 | 0.54 | 7.98 | 0.51 | 6.13 | 0.54 | 6.58 |
| SVM | 0.68 | 7.53 | 0.52 | 7.88 | 0.73 | 6.86 | 0.67 | 7.85 |
| RF | 0.75 | 7.33 | 0.56 | 7.84 | 0.76 | 6.82 | 0.75 | 7.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Xue, J.; Peng, J.; Biswas, A.; He, Y.; Shi, Z. Integrating Remote Sensing and Landscape Characteristics to Estimate Soil Salinity Using Machine Learning Methods: A Case Study from Southern Xinjiang, China. Remote Sens. 2020, 12, 4118. https://doi.org/10.3390/rs12244118
Wang N, Xue J, Peng J, Biswas A, He Y, Shi Z. Integrating Remote Sensing and Landscape Characteristics to Estimate Soil Salinity Using Machine Learning Methods: A Case Study from Southern Xinjiang, China. Remote Sensing. 2020; 12(24):4118. https://doi.org/10.3390/rs12244118
Chicago/Turabian StyleWang, Nan, Jie Xue, Jie Peng, Asim Biswas, Yong He, and Zhou Shi. 2020. "Integrating Remote Sensing and Landscape Characteristics to Estimate Soil Salinity Using Machine Learning Methods: A Case Study from Southern Xinjiang, China" Remote Sensing 12, no. 24: 4118. https://doi.org/10.3390/rs12244118